Characterization of Iflavirus in the Red Flour Beetle, Tribolium castaneum (Coleoptera; Tenebrionidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Insects
2.2. Cell Culture
2.3. SRA Data Search
2.4. RT-qPCR Assays
2.5. TcIV Genome Annotation and Phylogenetic Analysis
2.6. TcIV in Different Tissues and in Vertical Transmission
2.7. Immunocytochemistry (ICC)
3. Results
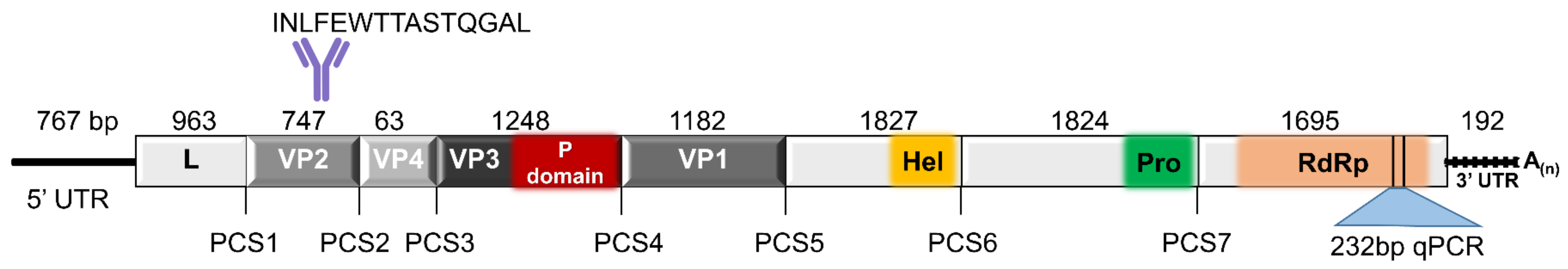
3.1. TcIV Genome Organization
3.2. TcIV Prevalence
3.3. TcIV Phylogenetic Analysis
3.4. TcIV in Sequence Read Archive Data (SRA)
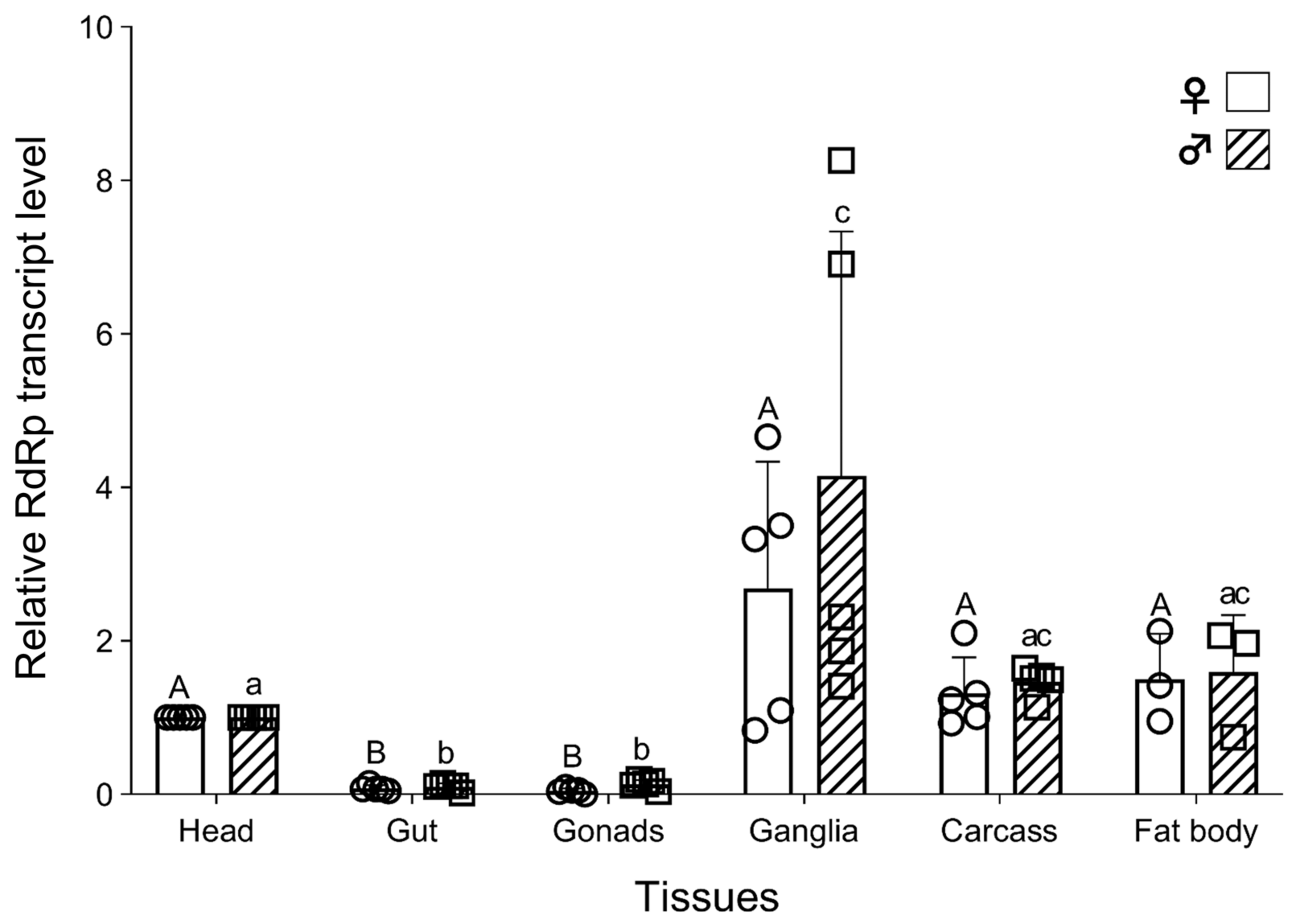
3.5. Tissue-Specific Distribution of TcIV
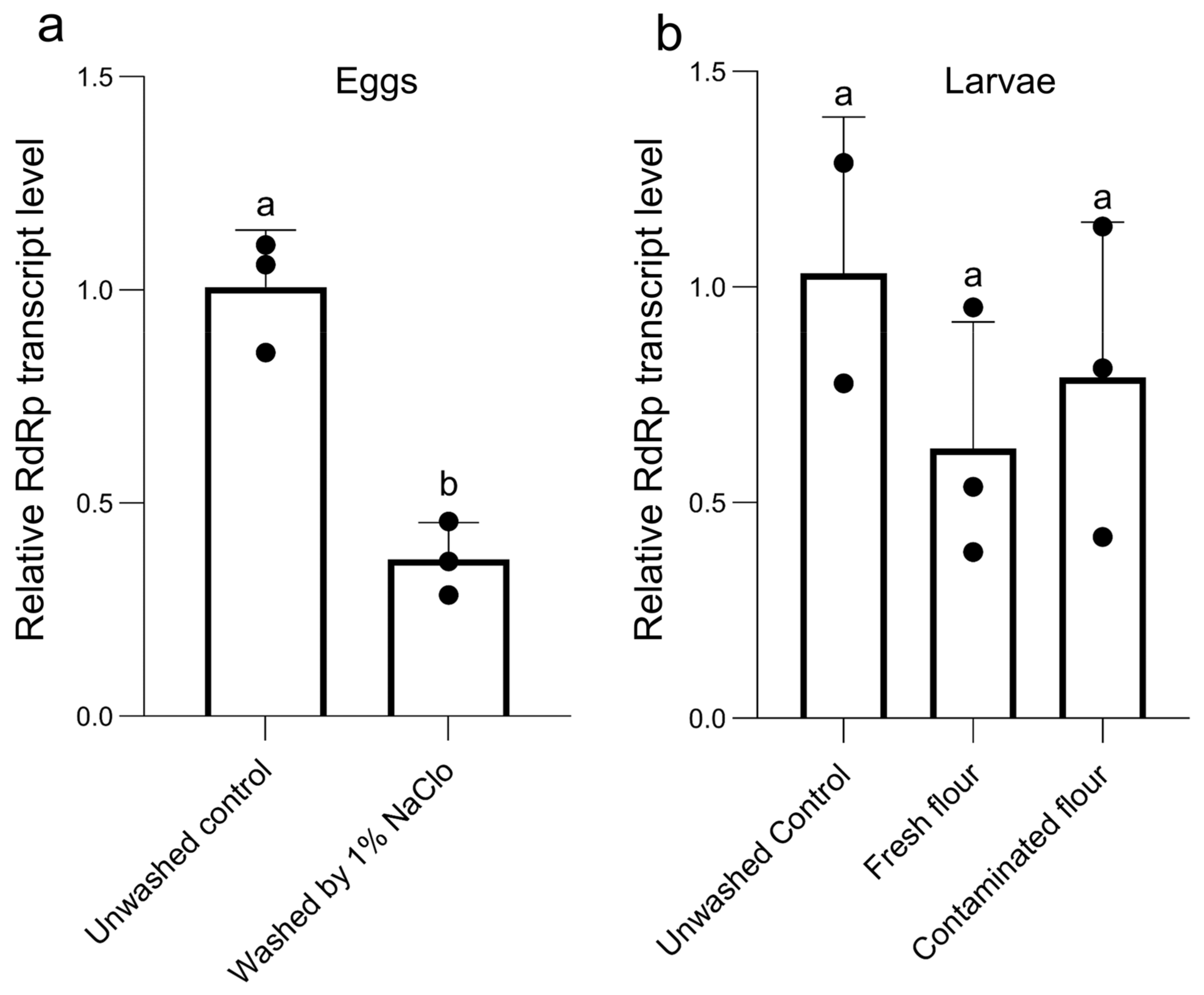
3.6. Vertical Transmission of TcIV
3.7. TcIV Infections in TcA Cell
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Consortium, T.G.S.; Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Beeman, R.W.; Brown, S.J.; et al. The Genome of the Model Beetle and Pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar] [CrossRef]
- Kim, H.S.; Murphy, T.; Xia, J.; Caragea, D.; Park, Y.; Beeman, R.W.; Lorenzen, M.D.; Butcher, S.; Manak, J.R.; Brown, S.J. BeetleBase in 2010: Revisions to Provide Comprehensive Genomic Information for Tribolium castaneum. Nucleic Acids Res. 2010, 38, D437–D442. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.L.; Stanley, D.; Ringbauer, J.A.; Beeman, R.W.; Silver, K.; Park, Y. A Cell Line Derived from the Red Flour Beetle Tribolium castaneum (Coleoptera: Tenebrionidae). Vitr. Cell Dev. Biol. Anim. 2012, 48, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Silver, K.; Jiang, H.; Fu, J.; Phillips, T.W.; Beeman, R.W.; Park, Y. The Tribolium castaneum Cell Line TcA: A New Tool Kit for Cell Biology. Sci. Rep. 2014, 4, 6840. [Google Scholar] [CrossRef]
- Miller, S.C.; Brown, S.J.; Tomoyasu, Y. Larval RNAi in Drosophila? Dev. Genes Evol. 2008, 218, 505–510. [Google Scholar] [CrossRef]
- Tomoyasu, Y.; Denell, R.E. Larval RNAi in Tribolium (Coleoptera) for Analyzing Adult Development. Dev. Genes Evol. 2004, 214, 575–578. [Google Scholar] [CrossRef]
- Bucher, G.; Scholten, J.; Klingler, M. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 2002, 12, R85–R86. [Google Scholar] [CrossRef]
