Integrative Taxonomy Approach Reveals Cryptic Diversity within the Phoretic Pseudoscorpion Genus Lamprochernes (Pseudoscorpiones: Chernetidae) †
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Taxonomic Sampling and Species Delimitation Workflow
2.2. Molecular Protocols and Phylogenetic Analyses
2.3. Molecular Species Delimitation
2.4. Divergence Time Analyses
2.5. Cytogenetic Analyses
2.6. Morphological Analyses
2.7. Morphometric Analyses
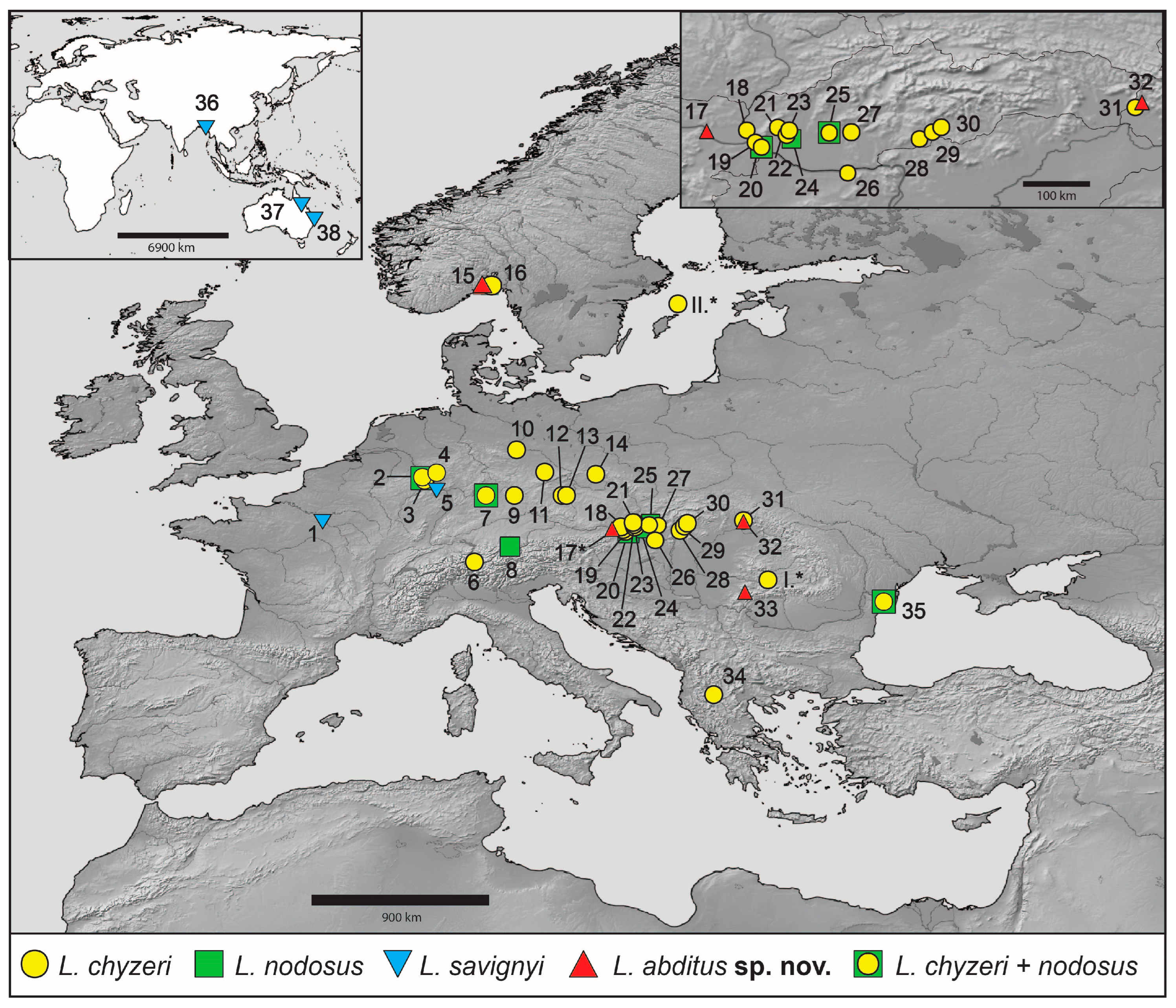
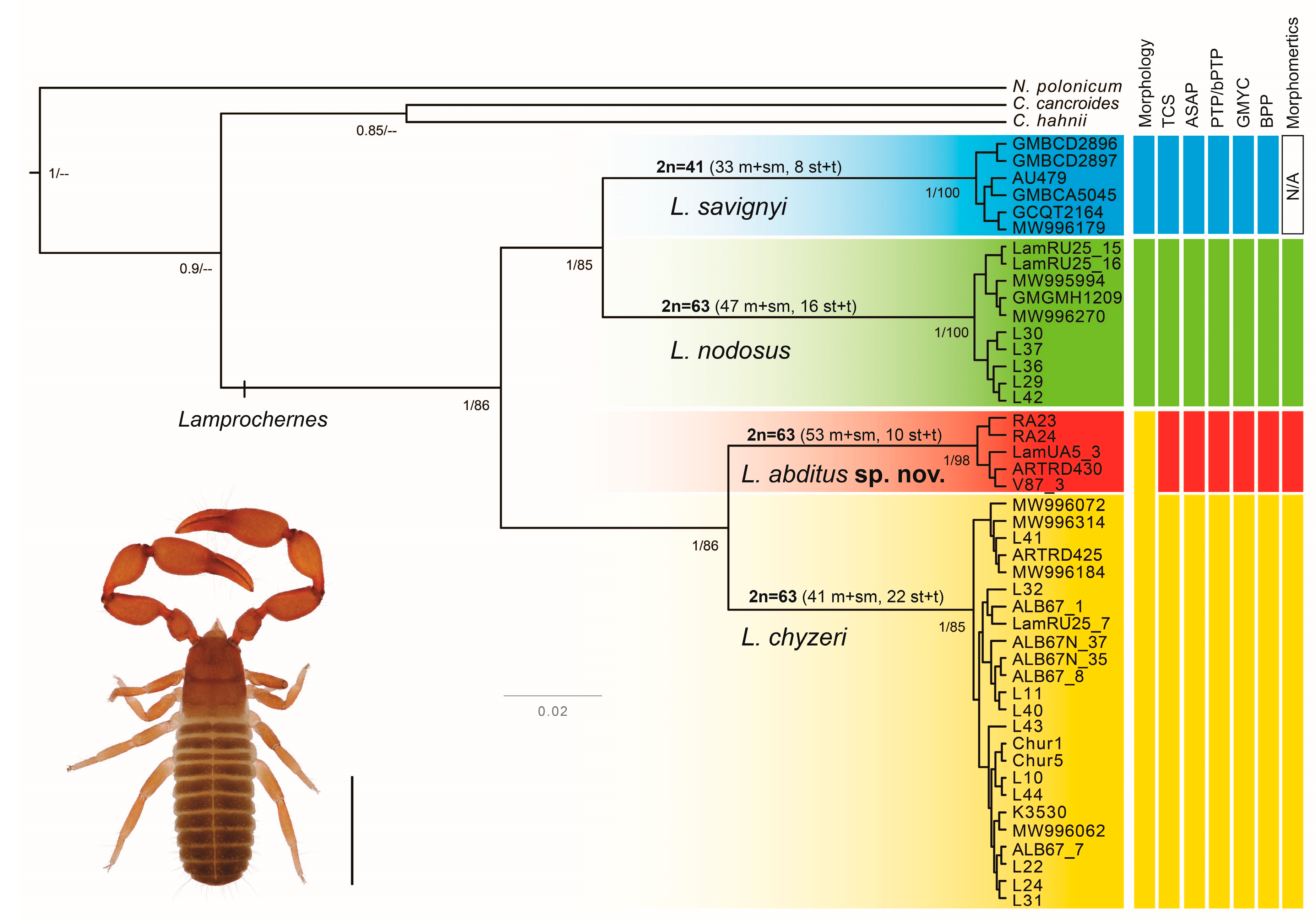
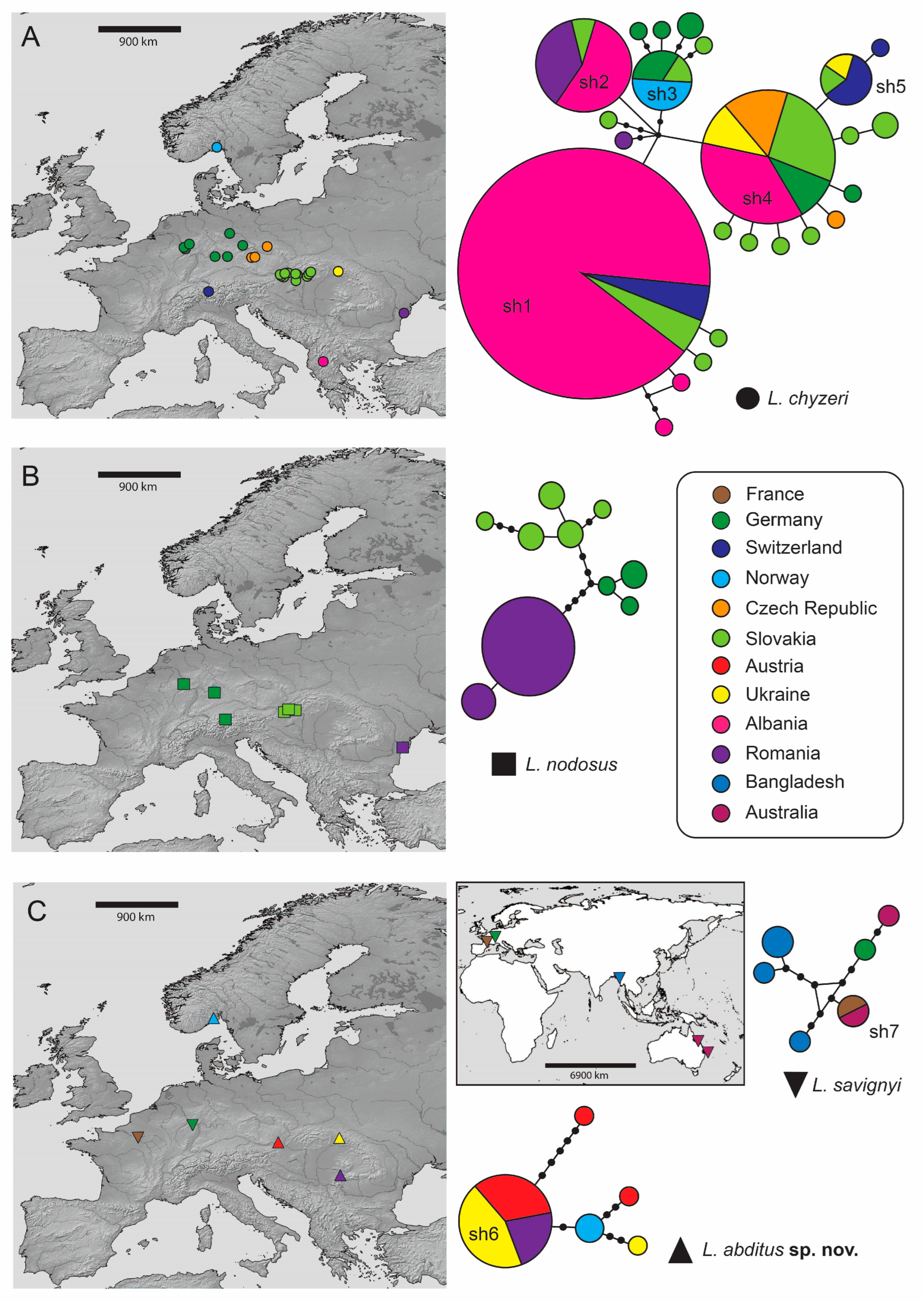
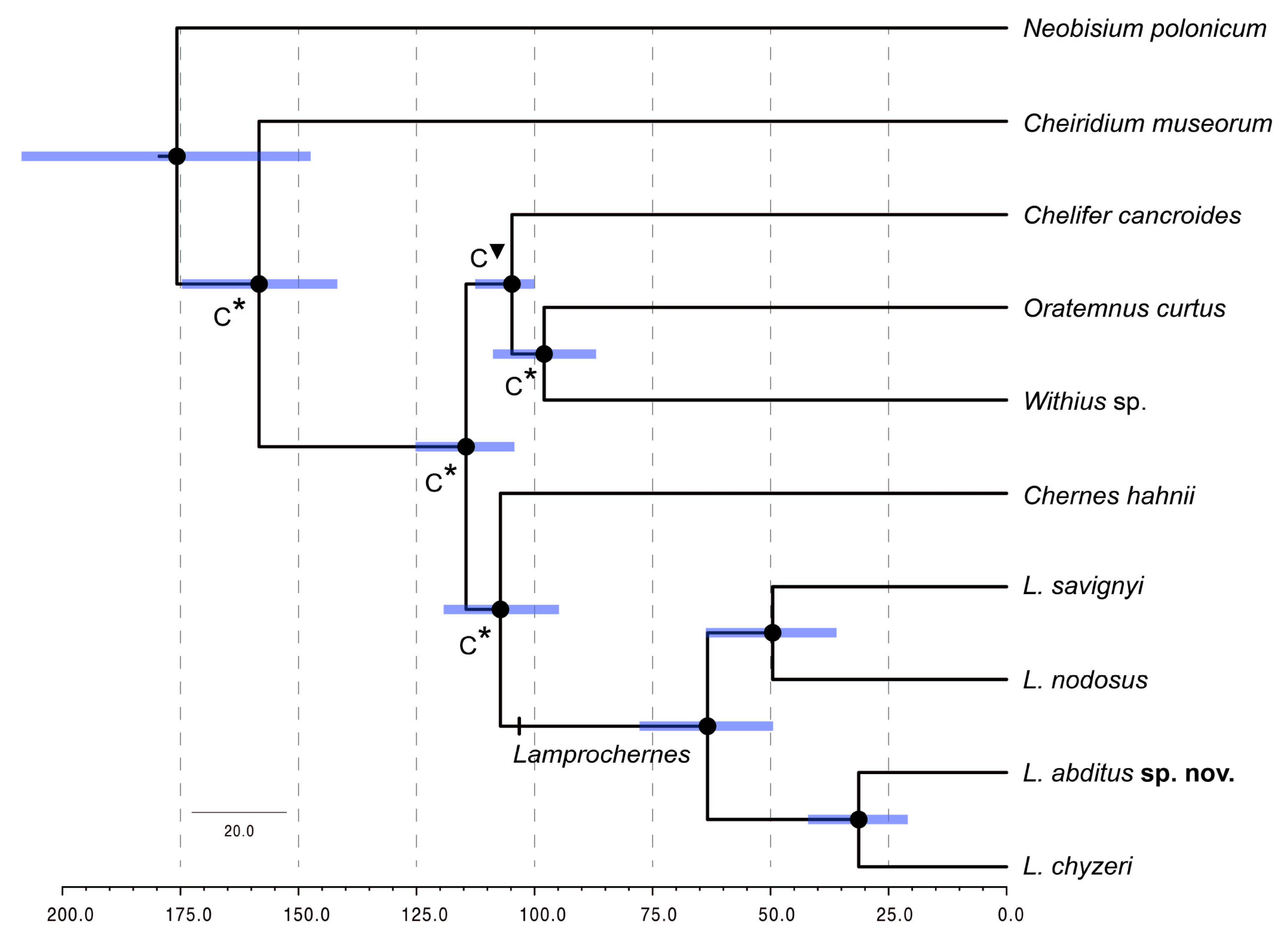
3. Results
3.1. Molecular Protocols and Phylogenetic Analyses
3.2. Molecular Species Delimitation and Lamprochernes Haplotype Network
3.3. Divergence Time Analyses
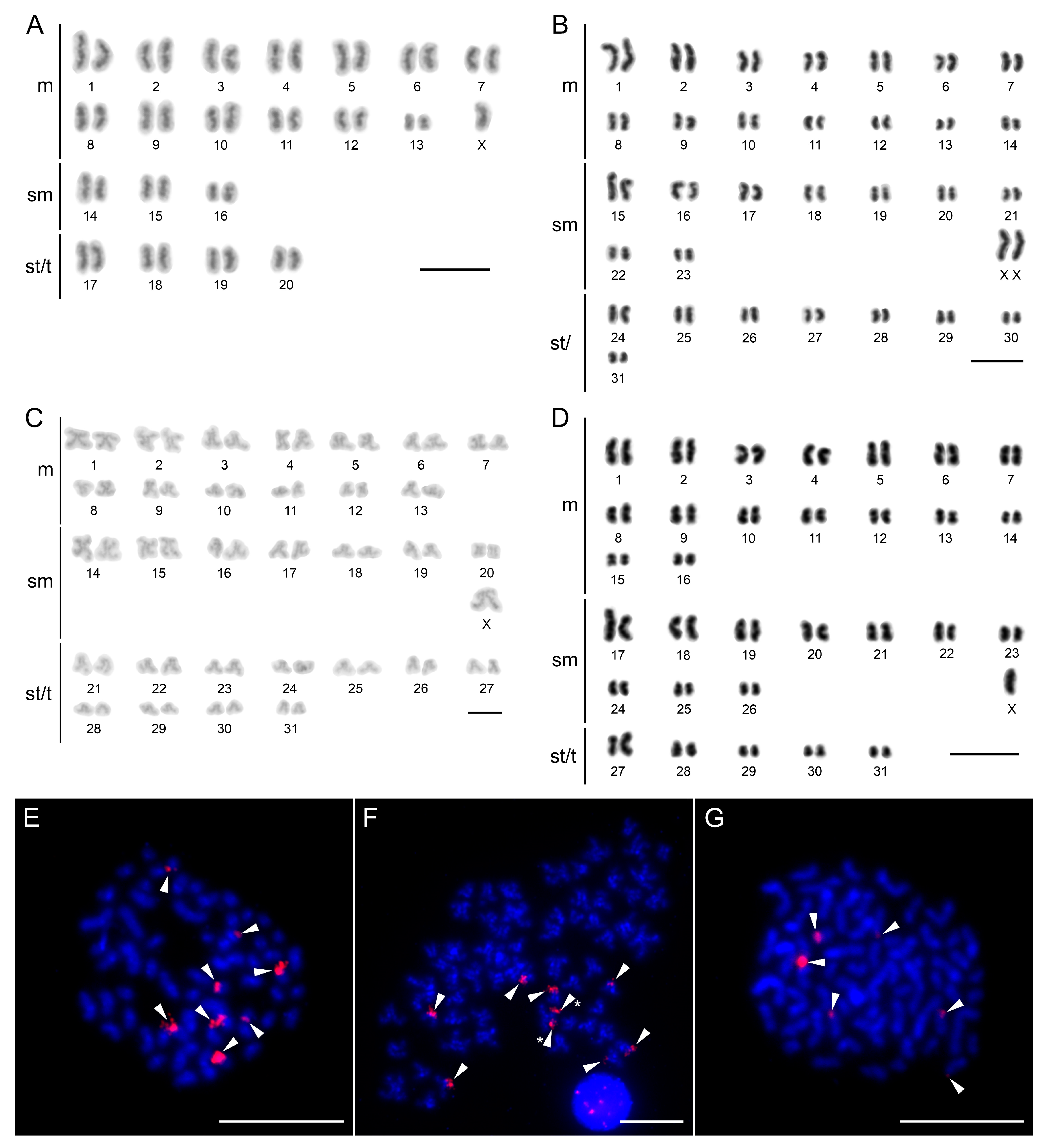
3.4. Cytogenetic Analyses
3.5. Multivariate Morphometrics
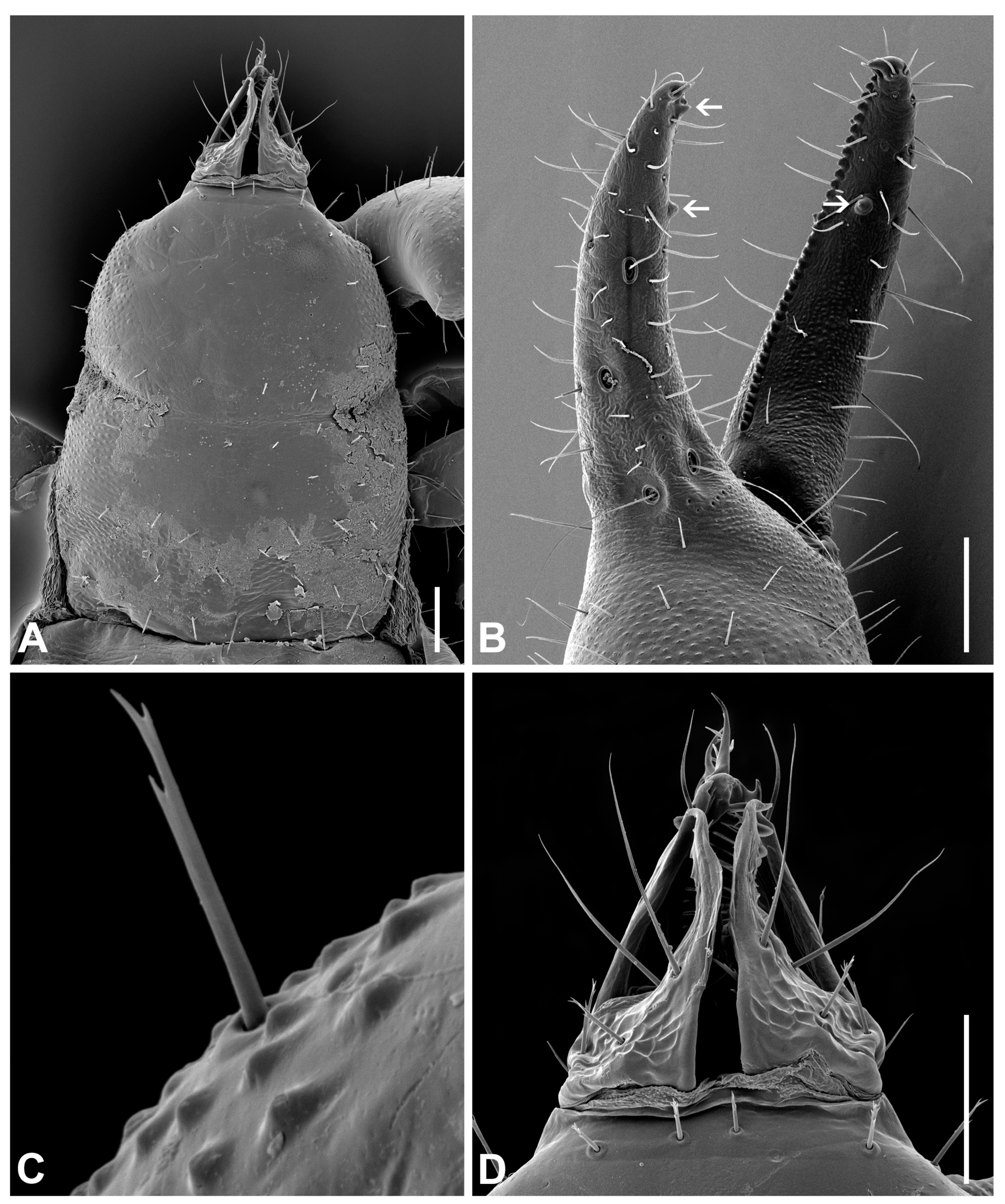
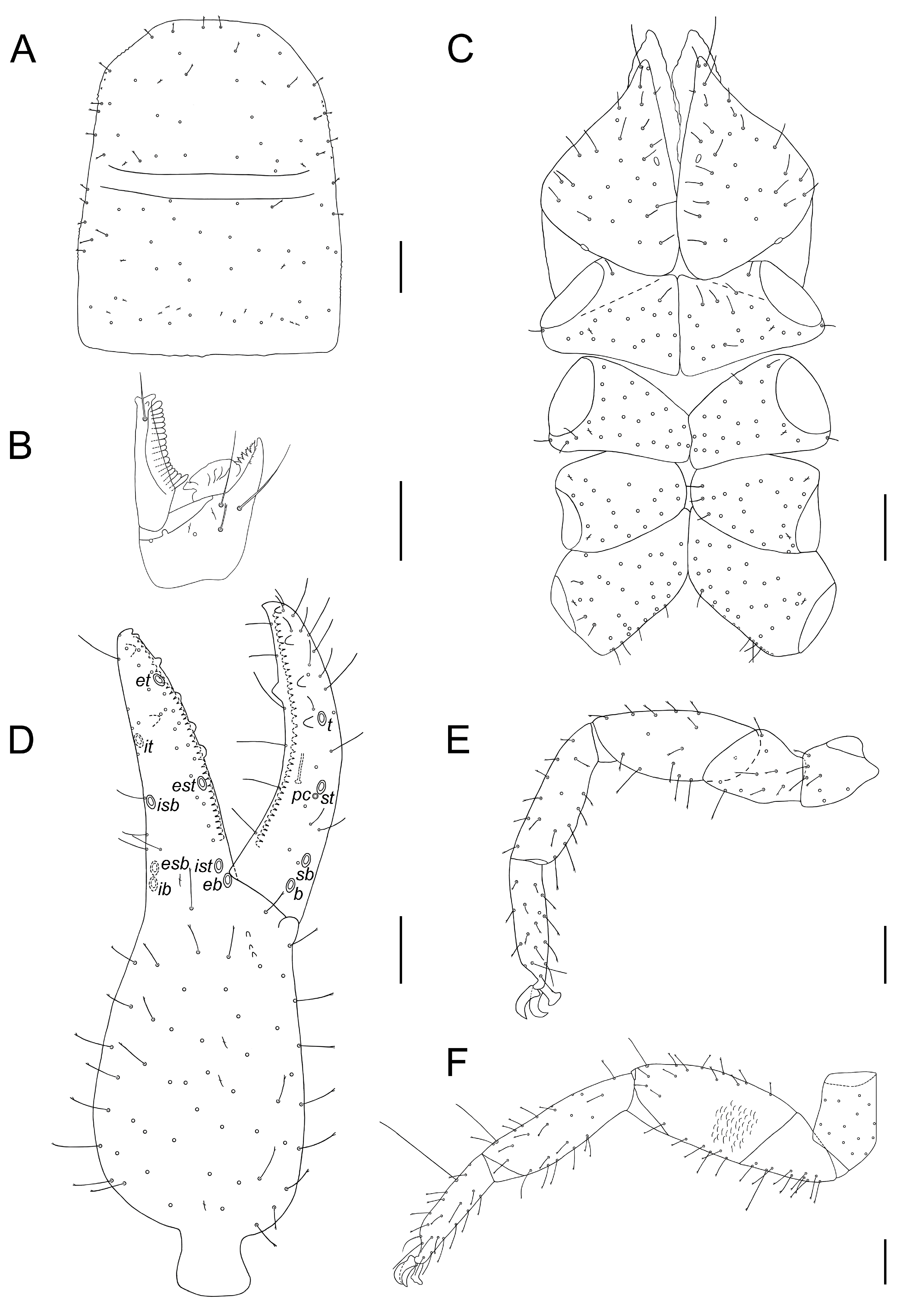
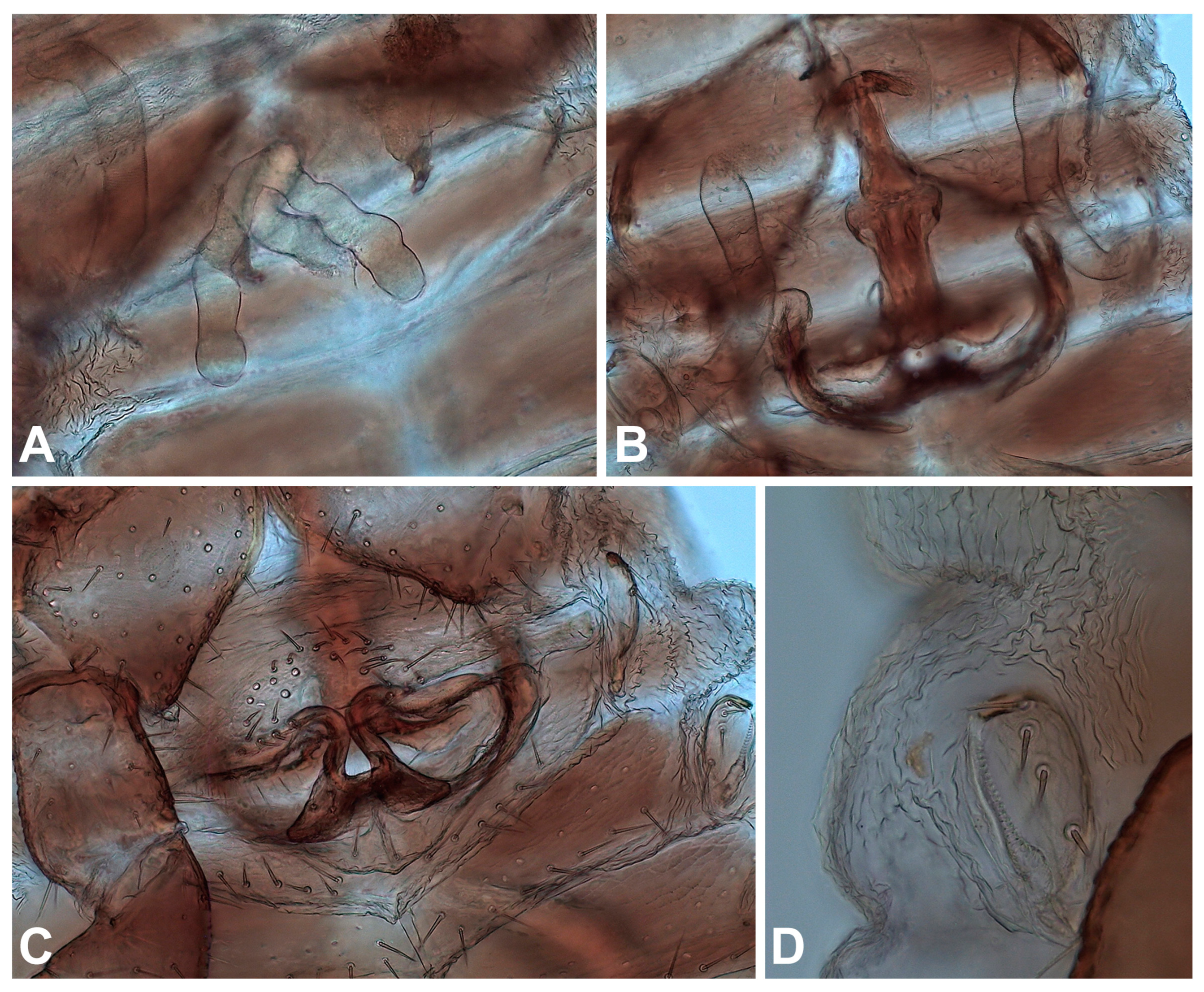
3.6. Morphological Analyses
- Chernetidae Menge, 1855
- Lamprochernetinae Beier, 1932
- Lamprochernes Tömösváry, 1883
- Diagnosis
- Description. Adult male (4♂) and female (7♀).
4. Discussion
4.1. Species Delimitation in Lamprochernes
4.2. Morphological Stasis in Lamprochernes Evolution
4.3. Distribution and Dispersal Capabilities of Lamprochernes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Alastruey-Izquierdo, A.; Alcazar-Fuoli, L.; Cuenca-Estrella, M. Antifungal susceptibility profile of cryptic species of Aspergillus. Mycopathologia 2014, 178, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.J. Multi-resistant aspergillosis due to cryptic species. Mycopathologia 2014, 178, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.; Cristofaro, M.; Bon, M.-C.; De Biase, A.; Petanović, R.; Vidović, B. The importance of cryptic species and subspecific populations in classic biological control of weeds: A North American perspective. BioControl 2018, 63, 417–425. [Google Scholar] [CrossRef]
- Neves, J.M.; Almeida, J.P.; Sturaro, M.J.; Fabre, N.N.; Pereira, R.J.; Mott, T. Deep genetic divergence and paraphyly in cryptic species of Mugil fishes (Actinopterygii: Mugilidae). Syst. Biodivers. 2020, 18, 116–128. [Google Scholar] [CrossRef]
- Davidson-Watts, I.; Walls, S.; Jones, G. Differential habitat selection by Pipistrellus pipistrellus and Pipistrellus pygmaeus identifies distinct conservation needs for cryptic species of echolocating bats. Biol. Conservat. 2006, 133, 118–127. [Google Scholar] [CrossRef]
- Delić, T.; Trontelj, P.; Rendoš, M.; Fišer, C. The importance of naming cryptic species and the conservation of endemic subterranean amphipods. Sci. Rep. 2017, 7, 3391. [Google Scholar] [CrossRef]
- Brochu, C.A.; Sumrall, C.D. Modern cryptic species and crocodylian diversity in the fossil record. Zool. J. Linn. Soc. 2020, 189, 700–711. [Google Scholar] [CrossRef]
- Fišer, C.; Robinson, C.T.; Malard, F. Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 2018, 27, 613–635. [Google Scholar] [CrossRef] [Green Version]
- Struck, T.H.; Feder, J.L.; Bendiksby, M.; Birkeland, S.; Cerca, J.; Gusarov, V.I.; Kistenich, S.; Larsson, K.-H.; Liow, L.H.; Nowak, M.D. Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol. 2018, 33, 153–163. [Google Scholar] [CrossRef]
