Molecular Phylogeny of Cimicoidea (Heteroptera: Cimicomorpha) Revisited: Increased Taxon Sampling Reveals Evolution of Traumatic Insemination and Paragenitalia
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Phylogenetic Relationships
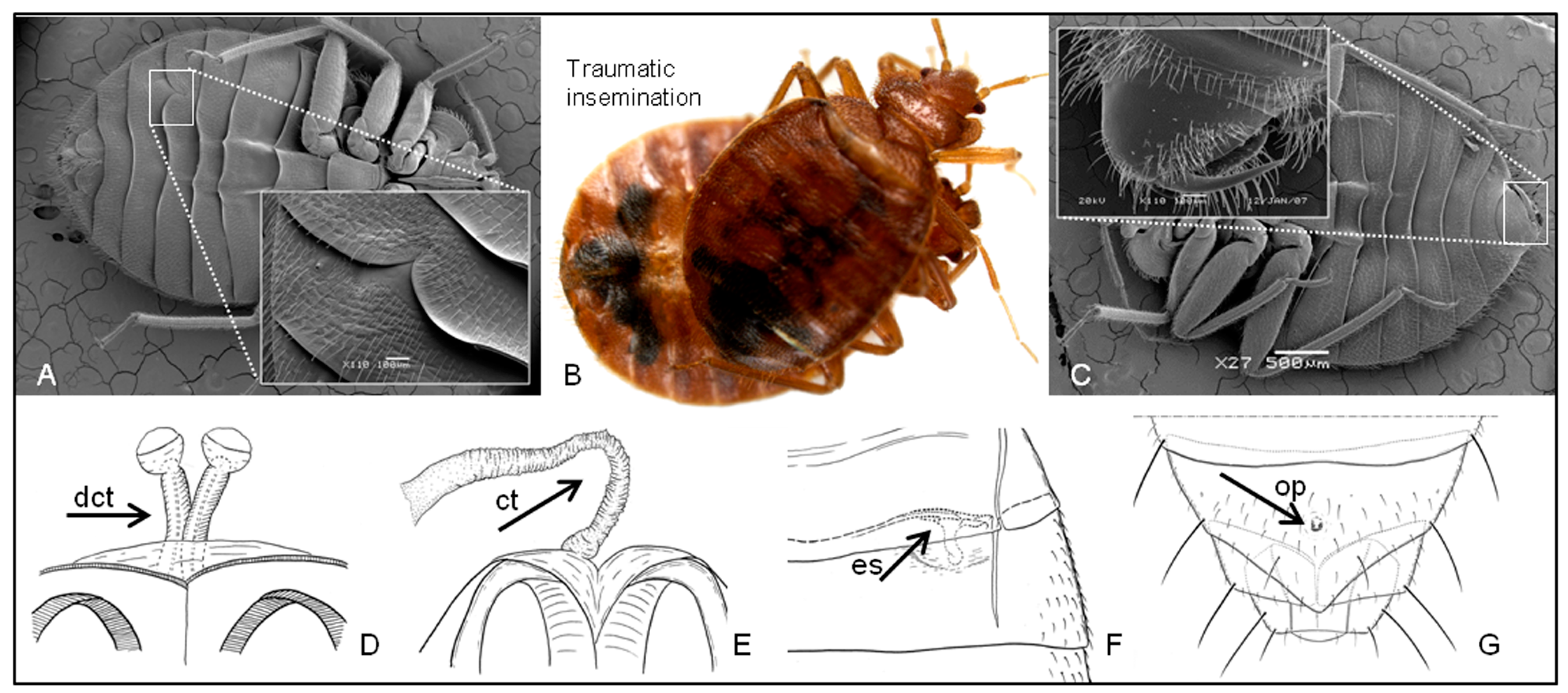
1.2. Traumatic Insemination and Paragenitalia
2. Materials and Methods
2.1. Taxon Sampling
2.2. Molecular Markers, DNA Extraction, and Other Molecular Protocols
2.3. Alignments and Phylogenetic Analyses
2.4. Reconstructing Ancestral Character States
2.4.1. Copulating Types
2.4.2. Reconstructions of Ancestral Character States
2.5. Testing Correlations
2.5.1. Paragenitalia and Copulation Types
2.5.2. Correlation Tests
3. Results
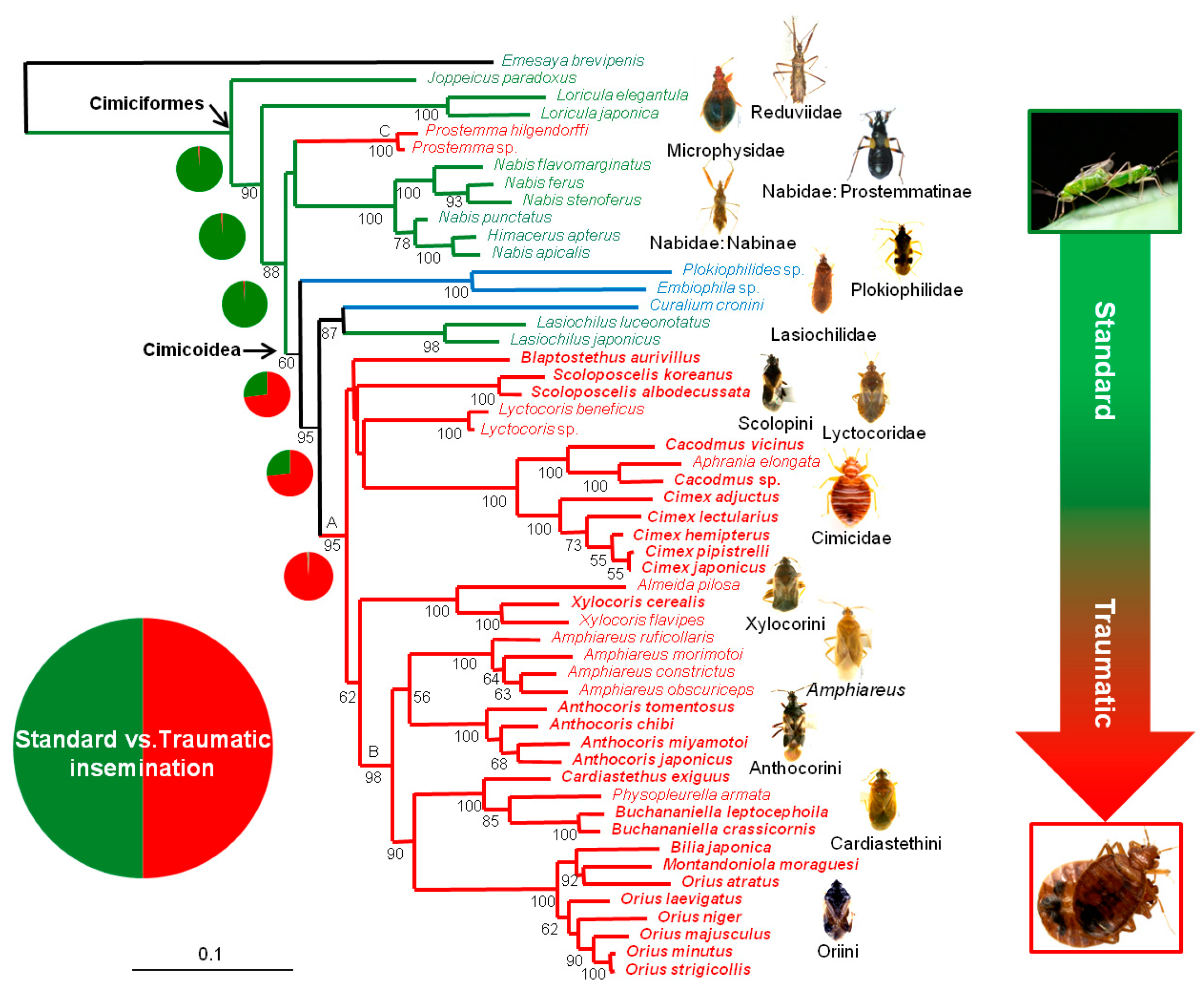
3.1. Phylogenetic Relationships
3.2. Reconstructions of Ancestral TI States
3.3. Bivariate Evolutionary Correlations
4. Discussion
4.1. Phylogenetic Incongruence among Analyses
4.2. Phylogenetic Relationships within Cimicoidea
4.3. Evolution of Traumatic Insemination with Special Reference to Paragenitalia
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Schuh, R.T.; Slater, J.A. True Bugs of the World (Hemiptera: Heteroptera), Classification and Natural History; Comstock Publishing Associates, Cornell University Press: Ithaca, NY, USA; London, UK, 1995. [Google Scholar]
- Schuh, R.T.; Weirauch, C.; Wheeler, W.C. Phylogenetic relationships within the Cimicomorpha (Hemiptera: Heteroptera): A total-evidence analysis. Syst. Entomol. 2009, 34, 15–48. [Google Scholar] [CrossRef]
- Jung, S.; Kim, H.; Yamada, K.; Lee, S. Molecular phylogeny and evolutionary habitat transition of the flower bugs (Heteroptera: Anthocoridae). Mol. Phylogenet. Evol. 2010, 57, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Usinger, R.L. Monograph of the Cimicidae (Hemiptera–Heteroptera); Entomological Society of America: College Park, MD, USA, 1966. [Google Scholar]