- Gilles, A.F.; Schinko, J.B.; Averof, M. Efficient CRISPR-Mediated Gene Targeting and Transgene Replacement in the Beetle Tribolium castaneum. Development 2015, 142, 2832–2839. [Google Scholar] [CrossRef]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frézal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-Arthropod Analysis Reveals Somatic PiRNAs as an Ancestral Defence against Transposable Elements. Nat. Ecol. Evol. 2018, 2, 174–181. [Google Scholar] [CrossRef]
- Van Oers, M.M. Genomics and Biology of Iflaviruses; Caister Academic Press: Poole, UK, 2010; pp. 227–250. [Google Scholar]
- Škubník, K.; Sukeník, L.; Buchta, D.; Füzik, T.; Procházková, M.; Moravcová, J.; Šmerdová, L.; Přidal, A.; Vácha, R.; Plevka, P. Capsid Opening Enables Genome Release of Iflaviruses. Sci. Adv. 2021, 7, eabd7130. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Christmon, K.; Heerman, M.C.; Posada-Florez, F.; Harrison, R.L.; Chen, Y.; Evans, J.D. Development of a Honey Bee RNA Virus Vector Based on the Genome of a Deformed Wing Virus. Viruses 2020, 12, 374. [Google Scholar] [CrossRef] [PubMed]
- Organtini, L.J.; Shingler, K.L.; Ashley, R.E.; Capaldi, E.A.; Durrani, K.; Dryden, K.A.; Makhov, A.M.; Conway, J.F.; Pizzorno, M.C.; Hafenstein, S. Honey Bee Deformed Wing Virus Structures Reveal That Conformational Changes Accompany Genome Release. J. Virol. 2017, 91, e01716–e01795. [Google Scholar] [CrossRef] [PubMed]
- Benaets, K.; van Geystelen, A.; Cardoen, D.; De Smet, L.; de Graaf, D.C.; Schoofs, L.; Larmuseau, M.H.D.; Brettell, L.E.; Martin, S.J.; Wenseleers, T. Covert Deformed Wing Virus Infections Have Long-Term Deleterious Effects on Honeybee Foraging and Survival. Proc. R. Soc. Biol. Sci. 2017, 284, 20162149. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Genersch, E. Deformed Wing Virus. J. Invertebr. Pathol. 2010, 103 (Suppl. 1), S48–S61. [Google Scholar] [CrossRef]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and Biological Characterization of Deformed Wing Virus of Honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef]