- de León, G.P.-P.; Nadler, S.A. What we don’t recognize can hurt us: A plea for awareness about cryptic species. J. Parasitol. 2010, 96, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Heethoff, M. Cryptic species–conceptual or terminological chaos? A response to Struck et al. Trends Ecol. Evol. 2018, 33, 310. [Google Scholar] [CrossRef] [PubMed]
- Swift, H.F.; Daglio, L.G.; Dawson, M. Three routes to crypsis: Stasis, convergence, and parallelism in the Mastigias species complex (Scyphozoa, Rhizostomeae). Mol. Phylogenet. Evol. 2016, 99, 103–115. [Google Scholar] [CrossRef] [Green Version]
- DeSalle, R.; Goldstein, P. Review and interpretation of trends in DNA barcoding. Front. Ecol. Evol. 2019, 7, 302. [Google Scholar] [CrossRef] [Green Version]
- Crespo, L.; Domènech, M.; Malumbres-Olarte, J.; Cardoso, P.; Moya-Laraño, J.; Macías-Hernández, N.; Opatova, V.; Arnedo, M. A DNA barcode-assisted annotated checklist of the spider (Arachnida, Araneae) communities associated to white oak woodlands in Spanish National Parks. Biodivers. Data J. 2018, 6, e29443. [Google Scholar] [CrossRef]
- Gill, B.A.; Musili, P.M.; Kurukura, S.; Hassan, A.A.; Goheen, J.R.; Kress, W.J.; Kuzmina, M.; Pringle, R.M.; Kartzinel, T.R. Plant DNA-barcode library and community phylogeny for a semi-arid East African savanna. Mol. Ecol. Resour. 2019, 19, 838–846. [Google Scholar] [CrossRef]
- Maggia, M.-E.; Decaëns, T.; Lapied, E.; Dupont, L.; Roy, V.; Schimann, H.; Orivel, J.; Murienne, J.; Baraloto, C.; Cottenie, K. At each site its diversity: DNA barcoding reveals remarkable earthworm diversity in neotropical rainforests of French Guiana. Appl. Soil Ecol. 2021, 164, 103932. [Google Scholar] [CrossRef]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef]
- Marthinsen, G.; Rui, S.; Timdal, E. OLICH: A reference library of DNA barcodes for Nordic lichens. Biodivers. Data J. 2019, 7, e36252. [Google Scholar] [CrossRef] [Green Version]
- Janzen, F.H.; Crampton, W.G.; Lovejoy, N.R. A new taxonomist-curated reference library of DNA barcodes for Neotropical electric fish (Teleostei: Gymnotiformes). Zool. J. Linn. Soc. 2022, XX, 1–25. [Google Scholar] [CrossRef]
- Grandcolas, P. Biodiversity in France. In Global Biodiversity: Selected Countries in Europe, 1st ed.; Pullaiah, P., Ed.; Apple Academic Press: New York, NY, USA, 2018; Volume 2, pp. 25–32. [Google Scholar]
- Bevanger, K. Biodiversity in Norway. In Global Biodiversity: Selected Countries in Europe, 1st ed.; Pullaiah, P., Ed.; Apple Academic Press: New York, NY, USA, 2018; Volume 2, pp. 223–260. [Google Scholar]
- Prakash, R.O.; Rumsey, F. Biodiversity in United Kingdom. In Global Biodiversity: Selected Countries in Europe, 1st ed.; Pullaiah, P., Ed.; Apple Academic Press: New York, NY, USA, 2018; Volume 2, pp. 379–405. [Google Scholar]
- Ulrych, L.; Guttová, A.; Urban, P.; Hindáková, A.; Kučera, J.; Kučera, V.; Šibík, J.; Mútňanová, M. Biodiversity in Slovakia. In Global Biodiversity: Selected Countries in Europe, 1st ed.; Pullaiah, P., Ed.; Apple Academic Press: New York, NY, USA, 2018; Volume 2, pp. 325–375. [Google Scholar]