- Weirauch, C.; Schuh, R.T. Systematics and evolution of Heteroptera: 25 Years of Progress. Annu. Rev. Entomol. 2011, 56, 487–510. [Google Scholar] [CrossRef] [PubMed]
- Ford, L.J. The Phylogeny and Biogeography of the Cimicoidea (Insecta, Hemiptera). Master’s Thesis, University of Connecticut, Storrs, CT, USA, 1979. [Google Scholar]
- Schuh, R.T.; Štys, P. Phylogenetic analysis of cimicomorphan family relationships (Heteroptera). J. N.Y. Entomol. Soc. 1991, 99, 298–350. [Google Scholar]
- Carayon, J. Insemination traumatique heterosexuelle et homosexuelle chez Xylocoris maculipennis (Hemiptera: Anthocoridae). C. R. Acad. Sci. 1974, 278, 2803–2806. [Google Scholar]
- Carayon, J. Insémination extra-génitale traumatique. In Traité de Zoologie; Grassé, P.P., Ed.; Masson: Paris, France, 1977; Volume 8, pp. 351–390. [Google Scholar]
- Arnqvist, G.; Rowe, L. Sexual Conflict. Monographs in Behaviour and Ecology; Princeton University Press: Princeton, NJ, USA, 2005. [Google Scholar]
- Reinhardt, K.; Siva-Jothy, M.T. Biology of the Bed Bugs (Cimicidae). Annu. Rev. Entomol. 2006, 52, 151–374. [Google Scholar] [CrossRef] [PubMed]
- Abele, L.G.; Gilchrist, S. Homosexual rape and sexual selection in acanthocephalan worms. Science 1977, 197, 81–83. [Google Scholar] [CrossRef]
- Reinhardt, K.; Harney, E.; Naylor, R.; Gorb, S.; Siva-Jothy, M.T. Female-Limited Polymorphism in the Copulatory Organ of a Traumatically Inseminating Insect. Am. Nat. 2007, 170, 931–935. [Google Scholar] [CrossRef]
- Lange, R.; Reinhardt, K.; Michiels, N.K.; Anthes, N. Functions, diversity, and evolution of traumatic mating. Biol. Rev. 2013, 88, 585–601. [Google Scholar] [CrossRef]
- Carayon, J. Traumatic insemination and the paragenital system. In Monograph of Cimicidae (Hemiptera, Heteroptera); Usinger, R.L., Ed.; Entomological Society of America: College Park, MD, USA, 1966; pp. 81–166. [Google Scholar]
- Tatarnic, N.J.; Cassis, G.; Hochuli, D.F. Traumatic insemination in the plant bug genus Coridromius Signoret (Heteroptera: Miridae). Biol. Lett. 2006, 2, 58–61. [Google Scholar] [CrossRef]