- Chen, Y.P.; Higgins, J.A.; Feldlaufer, M.F. Quantitative Real-Time Reverse Transcription-PCR Analysis of Deformed Wing Virus Infection in the Honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 2005, 71, 436–441. [Google Scholar] [CrossRef]
- Ongus, J.R.; Peters, D.; Bonmatin, J.-M.; Bengsch, E.; Vlak, J.M.; van Oers, M.M. Complete Sequence of a Picorna-like Virus of the Genus Iflavirus Replicating in the Mite Varroa destructor. J. Gen. Virol. 2004, 85, 3747–3755. [Google Scholar] [CrossRef]
- Carrau, T.; Lamp, B.; Reuscher, C.M.; Vilcinskas, A.; Lee, K.-Z. Organization of the Structural Protein Region of La Jolla Virus Isolated from the Invasive Pest Insect Drosophila suzukii. Viruses 2021, 13, 740. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Granberg, F.; Onorati, P.; Jansson, A.; Berggren, Å. Virus Prospecting in Crickets-Discovery and Strain Divergence of a Novel Iflavirus in Wild and Cultivated Acheta Domesticus. Viruses 2021, 13, 364. [Google Scholar] [CrossRef]
- Yuan, H.; Xu, P.; Yang, X.; Graham, R.I.; Wilson, K.; Wu, K. Characterization of a Novel Member of Genus Iflavirus in Helicoverpa armigera. J. Invertebr. Pathol. 2017, 144, 65–73. [Google Scholar] [CrossRef]
- Ye, Z.-X.; Li, Y.-H.; Chen, J.-P.; Zhang, C.-X.; Li, J.-M. Complete Genome Analysis of a Novel Iflavirus from a Leaf Beetle, Aulacophora lewisii. Arch. Virol. 2021, 166, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, D.; Isawa, H.; Fujita, R.; Murota, K.; Itokawa, K.; Higa, Y.; Katayama, Y.; Sasaki, T.; Mizutani, T.; Iwanaga, S.; et al. Isolation and Characterization of a New Iflavirus from Armigeres Spp. Mosquitoes in the Philippines. J. Gen. Virol. 2017, 98, 2876–2881. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-L.; Ye, Z.-X.; Chen, J.-P.; Zhang, C.-X.; Huang, H.-J.; Li, J.-M. Characterization of Two Novel Insect-Specific Viruses Discovered in the Green Leafhopper, Cicadella viridis. Insects 2022, 13, 378. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.A.; Ardisson-Araujo, D.M.P.; Tinoco, R.S.; Fernandes, O.A.; Melo, F.L.; Ribeiro, B.M. Complete Genome Sequence and Structural Characterization of a Novel Iflavirus Isolated from Opsiphanes invirae (Lepidoptera: Nymphalidae). J. Invertebr. Pathol. 2015, 130, 136–140. [Google Scholar] [CrossRef]