- Lopez-Vaamonde, C.; Kirichenko, N.; Cama, A.; Doorenweerd, C.; Godfray, H.C.J.; Guiguet, A.; Gomboc, S.; Huemer, P.; Landry, J.-F.; Laštůvka, A.; et al. Evaluating DNA Barcoding for Species Identification and Discovery in European Gracillariid Moths. Front. Ecol. Evol. 2021, 9, 626752. [Google Scholar] [CrossRef]
- Raupach, M.J.; Rulik, B.; Spelda, J. Surprisingly high genetic divergence of the mitochondrial DNA barcode fragment (COI) within Central European woodlice species (Crustacea, Isopoda, Oniscidea). ZooKeys 2022, 1082, 103–125. [Google Scholar] [CrossRef] [PubMed]
- Chimeno, C.; Hausmann, A.; Schmidt, S.; Raupach, M.J.; Doczkal, D.; Baranov, V.; Hübner, J.; Höcherl, A.; Albrecht, R.; Jaschhof, M. Peering into the Darkness: DNA Barcoding Reveals Surprisingly High Diversity of Unknown Species of Diptera (Insecta) in Germany. Insects 2022, 13, 82. [Google Scholar] [CrossRef] [PubMed]
- Astrin, J.J.; Höfer, H.; Spelda, J.; Holstein, J.; Bayer, S.; Hendrich, L.; Huber, B.A.; Kielhorn, K.-H.; Krammer, H.-J.; Lemke, M. Towards a DNA barcode reference database for spiders and harvestmen of Germany. PLoS ONE 2016, 11, e0162624. [Google Scholar] [CrossRef] [Green Version]
- Muster, C.; Spelda, J.; Rulik, B.; Thormann, J.; von der Mark, L.; Astrin, J.J. The dark side of pseudoscorpion diversity: The German Barcode of Life campaign reveals high levels of undocumented diversity in European false scorpions. Ecol. Evol. 2021, 11, 13815–13829. [Google Scholar] [CrossRef]
- Troudet, J.; Grandcolas, P.; Blin, A.; Vignes-Lebbe, R.; Legendre, F. Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 2017, 7, 9132. [Google Scholar] [CrossRef] [Green Version]
- Řezáč, M.; Arnedo, M.A.; Opatova, V.; Musilová, J.; Řezáčová, V.; Král, J. Taxonomic revision and insights into the speciation mode of the spider Dysdera erythrina species-complex (Araneae: Dysderidae): Sibling species with sympatric distributions. Invertebr. Syst. 2018, 32, 10–54. [Google Scholar] [CrossRef]
- Edwards, D.L.; Knowles, L.L. Species detection and individual assignment in species delimitation: Can integrative data increase efficacy? Proc. R. Soc. B. 2014, 281, 20132765. [Google Scholar] [CrossRef] [Green Version]
- Sukumaran, J.; Knowles, L.L. Multispecies coalescent delimits structure, not species. Proc. Natl. Acad. Sci. USA 2017, 114, 1607–1612. [Google Scholar] [CrossRef] [Green Version]
- WPC. World Pseudoscorpiones Catalog Natural History Museum Bern. Available online: http://wac.nmbe.ch/ (accessed on 5 May 2022).
- Weygoldt, P. The Biology of Pseudoscorpions; Harvard University Press: Cambridge, MA, USA, 1969; pp. 1–145. [Google Scholar]
- Legg, G.; Jones, R.E. Pseudoscorpions (Arthropoda, Arachnida). Keys and notes for the identification of the species. In Synopses of the British Fauna (New Series); Kermack, D.M., Barnes, R.S.K., Eds.; no. 40.; The Linnean Society of London and the Estuarine and Brackish-Water Sciences Association: Leiden, The Netherlands; New York, NY, USA; København, Denmark; Köln, Germany, 1988; pp. 1–159. [Google Scholar]
- Benavides, L.R.; Cosgrove, J.G.; Harvey, M.S.; Giribet, G. Phylogenomic interrogation resolves the backbone of the Pseudoscorpiones tree of life. Mol. Phylogenet. Evol. 2019, 139, 106509. [Google Scholar] [CrossRef]