- Yamada, K. Blaptostethus aurivillus, a new flower bug (Heteroptera, Anthocoridae) from West Malaysia. Jpn. J. Syst. Entomol. 2008, 14, 7–11. [Google Scholar]
- Yamada, K.; Hirowatari, T. Two new species of the flower bug genus Scoloposcelis (Insecta: Heteroptera: Anthocoridae) from Japan. Species Divers. 2005, 10, 125–133. [Google Scholar] [CrossRef]
- Yamada, K.; Yasunaga, T.; Nakatani, Y.; Hirowatari, T. The minute pirate-bug genus Xylocoris dufour (Hemiptera: Heteroptera: Anthocoridae) from rice mills in Thailand. Proc. Entomol. Soc. Wash. 2006, 108, 525–533. [Google Scholar]
- Yamada, K.; Yasunaga, T. Species of the minute pirate bug genus Buchananiella Reuter from Thailand (Heteroptera: Anthocoridae). Nouv. Rev. Entomol. 2009, 25, 273–280. [Google Scholar]
- Carayon, J. Insémination par ‘spermalège’ et cordon conducteur de spermatozoids chez Stricticimex brevispinosus Usinger (Heteroptera, Cimicidae). Rev. Zool. Bot. Afr. 1959, 60, 81–104. [Google Scholar]
- Reinhardt, K.; Naylor, R.; Siva–Jothy, M.T. Reducing a cost of traumatic insemination: Female bedbugs evolve a unique organ. Proc. R. Soc. B Biol. Sci. 2003, 270, 2371–2375. [Google Scholar] [CrossRef]
- Horton, D.R.; Lewis, T.M. Variation in male and female genitalia among ten species of North American Anthocoris (Hemiptera: Heteroptera: Anthocoridae). Ann. Entomol. Soc. Am. 2011, 104, 1260–1278. [Google Scholar] [CrossRef]
- Jung, S.; Lee, S. Systematic and phylogenetic study of the Korean Orius species based on morphological and molecular data (Heteroptera: Anthocoridae). Appl. Entomol. Zool. 2011, 46, 153–164. [Google Scholar] [CrossRef]
- Jung, S.; Lee, S. Taxonomic review of the genus Amphiareus (Heteroptera: Anthocoridae) in the Korean Peninsula. J. Asia Pac. Entomol. 2011, 14, 335–340. [Google Scholar] [CrossRef]
- Jung, S.; Lee, S. Two new records of the tribe Cardiastethini (Hemiptera: Heteroptera: Anthocoridae) from the Korean Peninsula. Zootaxa 2011, 2931, 59–64. [Google Scholar] [CrossRef]