- Beeman, R.W.; Stuart, J.J. A Gene for Lindane + Cyclodiene Resistance in the Red Flour Beetle (Coleoptera: Tenebrionidae). J. Econ. Entomol. 1990, 83, 1745–1751. [Google Scholar] [CrossRef]
- Juergens, K.B.; Huckabee, J.; Greninger, A.L. Two Novel Iflaviruses Discovered in Bat Samples in Washington State. Viruses 2022, 14, 994. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Park, Y.; Aikins, J.; Wang, L.J.; Beeman, R.W.; Oppert, B.; Lord, J.C.; Brown, S.J.; Lorenzen, M.D.; Richards, S.; Weinstock, G.M.; et al. Analysis of Transcriptome Data in the Red Flour Beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2008, 38, 380–386. [Google Scholar] [CrossRef]
- Procházková, M.; Füzik, T.; Škubník, K.; Moravcová, J.; Ubiparip, Z.; Přidal, A.; Plevka, P. Virion Structure and Genome Delivery Mechanism of Sacbrood Honeybee Virus. Proc. Natl. Acad. Sci. USA 2018, 115, 7759–7764. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, S.; Shan, T.; Hou, R.; Liu, Z.; Li, W.; Guo, L.; Wang, Y.; Chen, P.; Wang, X.; et al. Virome Comparisons in Wild-Diseased and Healthy Captive Giant Pandas. Microbiome 2017, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.H.; Che, X.; Garcia, J.A.; Klena, J.D.; Lee, B.; Muller, D.; Ulrich, W.; Corrigan, R.M.; Nichol, S.; Jain, K.; et al. Viral Diversity of House Mice in New York City. mBio 2018, 9, e01354-17. [Google Scholar] [CrossRef] [PubMed]
- Naoi, Y.; Kishimoto, M.; Masuda, T.; Ito, M.; Tsuchiaka, S.; Sano, K.; Yamasato, H.; Omatsu, T.; Aoki, H.; Furuya, T.; et al. Characterization and Phylogenetic Analysis of a Novel Picornavirus from Swine Feces in Japan. Arch. Virol. 2016, 161, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Dainat, B.; Locke, B.; Cordoni, G.; Berthoud, H.; Gauthier, L.; Neumann, P.; Budge, G.E.; Ball, B.V.; Stoltz, D.B. Genetic Characterization of Slow Bee Paralysis Virus of the Honeybee (Apis mellifera L.). J. Gen. Virol. 2010, 91, 2524–2530. [Google Scholar] [CrossRef]