- Wilcox, T.P.; Hugg, L.; Zeh, J.A.; Zeh, D.W. Mitochondrial DNA sequencing reveals extreme genetic differentiation in a cryptic species complex of neotropical pseudoscorpions. Mol. Phylogenet. Evol. 1997, 7, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Murienne, J.r.m.; Harvey, M.S.; Giribet, G. First molecular phylogeny of the major clades of Pseudoscorpiones (Arthropoda: Chelicerata). Mol. Phylogenet. Evol. 2008, 49, 170–184. [Google Scholar] [CrossRef] [PubMed]
- Pfeiler, E.; Bitler, B.G.; Castrezana, S.; Matzkin, L.M.; Markow, T.A. Genetic diversification and demographic history of the cactophilic pseudoscorpion Dinocheirus arizonensis from the Sonoran Desert. Mol. Phylogenet. Evol. 2009, 52, 133–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moulds, T.A.; Murphy, N.; Adams, M.; Reardon, T.; Harvey, M.S.; Jennings, J.; Austin, A.D. Phylogeography of cave pseudoscorpions in southern Australia. J. Biogeogr. 2007, 34, 951–962. [Google Scholar] [CrossRef]
- Zeh, J.; Zeh, D.; Bonilla, M. Phylogeography of the harlequin beetle-riding pseudoscorpion and the rise of the Isthmus of Panamá. Mol. Ecol. 2003, 12, 2759–2769. [Google Scholar] [CrossRef]
- Opatova, V.; Šťáhlavský, F. Phoretic or not? Phylogeography of the pseudoscorpion Chernes hahnii (Pseudoscorpiones: Chernetidae). J. Arachnol. 2018, 46, 104–113. [Google Scholar] [CrossRef]
- Harms, D.; Roberts, J.D.; Harvey, M.S. Climate variability impacts on diversification processes in a biodiversity hotspot: A phylogeography of ancient pseudoscorpions in south-western Australia. Zool. J. Linn. Soc. 2019, 186, 934–949. [Google Scholar] [CrossRef]
- Johnson, J.; Loria, S.F.; Joseph, M.M.; Harms, D. Biogeographical and Diversification Analyses of Indian Pseudoscorpions Reveal the Western Ghats as Museums of Ancient Biodiversity. Mol. Phylogenet. Evol. 2022, 175, 107495. [Google Scholar] [CrossRef]
- Muster, C.; Schmarda, T.; Blick, T. Vicariance in a cryptic species pair of European pseudoscorpions (Arachnida, Pseudoscorpiones, Chthoniidae). Zool. Anz. 2004, 242, 299–311. [Google Scholar] [CrossRef]
- Christophoryová, J.; Krajčovičová, K.; Henderickx, H.; Španiel, S. A multivariate study of differentiating characters between three European species of the genus Lasiochernes Beier, 1932 (Pseudoscorpiones, Chernetidae). ZooKeys 2016, 629, 51–81. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Král, J. Karyotype analysis and achiasmatic meiosis in pseudoscorpions of the family Chthoniidae (Arachnida: Pseudoscorpiones). Hereditas 2004, 140, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Šťáhlavský, F.; Henderickx, H.; Král, J. Karyotype study on pseudoscorpions of the genus Lasiochernes Beier (Pseudoscorpiones, Chernetidae). Folia Biol. 2005, 53, 69–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šťáhlavský, F.; Král, J.; Harvey, M.S.; Haddad, C.R. A karyotype study on the pseudoscorpion families Geogarypidae, Garypinidae and Olpiidae (Arachnida: Pseudoscorpiones). Eur. J. Entomol. 2006, 103, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Šťáhlavský, F.; Král, J.; Harvey, M.S.; Haddad, C. The first cytogenetic characterization of atemnids: Pseudoscorpions with the highest chromosome numbers (Arachnida: Pseudoscorpiones). Cytogenet. Genome Res. 2012, 137, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Šťáhlavský, F.; Christophoryová, J.; Henderickx, H. A karyological study of four European species of Roncus (Pseudoscorpiones: Neobisiidae). Eur. J. Entomol. 2013, 110, 393–399. [Google Scholar] [CrossRef] [Green Version]