- Jung, S.; Yamada, K.; Lee, S. A new species of Scoloposcelis Fieber (Hemiptera: Heteroptera: Anthocoridae: Scolopini) from the Korean Peninsula, with a key to the Palaearctic species. Zootaxa 2011, 2766, 64–68. [Google Scholar]
- Jung, S.; Yasunaga, T.; Lee, S. Taxonomic review of the genus Orius (Heteroptera: Anthocoridae) in the Korean Peninsula. J. Asia Pac. Entomol. 2011, 14, 64–74. [Google Scholar] [CrossRef]
- Jung, S.; Duwal, R.K.; Lee, S. COI barcoding of true bugs (Insecta, Heteroptera). Mol. Ecol. Resour. 2011, 11, 266–270. [Google Scholar] [CrossRef]
- Jung, S.; Lee, S. Correlated evolution and Bayesian divergence time estimates of the Cimicoidea (Heteroptera: Cimicomorpha) reveal the evolutionary history. Syst. Entomol. 2012, 37, 22–31. [Google Scholar] [CrossRef]
- Jung, S.; Lee, S. Molecular phylogeny of the plant bugs (Heteroptera: Miridae) and the evolution of feeding habits. Cladistics 2012, 28, 50–79. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primer for ampllification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Campbell, B.C.; Ross, G.F.; Woodward, T.E. Paraphyly of Homoptera and Auchenorrhyncha inferred from 18S rDNA nucleotide sequences. Syst. Entomol. 1995, 20, 175–194. [Google Scholar] [CrossRef]
- Hillis, D.M.; Dixon, M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991, 66, 411–453. [Google Scholar] [CrossRef]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Kambhampati, S.; Smith, P.T. PCR primers for the amplification of four insect mitochondrial gene fragments. Insect Mol. Biol. 1995, 4, 233–236. [Google Scholar] [CrossRef]
- Rambaut, A. Se–Al. Sequence Alignment Editor, Version 2.0 Alpha 11; University of Oxford, Department of Zoology: Oxford, UK, 2002. Available online: http://tree.bio.ed.ac.uk/software/seal/ (accessed on 8 December 2022).