- Cato, A.J.; Elliott, B.; Nayak, M.K.; Phillips, T.W. Geographic Variation in Phosphine Resistance Among North American Populations of the Red Flour Beetle (Coleoptera: Tenebrionidae). J. Econ. Entomol. 2017, 110, 1359–1365. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.-D.; Tian, J.-H.; Chen, L.-J.; Chen, X.; Li, C.-X.; Qin, X.-C.; Li, J.; Cao, J.-P.; Eden, J.-S.; et al. Redefining the Invertebrate RNA Virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Bailey, L.; Fernando, E.F.W. Effects of Sacbrood Virus on Adult Honey-Bees. Ann. Appl. Biol. 1972, 72, 27–35. [Google Scholar] [CrossRef]
- Carballo, A.; Murillo, R.; Jakubowska, A.; Herrero, S.; Williams, T.; Caballero, P. Co-Infection with Iflaviruses Influences the Insecticidal Properties of Spodoptera Exigua Multiple Nucleopolyhedrovirus Occlusion Bodies: Implications for the Production and Biosecurity of Baculovirus Insecticides. PLoS ONE 2017, 12, e0177301. [Google Scholar] [CrossRef]
- Carballo, A.; Williams, T.; Murillo, R.; Caballero, P. Iflavirus Covert Infection Increases Susceptibility to Nucleopolyhedrovirus Disease in Spodoptera exigua. Viruses 2020, 12, 509. [Google Scholar] [CrossRef]
- Yuan, H.; Xu, P.; Xiao, Y.; Yang, L.; Yang, X.; Wu, K. Infection of Cotton Bollworm by Helicoverpa armigera Iflavirus Decreases Larval Fitness. J. Invertebr. Pathol. 2020, 173, 107384. [Google Scholar] [CrossRef]
- Carrillo-Tripp, J.; Krueger, E.N.; Harrison, R.L.; Toth, A.L.; Miller, W.A.; Bonning, B.C. Lymantria dispar Iflavirus 1 (LdIV1), a New Model to Study Iflaviral Persistence in Lepidopterans. J. Gen. Virol. 2014, 95, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, Y.; Bonning, B.C. RNA Virus Discovery in Insects. Curr. Opin. Insect Sci. 2015, 8, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Fabricant, C.G.; Gillespie, J.H.; Krook, L. Intracellular and extracellular mineral crystal formation induced by viral infection of cell cultures. Infect Immun. 1971, 3, 416–419. [Google Scholar] [CrossRef] [PubMed]