- Kotrbová, J.; Opatova, V.; Gardini, G.; Šťáhlavský, F. Karyotype diversity of pseudoscorpions of the genus Chthonius (Pseudoscorpiones, Chthoniidae) in the Alps. Comp. Cytogenet. 2016, 10, 325–345. [Google Scholar]
- Zaragoza, J.A.; Šťáhlavský, F. A new Roncus species (Pseudoscorpiones: Neobisiidae) from Montseny Natural Park (Catalonia, Spain), with remarks on karyology. Zootaxa 2008, 1693, 27–40. [Google Scholar] [CrossRef]
- Harvey, M.S.; Ratnaweerae, P.B.; Udagamae, P.V.; Wijesinghee, M.R. A new species of the pseudoscorpion genus Megachernes (Pseudoscorpiones: Chernetidae) associated with a threatened Sri Lankan rainforest rodent, with a review of host associations of Megachernes. J. Nat. Hist. 2012, 46, 2519–2535. [Google Scholar] [CrossRef]
- Christophoryová, J.; Gilgado, J.D.; Bobbitt, I.; Krajčovičová, K. Lamprochernes savignyi (Simon, 1881) (Arachnida, Pseudoscorpiones) recorded in Central Europe for the first time. Check List 2021, 17, 497–501. [Google Scholar] [CrossRef]
- Beier, M. Phoresie und phagophilie bei pseudoscorpionen. Oesterr. zool. Zeitschr. 1948, 1, 441–497. [Google Scholar]
- Jones, P.E. Phoresy and commensalism in British pseudoscropions. Proc. Trans. Br. Ent. Nat. Hist. Soc. 1978, 11, 90–96. [Google Scholar]
- Kaňuchová, A.; Christophoryová, J.; Krajčovičová, K. Pseudoscorpions (Arachnida) collected from the heaps with decomposing material in Slovakia. Fragm. Faunist. 2015, 58, 111–122. [Google Scholar] [CrossRef]
- Christophoryová, J.; Jajcayová, D.; Krajčovičová, K. Pseudoscorpions (Arachnida: Pseudoscorpiones) living in tree microhabitats in Slovakia. Klapalekiana 2017, 53, 283–297. [Google Scholar]
- Christophoryová, J.; Kaňuchová, A.; Krajčovičová, K. Faunistic survey of pseudoscorpions (Arachnida: Pseudoscorpiones) collected from compost heaps in Slovakia. Klapalekiana 2017, 53, 11–19. [Google Scholar]
- Harvey, M.S. Redescription and new synonyms of the cosmopolitan species Lamprochernes savignyi (Simon) (Chernetidae: Pseudoscorpionida). Bull. Br. arachnol. Soc. 1987, 7, 111–116. [Google Scholar]
- Drogla, R.; Lippold, K. Zur Kenntnis der Pseudoskorpion-Fauna von Ostdeutschland (Arachnida, Pseudoscorpiones). Arachnol. Mitt. 2004, 27, 1–54. [Google Scholar] [CrossRef]
- Beier, M. Pseudoscorpionidea II: Subord. C. Cheliferinea. Tierreich 1932, 58, i–xxi, 1–294. [Google Scholar]
- Navás, L. Algunos Quernetos (Arácnidos) de la provincia de Zaragoza. Bol. Soc. Ent. Est. 1918, 1, 83–90. [Google Scholar]
- Beier, M. Zur Kenntnis der Lamprochernetinae (Pseudoscorp.). Zool. Anz. 1932, 97, 258–267. [Google Scholar]
- Beier, M. Zoologische Forschungsreise nach den Jonischen Inseln und dem Peloponnes. Sitz.-Ber. Akad. Wiss. 1929, 128, 425–456. [Google Scholar]
- Beier, M. Ordnung Pseudoscorpionidea (Afterskorpione). In Bestimmungsbücher zur Bodenfauna Europas, 1st ed.; Aguilar, J., Beier, M., Franz, H., Raw, F., Eds.; Akademie-Verlag: Berlin, Germany, 1963; pp. 1–313. [Google Scholar]