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis; Pasada, D., Ed.; Humana Press: New York, NY, USA, 2009; pp. 39–64. [Google Scholar]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Maddison, D.R.; Maddison, W.P. MacClade 4: Analysis of Phylogeny and Character Evolution; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Farris, J.S.; Källersjö, M.; Kluge, A.G.; Bult, C. Testing significance of incongruence. Cladistics 1994, 10, 315–319. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). (Version 4.0b10); Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Goloboff, P.A. Analyzing large data sets in reasonable times, solutions for composite optima. Cladistics 1999, 15, 415–428. [Google Scholar] [CrossRef]
- Nixon, K.C. The Parsimony Ratchet, a new method for rapid parsimony analysis. Cladistics 1999, 15, 407–414. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web–servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Schuh, R.T.; Weirauch, C.; Henry, T.J.; Halbert, S.E. Curaliidae, a new family of Heteroptera (Insecta: Hemiptera) from the eastern United States. Ann. Entomol. Soc. Am. 2008, 101, 20–29. [Google Scholar] [CrossRef]
- Schuh, R.T. Heissophila macrotheleae, a new genus and new species of Plokiophilidae from Thailand (Hemiptera, Heteroptera), with comments on the family diagnosis. Denisia 2006, 19, 637–645. [Google Scholar]
- Pagel, M.; Meade, A. BaysTrait Version 2.0–Draft Manual. Available online: http://www.evolution.reading.ac.uk/BayesTraitsV2.html (accessed on 8 December 2022).
- Pagel, M. Detecting correlated evolution on phylogenies: A general method for the comparative analysis of discrete characters. Proc. R. Soc. B Biol. Sci. 1994, 255, 37–45. [Google Scholar]
- Pagel, M.; Meade, A. Bayesian analysis of correlated evolution of discrete characters by reversible–jump Markov chain Monte Carlo. Am. Nat. 2006, 167, 808–825. [Google Scholar] [CrossRef] [PubMed]
- Maddison, D.R.; Maddison, W.P. Mesquite: A Modular System for Evolutionary Analysis. (Version 2.6). Available online: http://mesquiteproject.org (accessed on 8 December 2022).