- Fannon, J.M.; Ryabov, E.V. Iflavirus (Deformed Wing Virus). Routledge Handbooks; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Xu, P.; Liu, Y.; Graham, R.I.; Wilson, K.; Wu, K. Densovirus Is a Mutualistic Symbiont of a Global Crop Pest (Helicoverpa armigera) and Protects against a Baculovirus and Bt Biopesticide. PLoS Pathog. 2014, 10, e1004490. [Google Scholar] [CrossRef]
- Bitondi, M.M.G.; Nascimento, A.M.; Cunha, A.D.; Guidugli, K.R.; Nunes, F.M.F.; Simões, Z.L.P. Characterization and Expression of the Hex 110 Gene Encoding a Glutamine-Rich Hexamerin in the Honey Bee, Apis mellifera. Arch. Insect Biochem. Physiol. 2006, 63, 57–72. [Google Scholar] [CrossRef]
- Park, C.; Kang, H.S.; Jeong, J.; Kang, I.; Choi, K.; Yoo, M.-S.; Kim, Y.-H.; Kang, S.-W.; Lim, H.-Y.; Yoon, B.-S.; et al. In-Situ Hybridization for the Detection of Sacbrood Virus in Infected Larvae of the Honey Bee (Apis cerana). J. Comp. Pathol. 2016, 154, 258–262. [Google Scholar] [CrossRef]



| Conserved Protease Cleavage Sites in Different Iflaviruses | |||||
|---|---|---|---|---|---|
| PCS1 | PCS2 | PCS3 | PCS4 | PCS5 | |
| LP; VP2 | VP2; VP4 | VP4; VP3 | VP3; VP1 | VP1; C-Terminus | |
| TcIV | ALPE321MD | PYPE570MD | DNNR591DN | AYAE1007GE | VSPE1401AL |
| DWV | AKPE211MD | AKPE464MD | GNNM485DN | AIPE901GE | AVPE1288AP |
| SBPV | AQPE176MD | ALPE437MD | DVNC458DN | AEPE888ME | AMPE1303GF |
| DsLJV | PQPQ171ME | PVVQ408ME | IKNM432DK | AFAQ856MD | PSSQ1113ME |
| SRX Accession # | Geographic Location | Strain | Total # of Reads (Range) | % Transcripts |
|---|---|---|---|---|
| SRX1336374..SRX1381752 | USDA ARS | Unknown | 1.06~19.81 M | 0~0.015% |
| SRX1381239..SRX1433712 | USDA ARS | Unknown | 2.72~36.36 M | 0~0.07% |
| SRX2591944..SRX2591991 | Vanderbilt University (GEO) | Unknown | 8.99~24.77 M | 0.0002~0.05% |
| SRX390599..SRX391110 | University of Muenster | SB/Cro1 | 21.82~42.24 M | 0.0005~0.02% |
| SRX2695346 | USDA ARS left for Grain and Animal Health Research | Oppert lab | 154.78 M | 0.003% |
| SRX818546..SRX819651 | Wayne State University | GAI | 23.79~66.43 M | 0.005~0.02% |
| SRX3119712..SRX3118448 | Wayne State University | Unknown | 43.02~88.05 M | 0.006~0.06% |
| SRX7097353..SRX7140859 | Jiangsu University | Unknown | 63.4 M | 0~0.01% |