- Christophoryová, J.; Šťáhlavský, F.; Fedor, P. An updated identification key to the pseudoscorpions (Arachnida: Pseudoscorpiones) of the Czech Republic and Slovakia. Zootaxa 2011, 2876, 35–48. [Google Scholar] [CrossRef]
- Mahnert, V. Die Pseudoskorpione Österreichs (Arachnida, Pseudoscorpiones). Denisia 2004, 12, 459–471. [Google Scholar]
- Shorthouse, D.P. SimpleMappr, an online tool to produce publication-quality point maps. Available online: http://www.simplemappr.net/ (accessed on 5 May 2022).
- Ersts, P. Geographic Distance Matrix Generator (version 1.2.3). American Museum of Natural History, Center for Biodiversity and Conservation. Available online: http://biodiversityinformatics.amnh.org/open_source/gdmg (accessed on 5 May 2022).
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Drummond, A.; Ashton, B.; Buxton, S.; Cheung, M.; Cooper, A.; Duran, C.; Field, M.; Heled, J.; Kearse, M.; Markowitz, S.; et al. Geneious v5.6. Available online: http://www.geneious.com/ (accessed on 5 May 2022).
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, W7–13. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J. TRACER v.1.5, 1.4. Available online: http://tree.bio.ed.ac.uk/software/tracer/ (accessed on 5 May 2022).
- Rambaut, A. FigTree v. 1.3.1. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 5 May 2022).
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [Green Version]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Kamoun, S.; Sumlin, W.D.; Vogler, A.P. Sequence-Based Species Delimitation for the DNA Taxonomy of Undescribed Insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezard, T.; Fujisawa, T.; Barraclough, T.G. splits: SPecies’ LImits by Threshold Statistics. Rpackage version 1.0-11/r29. Available online: http://r-forge.r-project.org/projects/splits/ (accessed on 5 May 2022).
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour 2021, 21, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base susbtitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Flouri, T.; Jiao, X.; Rannala, B.; Yang, Z. Species tree inference with BPP using genomic sequences and the multispecies coalescent. Mol. Biol. Evol. 2018, 35, 2585–2593. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Rannala, B. Bayesian species delimitation using multilocus sequence data. PNAS 2010, 107, 9264–9269. [Google Scholar] [CrossRef] [Green Version]
- Rannala, B.; Yang, Z. Improved Reversible Jump Algorithms for Bayesian Species Delimitation. Genetics 2013, 194, 245–253. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Brower, A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. PNAS 1994, 91, 6491–6495. [Google Scholar] [CrossRef] [Green Version]
- Judson, M.L. Cheliferoid pseudoscorpions (Arachnida, Chelonethi) from the Lower Cretaceous of France. Geodiversitas 2009, 31, 61–71. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Zeh, J.A.; Zeh, D.W.; Král, J. Karyotypes of the neotropical pseudoscorpions Semeiochernes armiger and Cordylochernes scorpioides (Pseudoscorpiones: Chernetidae). J. Arachnol. 2009, 37, 287–291. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Zacaro, A. LEVAN, an ImageJ plugin for morphological cytogenetic analysis of mitotic and meiotic chromosomes. Initial version. Available online: http://rsbweb.nih.gov/ij/ (accessed on 5 May 2022).