- Carayon, J. Caractères systématiques et classification des Anthocoridae (Hemipt.). Ann. Soc. Entomol. Fr. 1972, 7, 737–770. [Google Scholar]
- Barker, D.; Pagel, M. Predicting functional gene links from phylogenetic-statistical analyses of whole genomes. PLoS Comput. Biol. 2005, 1, 24–31. [Google Scholar] [CrossRef]
- Kim, J.; Roca-Cusachs, M.; Jung, S. Molecular phylogeny of Nabidae (Hemiptera: Heteroptera: Cimicomorpha): Insight into relationships and reclassification with the proposal of the new tribe Stenonabini. Syst. Entomol. 2022, 47, 1–12. [Google Scholar] [CrossRef]
- Štys, P.; Kerzhner, I.M. Rank and nomenclature of higher taxa in recent Heteroptera. Act. Entomol. Bohemos. 1975, 72, 65–79. [Google Scholar]
- Cassis, G.; Gross, G.F. Hemiptera: Heteroptera (Coleorrhyncha to Cimicomorpha). In Zoological Catalogue of Australia; Houston, W.W.K., Maynard, G.V., Eds.; CSIRO: Melbourne, Australia, 1995; Volume 37. [Google Scholar]
- Péricart, J. Family Anthocoridae fieber, 1836-flower bugs, minute pirate bugs. In Catalogue of the Heteroptera of the Palaearctic Region. Vol. 2. Cimicomorpha I; Aukema, B., Rieger, C., Eds.; The Netherlands Entomological Society: Amsterdam, The Netherlands, 1996; Volume 2, pp. 108–140. [Google Scholar]
- Schuh, R.T.; Weirauch, C. True Bugs of the World (Hemiptera: Heteroptera): Classification and Natural History, 2nd ed.; Siri Scientific Press: Manchester, UK, 2020. [Google Scholar]
- Tatarnic, N.J.; Cassis, G. Sexual coevolution in the traumatically inseminating plant bug genus Coridromius. J. Evol. Biol. 2010, 23, 1321–1326. [Google Scholar] [CrossRef]

| Family | Subfamily/Tribe | Species | Copulation Type | Paragenitalia Type | Collecting Country | Accession Number | |||
|---|---|---|---|---|---|---|---|---|---|
| 18S rRNA | 28S rRNA | 16S rRNA | COI | ||||||
| Ingroup | |||||||||
| Anthocoridae | Oriini | Orius atratus | TI | CT | Japan | GQ258414 | GQ258449 | GQ258387 | GQ292177 |
| Orius laevigatus | TI | CT | The Netherlands | GQ258416 | GQ258451 | GQ258371 | GQ292148 | ||
| Orius niger | TI | CT | Nepal | GQ258418 | GQ258453 | GQ258392 | GQ292182 | ||
| Orius majusculus | TI | CT | USA | JQ782789 | JQ782811 | JQ782758 | JQ782832 | ||
| Orius minutus | TI | CT | Republic of Korea | GQ258417 | GQ258452 | GQ258372 | GQ292157 | ||
| Orius strigicollis | TI | CT | Republic of Korea | GQ258420 | GQ258455 | GQ258374 | GQ292146 | ||
| Bilia japonica | TI | CT | Nepal | GQ258406 | GQ258439 | GQ258363 | JQ782816 | ||
| Montandoniola moraguesi | TI | CT | Republic of Korea | GQ258413 | GQ258448 | GQ258370 | - | ||
| Anthocorini | Anthocoris chibi | TI | CT | Republic of Korea | GQ258403 | GQ258437 | GQ258362 | GQ292164 | |
| Anthocoris miyamotoi | TI | CT | Republic of Korea | GQ258405 | GQ258438 | GQ258361 | GQ292152 | ||
| Anthocoris tomentosus | TI | CT | USA | JQ782774 | JQ782810 | JQ782755 | JQ782815 | ||