| SRX2390214.. SRX2390215 | USDA ARS | Unknown | 31.40~32.62 M | 0.015% |
| SRX2265569..SRX2265586 | University of Muenster | Cro1 | 19.60~29.83 M | 0.02% |
| SRX1925245..SRX1925248 | Nanjing University (GEO) | GA-1 | 5.85~6.11 M | 0.02~0.08% |
| SRX5103179..SRX5103190 | South China Agriculture University | Unknown | 7.89~10.38 M | 0.05~0.06% |
| SRX3406391 | Nanjing Normal University | Unknown | 6.85 M | 0.07% |
| SRX5361486..SRX5361487 | Nanjing Normal University | Unknown | 70.59~74.60 M | 0.07% |
| SRX2944938 | Nanjing Normal University | GA-1 | 6.83 M | 0.07% |
| SRX1513481 | Nanjing Normal University | GA-1 | 7.31 M | 0.07% |
| SRX476140-SRX699016 | Nanjing Normal University | GA-1 | 7.12~7.58 M | 0.07% |
| SRX1081871 | Nanjing Normal University | GA-1 | 6.96 M | 0.07% |
| SRX6408778..SRX6408837 | University of Münster (GEO) | Cro1 | 13.28~21.57 M | 0.01~93.19% |
| SRX6808996..SRX6812159 | Harvard University | Unknown | 7.52~46.48 M | 0.12~0.59% |
| SRX8875633..SRX8875638 | Yangzhou University | GA-1 | 24.7~25.97 M | 0.19~70.67% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatehi, S.; Aikins, M.; Philips, T.W.; Brown, S.; Zhu, K.Y.; Scully, E.D.; Park, Y. Characterization of Iflavirus in the Red Flour Beetle, Tribolium castaneum (Coleoptera; Tenebrionidae). Insects 2023, 14, 220. https://doi.org/10.3390/insects14030220
Fatehi S, Aikins M, Philips TW, Brown S, Zhu KY, Scully ED, Park Y. Characterization of Iflavirus in the Red Flour Beetle, Tribolium castaneum (Coleoptera; Tenebrionidae). Insects. 2023; 14(3):220. https://doi.org/10.3390/insects14030220
Chicago/Turabian StyleFatehi, Soheila, Michael Aikins, Thomas W. Philips, Susan Brown, Kun Yan Zhu, Erin D. Scully, and Yoonseong Park. 2023. "Characterization of Iflavirus in the Red Flour Beetle, Tribolium castaneum (Coleoptera; Tenebrionidae)" Insects 14, no. 3: 220. https://doi.org/10.3390/insects14030220
APA StyleFatehi, S., Aikins, M., Philips, T. W., Brown, S., Zhu, K. Y., Scully, E. D., & Park, Y. (2023). Characterization of Iflavirus in the Red Flour Beetle, Tribolium castaneum (Coleoptera; Tenebrionidae). Insects, 14(3), 220. https://doi.org/10.3390/insects14030220