- Levan, A.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Green, D.M.; Sessions, S.K. Nomenclature for chromosomes. In Amphibian Cytogenetics and Evolution, 1st ed.; Green, D.M., Sessions, S.K., Eds.; Academic Press Inc.: London, UK, 1991; pp. 431–432. [Google Scholar]
- Šťáhlavský, F.; Štundlová, J.; Lowe, G.; Stockmann, M.; Kovařík, F. Application of cytogenetic markers in the taxonomy of flat rock scorpions (Scorpiones: Hormuridae), with the description of Hadogenes weygoldti sp. n. Zool. Anz. 2018, 273, 173–182. [Google Scholar] [CrossRef]
- Sadílek, D.; Nguyen, P.; Koç, H.; Kovařík, F.; Yağmur, E.A.; Šťáhlavský, F. Molecular cytogenetics of Androctonus scorpions: An oasis of calm in the turbulent karyotype evolution of the diverse family Buthidae. Biol. J. Linn. Soc. 2015, 115, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Chamberlin, J.C. The Arachnid Order Chelonethida; Stanford University Press: Stanford, CA, USA, 1931; pp. 1–284. [Google Scholar]
- Harvey, M.S. The phylogeny and classification of the Pseudoscorpionida (Chelicerata: Arachnida). Invertebr. Syst. 1992, 6, 1373–1435. [Google Scholar] [CrossRef]
- Judson, M.L. A new and endangered species of the pseudoscorpion genus Lagynochthonius from a cave in Vietnam, with notes on chelal morphology and the composition of the Tyrannochthoniini (Arachnida, Chelonethi, Chthoniidae). Zootaxa 2007, 1627, 53–68. [Google Scholar] [CrossRef]
- Marhold, K. Multivariate morphometrics and its application to monography at specific and infraspecific levels. In Monographic plant systematics: Fundamental assessment of plant biodiversity; Stuessy, T.F., Lack, H.W., Eds.; A.R.G. Gantner Verlag K.G.: Ruggell, Liechtenstein, 2011; pp. 73–99. [Google Scholar]
- Pearson, K. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1900, 50, 157–175. [Google Scholar] [CrossRef] [Green Version]
- Zar, J.H. Biostatistical analysis, 4th ed.; Upper Saddle River, N.J., Ed.; Prentice Hall: Englewood Cliffts, NJ, USA, 1999; pp. 1–663. [Google Scholar]
- Klecka, W.R. Discriminant analysis, Series: Quantitative Applications in the Social Sciences, 1st ed.; Sage Publications, Inc.: Beverly Hills, CA, USA, 1980; pp. 1–76. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Šlenker, M.; Koutecký, P.; Marhold, K. MorphoTools2: An R package for the multivariate morphometric analysis. Bioinformatics 2022, 38, 2954–2955. [Google Scholar] [CrossRef]
- Beier, M. Türkiye Psevdoscorpion’lari hakkinda. Türkische Pseudoscorpione. Rev. Fac. Scien. Univ. Istanbul B. 1949, 14, 1–20. [Google Scholar]
- Mahnert, V. Die Pseudoskorpione (Arachnida) Kenyas V. Chernetidae. Rev. Suisse Zool. 1982, 89, 691–712. [Google Scholar] [CrossRef]
- Callaini, G. Pseudoscorpioni dell’isola di Montecristo (Arachnida). Notulae Chernetologicae IX. Redia 1983, 66, 147–165. [Google Scholar]
- Petrov, B.P. The false scorpions (Arachnida: Pseudoscorpiones) of the Eastern Rhodopes (Bulgaria and Greece). In Biodiversity of Bulgaria. 2. Biodiversity of Eastern Rhodopes (Bulgaria and Greece); Beron, P., Popov, A., Eds.; Pensoft & National Museum of Natural History: Sofia, Bulgaria, 2004; pp. 153–166. [Google Scholar]
- Sezek, F.; Özkan, M. A new record for the Turkish pseudoscorpion fauna; Lamprochernes savignyi (Simon, 1881) (Arachnida, Pseudoscorpionida). Turk. J. Zool. 2006, 30, 255–259. [Google Scholar]
- Mahnert, V. Pseudoscorpions (Arthropoda, Arachnida). Order Pseudoscorpiones. In Arthropod fauna of the UAE; van Harten, A., Ed.; no. 2; Dar Al Ummah Printing: Adu Dhabi, United Arab Emirates, 2009; pp. 26–42. [Google Scholar]
- Chamberlin, J.C. New and little-known false scorpions (Arachnida, Chelonethida) from Monterey County, California. Bull. Am. Mus. Nat. Hist. 1952, 99, 259–312. [Google Scholar]
- Hoff, C.C. The pseudoscorpions of Illinois. Bull. Ill. Nat. Hist. Surv. 1949, 24, 407–498. [Google Scholar] [CrossRef]
- Ohira, H.; Kaneko, S.; Faulks, L.; Tsutsumi, T. Unexpected species diversity within Japanese Mundochthonius pseudoscorpions (Pseudoscorpiones: Chthoniidae) and the necessity for improved species diagnosis revealed by molecular and morphological examination. Invertebr. Syst. 2018, 32, 259–277. [Google Scholar] [CrossRef]
- Ohira, H.; Sato, K.; Tsutsumi, T.; Kaneko, S.; Choi, H.-J. DNA barcoding suggested the existence of cryptic species and high biodiversity of South Korean pseudoscorpions (Arachnida, Pseudoscorpiones). J. Asia Pac. Biodivers. 2018, 11, 399–407. [Google Scholar] [CrossRef]
- Tyagi, K.; Kumar, V.; Kundu, S.; Pakrashi, A.; Prasad, P.; Caleb, J.T.; Chandra, K. Identification of Indian spiders through DNA barcoding: Cryptic species and species complex. Sci. Rep. 2019, 9, 14033. [Google Scholar] [CrossRef]
- Mohammed-Geba, K.; Obuid-Allah, A.H.; El-Shimy, N.A.; Mahbob, M.A.E.-M.; Ali, R.S.; Said, S.M. DNA barcoding for scorpion species from New Valley Governorate in Egypt reveals different degrees of cryptic speciation and species misnaming. Conservation 2021, 1, 228–240. [Google Scholar] [CrossRef]
- Esselstyn, J.A.; Evans, B.J.; Sedlock, J.L.; Anwarali Khan, F.A.; Heaney, L.R. Single-locus species delimitation: A test of the mixed Yule-coalescent model, with an empirical application to Philippine round-leaf bats. Proc. Biol. Sci. 2012, 279, 3678–3686. [Google Scholar] [CrossRef]
- Kekkonen, M.; Hebert, P.D. DNA barcode-based delineation of putative species: Efficient start for taxonomic workflows. Mol. Ecol. Resour. 2014, 14, 706–715. [Google Scholar] [CrossRef] [Green Version]
- Lukhtanov, V. Species delimitation and analysis of cryptic species diversity in the XXI century. Entomol. Rev. 2019, 99, 463–472. [Google Scholar] [CrossRef]
- Milam, J.; Johnson, D.E.; Andersen, J.C.; Fassler, A.B.; Narango, D.L.; Elkinton, J.S. Validating morphometrics with DNA barcoding to reliably separate three cryptic species of Bombus Cresson (Hymenoptera: Apidae). Insects 2020, 11, 669. [Google Scholar] [CrossRef] [PubMed]
- Romero-Ortiz, C.; Flórez, D.E.; Sarmiento, C.E. Assessment of morphometric characters to delimit species of Apolpium Chamberlin, 1930 (Pseudoscorpiones, Olpiidae). J. Nat. Hist. 2020, 54, 1025–1043. [Google Scholar] [CrossRef]
- Morgan-Richards, M. A new species of tree weta from the North Island of New Zealand (Hemideina: Stenopelmatidae: Orthoptera). N. Z. Entomol. 1995, 18, 15–23. [Google Scholar] [CrossRef]
- Lukhtanov, V.A.; Dantchenko, A.V.; Vishnevskaya, M.S.; Saifitdinova, A.F. Detecting cryptic species in sympatry and allopatry: Analysis of hidden diversity in Polyommatus (Agrodiaetus) butterflies (Lepidoptera: Lycaenidae). Biol. J. Linn. Soc. 2015, 116, 468–485. [Google Scholar] [CrossRef] [Green Version]
- Suwannamit, S.; Baimai, V.; Otsuka, Y.; Saeung, A.; Thongsahuan, S.; Tuetun, B.; Apiwathnasorn, C.; Jariyapan, N.; Somboon, P.; Takaoka, H. Cytogenetic and molecular evidence for an additional new species within the taxon Anopheles barbirostris (Diptera: Culicidae) in Thailand. Parasitol. Res. 2009, 104, 905–918. [Google Scholar] [CrossRef]