| Anthocoris japonicus | TI | CT | Republic of Korea | GQ258404 | GQ258436 | GQ258360 | GQ292142 | ||
| Cardiastethini | Amphiareus ruficollaris | TI | None | Japan | GQ258394 | GQ258430 | GQ258383 | GQ292169 | |
| Amphiareus obscuriceps | TI | None | Republic of Korea | GQ258393 | GQ258429 | GQ258358 | GQ292178 | ||
| Amphiareus constrictus | TI | None | Japan | GQ258397 | GQ258427 | GQ258359 | GQ292170 | ||
| Amphiareus morimotoi | TI | None | Japan | GQ258398 | GQ258428 | GQ258361 | GQ292174 | ||
| Buchananiella crassicornis | TI | OP | Malaysia | GQ258407 | GQ258441 | GQ258364 | GQ292144 | ||
| Buchananiella leptocephala | TI | OP | Malaysia | GQ258408 | GQ258442 | GQ258365 | JQ782817 | ||
| Physopleurella armata | TI | None | Republic of Korea | GQ258421 | GQ258456 | GQ258375 | GQ292167 | ||
| Cardiastethus exiguus | TI | OP | Republic of Korea | GQ258409 | GQ258443 | GQ258366 | GQ292165 | ||
| Scolopini | Scoloposcelis albodecussata | TI | CT | Japan | GQ258422 | GQ258457 | GQ258376 | GQ292128 | |
| Scoloposcelis koreanus | TI | CT | Republic of Korea | GQ258423 | GQ258458 | GQ258377 | GQ292130 | ||
| Xylocorini | Xylocoris cerealis | TI | ES | Thailand | GQ258395 | GQ258459 | GQ258384 | GQ292172 | |
| Xylocoris flavipes | TI | None | Thailand | JQ782790 | JQ782795 | JQ782756 | JQ782835 | ||
| Almeidini | Almeida pilosa | TI | None | Thailand | JQ782793 | JQ782794 | JQ782754 | JQ782814 | |
| Blaptostethini | Blaptostethus aurivillus | TI | DCT | Malaysia | GQ258400 | GQ258440 | JQ782772 | - | |
| Lyctocoridae | Lyctocoris beneficus | TI | None | Republic of Korea | GQ258412 | GQ258447 | GQ258369 | JQ782826 | |
| Lyctocoris sp. | TI | None | Cambodia | JQ782786 | JQ782804 | JQ782757 | JQ782827 | ||
| Lasiochilidae | Lasiochilus japonicus | SI | None | Republic of Korea | GQ258410 | GQ258445 | GQ258367 | GQ292184 | |
| Lasiochilus luceonotatus | SI | None | Japan | GQ258408 | GQ258446 | GQ258368 | JQ782825 | ||
| Cimicidae | Cimicinae | Cimex lectularius | TI | ES | USA | JQ782782 | JQ782797 | JQ782771 | JQ782823 |
| Cimex adjunctus | TI | ES | USA | JQ782778 | JQ782801 | JQ782767 | JQ782820 | ||
| Cimex hemipterus | TI | ES | Malaysia | JQ782779 | JQ782802 | JQ782768 | JQ782821 | ||
| Cimex pipistrelli | TI | ES | Czech Republic | JQ782780 | JQ782803 | JQ782770 | JQ782824 | ||
| Cimex japonicus | TI | ES | Japan | JQ782781 | JQ782796 | JQ782769 | JQ782822 | ||
| Cacodminae | Cacodmus vicinus | TI | ES | Egypt | JQ782777 | JQ782800 | JQ782766 | JQ782819 | |
| Cacodmus sp. | TI | ES | Morocco | JQ782776 | JQ782799 | JQ782765 | JQ782818 | ||
| Aphrania elongata | TI | None | Algeria | JQ782775 | JQ782798 | JQ782764 | - | ||
| Curaliidae | Curalium cronini | uncertain | uncertain | USA | EU683128 | - | - | - | |
| Plokiophilidae | Plokiophilinae | Plokiophiloides sp. | uncertain | None | Laos | JQ782792 | JQ782813 | - | - |
| Plokiophilinae | Embiophila sp. | uncertain | None | Thailand | JQ782791 | JQ782812 | JQ782773 | - | |