- Ojanguren-Affilastro, A.A.; Adilardi, R.S.; Cajade, R.; Ramirez, M.J.; Ceccarelli, F.S.; Mola, L.M. Multiple approaches to understanding the taxonomic status of an enigmatic new scorpion species of the genus Tityus (Buthidae) from the biogeographic island of Paraje Tres Cerros (Argentina). PLoS ONE 2017, 12, e0181337. [Google Scholar] [CrossRef]
- Řezáč, M.; Král, J.; Pekár, S. The spider genus Dysdera (Araneae, Dysderidae) in Central Europe: Revision and natural history. J. Arachnol. 2007, 35, 432–462. [Google Scholar] [CrossRef]
- Štundlová, J.; Šmíd, J.; Nguyen, P.; Šťáhlavský, F. Cryptic diversity and dynamic chromosome evolution in Alpine scorpions (Euscorpiidae: Euscorpius). Mol. Phylogenet. Evol. 2019, 134, 152–163. [Google Scholar] [CrossRef]
- Červená, M.; Šťáhlavský, F.; Papáč, V.; Kováč, Ľ.; Christophoryová, J. Morphological and cytogenetic characteristics of Neobisium (Blothrus) slovacum Gulička, 1977 (Pseudoscorpiones, Neobisiidae), the northernmost troglobitic species of the subgenus Blothrus in Europe. ZooKeys 2019, 817, 113–130. [Google Scholar] [CrossRef] [Green Version]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ontano, A.Z.; Gainett, G.; Aharon, S.; Ballesteros, J.A.; Benavides, L.R.; Corbett, K.F.; Gavish-Regev, E.; Harvey, M.S.; Monsma, S.; Santibáñez-López, C.E. Taxonomic sampling and rare genomic changes overcome long-branch attraction in the phylogenetic placement of pseudoscorpions. Mol. Biol. Evol. 2021, 38, 2446–2467. [Google Scholar] [CrossRef] [PubMed]
- Harms, D. The origins of diversity in ancient landscapes: Deep phylogeographic structuring in a pseudoscorpion (Pseudotyrannochthoniidae: Pseudotyrannochthonius) reflects Plio-Pleistocene climate fluctuations. Zool. Anz. 2018, 273, 112–123. [Google Scholar] [CrossRef]
- Arabi, J.; Judson, M.L.; Deharveng, L.; Lourenço, W.R.; Cruaud, C.; Hassanin, A. Nucleotide composition of CO1 sequences in Chelicerata (Arthropoda): Detecting new mitogenomic rearrangements. J. Mol. Evol. 2012, 74, 81–95. [Google Scholar] [CrossRef]
- Colborn, J.; Crabtree, R.E.; Shaklee, J.B.; Pfeiler, E.; Bowen, B.W. The evolutionary enigma of bonefishes (Albula spp.): Cryptic species and ancient separations in a globally distributed shorefish. Evolution 2001, 55, 807–820. [Google Scholar] [CrossRef]
- Szudarek-Trepto, N.; Kazmierski, A.; Dabert, J. Long-term stasis in acariform mites provides evidence for morphologically stable evolution: Molecular vs. morphological differentiation in Linopodes (Acariformes; Prostigmata). Mol. Phylogenet. Evol. 2021, 163, 107237. [Google Scholar] [CrossRef]
- Cerca, J.; Meyer, C.; Stateczny, D.; Siemon, D.; Wegbrod, J.; Purschke, G.; Dimitrov, D.; Struck, T.H. Deceleration of morphological evolution in a cryptic species complex and its link to paleontological stasis. Evolution 2020, 74, 116–131. [Google Scholar] [CrossRef]
- Bond, J.E.; Hedin, M.C.; Ramirez, M.G.; Opell, B.D. Deep molecular divergence in the absence of morphological and ecological change in the Californian coastal dune endemic trapdoor spider Aptostichus simus. Mol. Ecol. 2001, 10, 899–910. [Google Scholar] [CrossRef]
- Adams, S.A.; Tsutsui, N.D. The evolution of species recognition labels in insects. Philos. Trans. R. Soc. B 2020, 375, 20190476. [Google Scholar] [CrossRef]
- Stemme, T.; Pfeffer, S.E. Anatomy of the nervous system in Chelifer cancroides (Arachnida: Pseudoscorpiones) with a distinct sensory pathway associated with the pedipalps. Insects 2021, 13, 25. [Google Scholar] [CrossRef]
- Zeh, D.W.; Zeh, J.A. When Morphology Misleads—Interpopulation Uniformity in Sexual Selection Masks Genetic-Divergence in Harlequin Beetle-Riding Pseudoscorpion Populations. Evolution 1994, 48, 1168–1182. [Google Scholar] [CrossRef]
- Poinar, G.O.; Ćurčić, B.P.; Cokendolpher, J.C. Arthropod phoresy involving pseudoscorpions in the past and present. Acta Arachnol. 1998, 47, 79–96. [Google Scholar] [CrossRef]
- Borges, R.M. Phoresy Involving Insects as Riders or Rides: Life History, Embarkation, and Disembarkation. Ann. Entomol. Soc. Am. 2022, 115, 219–231. [Google Scholar] [CrossRef]
- Harvey, M.S.; Lopes, P.C.; Goldsmith, G.R.; Halajian, A.; Hillyer, M.J.; Huey, J.A. A novel symbiotic relationship between sociable weaver birds (Philetairus socius) and a new cheliferid pseudoscorpion (Pseudoscorpiones: Cheliferidae) in southern Africa. Invertebr. Syst. 2015, 29, 444–456. [Google Scholar] [CrossRef]
- Krajčovičová, K.; Matyukhin, A.V.; Christophoryová, J. First comprehensive research on pseudoscorpions (Arachnida: Pseudoscorpiones) collected from bird nests in Russia. Turk. J. Zool. 2018, 42, 480–487. [Google Scholar] [CrossRef]

| Character/Species | L. abditus sp. nov. | L. chyzeri | L. nodosus | L. savignyi |
|---|---|---|---|---|
| Shape of palpal trochanter protuberance | pointed | pointed | rounded | rounded |
| Palpal chela, length | 0.92–1.02 | 0.77–1.04 | 0.71–0.91 | 0.69–0.85 |
| Palpal femur, length | 0.54–0.59 | 0.44–0.65 | 0.36–0.50 | 0.32–0.46 |
| Setae number on male genital operculum: anterior/posterior | 29–37/14–18 | 27–41/11–19 | 18–23/9–12 | 15–22/10–14 |
| Setae number on female genital operculum: anterior/posterior | 19–32/9–15 | 21–29/9–15 | 17–22/6–9 | 16–21/5–8 |
| Tarsus of leg IV length/distance of the tactile seta from the tarsus base, ratio | 2.71–3.29 | 2.88–3.54 | 2.72–3.68 | 2.40–2.77 |
| Distribution | Austria, Norway, Ukraine, Romania | Europe, Kazakhstan, Russia, Turkey | Europe, Central Africa, South Asia | Africa, Australia, America, New Zealand, South Asia, West to Switzerland and South Europe |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christophoryová, J.; Krajčovičová, K.; Šťáhlavský, F.; Španiel, S.; Opatova, V. Integrative Taxonomy Approach Reveals Cryptic Diversity within the Phoretic Pseudoscorpion Genus Lamprochernes (Pseudoscorpiones: Chernetidae). Insects 2023, 14, 122. https://doi.org/10.3390/insects14020122
Christophoryová J, Krajčovičová K, Šťáhlavský F, Španiel S, Opatova V. Integrative Taxonomy Approach Reveals Cryptic Diversity within the Phoretic Pseudoscorpion Genus Lamprochernes (Pseudoscorpiones: Chernetidae). Insects. 2023; 14(2):122. https://doi.org/10.3390/insects14020122
Chicago/Turabian StyleChristophoryová, Jana, Katarína Krajčovičová, František Šťáhlavský, Stanislav Španiel, and Vera Opatova. 2023. "Integrative Taxonomy Approach Reveals Cryptic Diversity within the Phoretic Pseudoscorpion Genus Lamprochernes (Pseudoscorpiones: Chernetidae)" Insects 14, no. 2: 122. https://doi.org/10.3390/insects14020122
APA StyleChristophoryová, J., Krajčovičová, K., Šťáhlavský, F., Španiel, S., & Opatova, V. (2023). Integrative Taxonomy Approach Reveals Cryptic Diversity within the Phoretic Pseudoscorpion Genus Lamprochernes (Pseudoscorpiones: Chernetidae). Insects, 14(2), 122. https://doi.org/10.3390/insects14020122