| Outgroup | |||||||||
| Microphysidae | Loricula pilosella | SI | None | Republic of Korea | GU194610 | GU194685 | GU194532 | GU194763 | |
| Loricula elegantula | SI | None | EU683151 | AY252577 | EU683098 | - | |||
| Nabidae | Nabinae | Nabis stenoferus | SI | None | Republic of Korea | GQ258426 | GQ258434 | GQ258379 | GQ292211 |
| Nabis apicallis | SI | None | Cambodia | JQ782783 | JQ782806 | JQ782761 | JQ782828 | ||
| Nabis punctatus | SI | None | Republic of Korea | JQ782785 | JQ782807 | JQ782763 | JQ782830 | ||
| Nabis ferus | SI | None | Cambodia | JQ782784 | JQ782805 | JQ782762 | JQ782829 | ||
| Nabis flavomarginatus | SI | None | Republic of Korea | GQ258424 | GQ258433 | GQ258380 | GQ292213 | ||
| Himacerus apterus | SI | None | Republic of Korea | GQ258425 | GQ258435 | GQ258381 | GQ292205 | ||
| Prostemmatinae | Prostemma sp. | TI | None | Cambodia | JQ782788 | JQ782809 | JQ782760 | JQ782834 | |
| Prostemma hilgendorfii | TI | None | Cambodia | JQ782787 | JQ782808 | JQ782759 | JQ782833 | ||
| Joppeicidae | Joppeicus paradoxus | SI | None | USA | EU683147 | EU683200 | EU683094 | AY252951 | |
| Reduviidae | Emesaya brevipennis | SI | None | USA | EU683139 | AY252560 | AY252796 | EU683231 | |
| Region | Primer | Sequence | Annealing Temp. | Reference |
|---|---|---|---|---|
| COI | LCO1490 HCO2198 | GGTCAACAAATCATAAAGATATTGG TAAACTTCAGGGTGACAAAAAATCA | 43.5–48 °C | [32] |
| 18S rRNA | 18S-1 18S-4 18S-2 18S-3 | CTGGTTGATCCTGCCAGTAGT GATCCTTCTGCAGGTTCACC AGATACCGCCCTAGTTCTAACC GGTTAGAACTAGGGCGGTATCT | 48–50 °C | [33] |
| 28S rRNA | 28S-DD 28S-FF | GGGACCCGTCTTGAAACAC TTACACACTCCTTAGCGGAT | 45–50 °C | [34] |
| 16S rRNA | 16S-A 16S-B | CGCCTGTTTAACAAAAACAT CCGGTTGAACTCAGATCA | 45–50 °C | [35,36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, S.; Kim, J.; Balvín, O.; Yamada, K. Molecular Phylogeny of Cimicoidea (Heteroptera: Cimicomorpha) Revisited: Increased Taxon Sampling Reveals Evolution of Traumatic Insemination and Paragenitalia. Insects 2023, 14, 267. https://doi.org/10.3390/insects14030267
Jung S, Kim J, Balvín O, Yamada K. Molecular Phylogeny of Cimicoidea (Heteroptera: Cimicomorpha) Revisited: Increased Taxon Sampling Reveals Evolution of Traumatic Insemination and Paragenitalia. Insects. 2023; 14(3):267. https://doi.org/10.3390/insects14030267
Chicago/Turabian StyleJung, Sunghoon, Junggon Kim, Ondřej Balvín, and Kazutaka Yamada. 2023. "Molecular Phylogeny of Cimicoidea (Heteroptera: Cimicomorpha) Revisited: Increased Taxon Sampling Reveals Evolution of Traumatic Insemination and Paragenitalia" Insects 14, no. 3: 267. https://doi.org/10.3390/insects14030267
APA StyleJung, S., Kim, J., Balvín, O., & Yamada, K. (2023). Molecular Phylogeny of Cimicoidea (Heteroptera: Cimicomorpha) Revisited: Increased Taxon Sampling Reveals Evolution of Traumatic Insemination and Paragenitalia. Insects, 14(3), 267. https://doi.org/10.3390/insects14030267






