Two Distinct Genotypes of Spissistilus festinus (Say, 1830) Reproduce and Differentially Transmit Grapevine Red Blotch Virus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. S. festinus Populations
2.3. Establishment of Mixed Mating Pairs
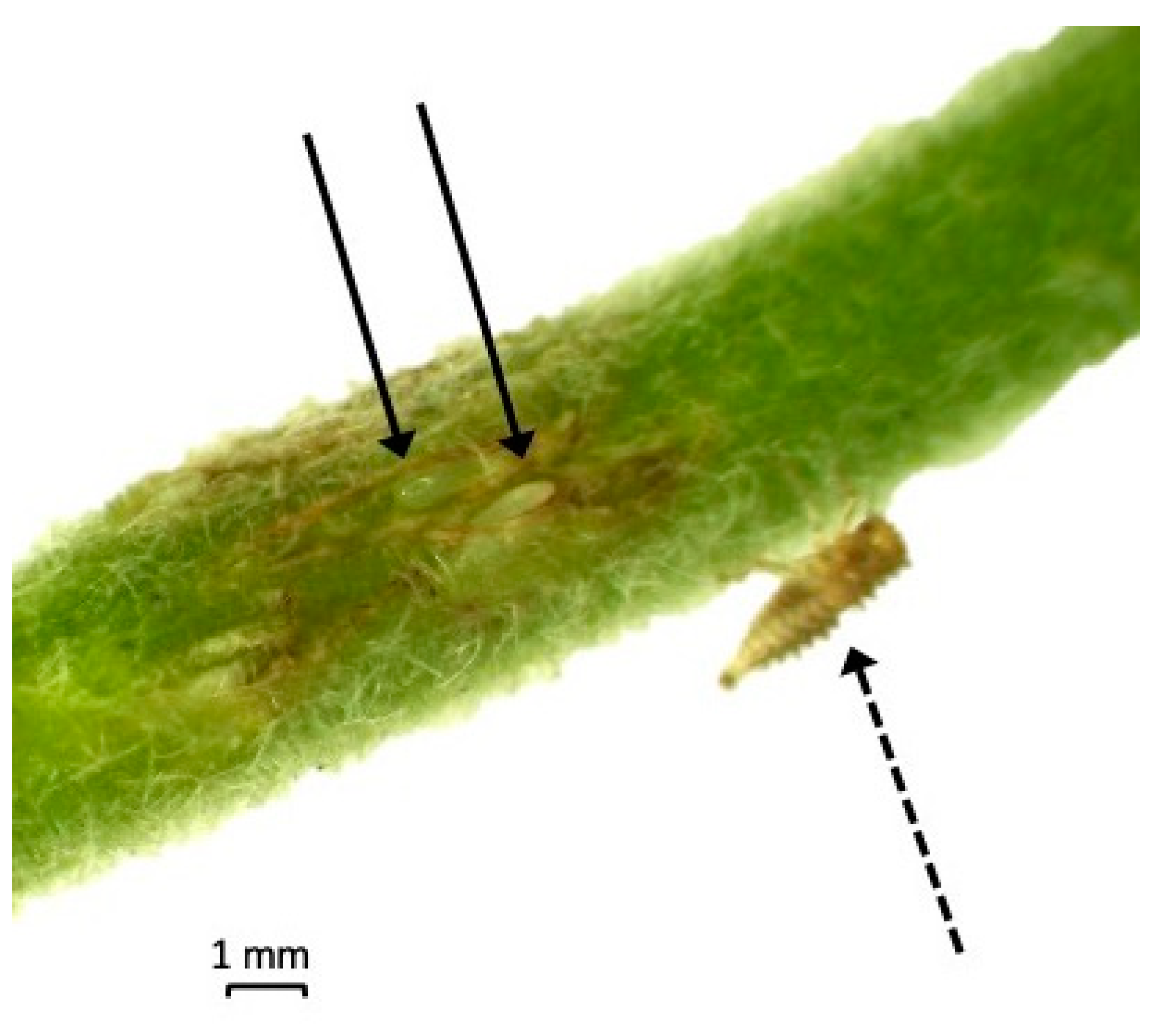
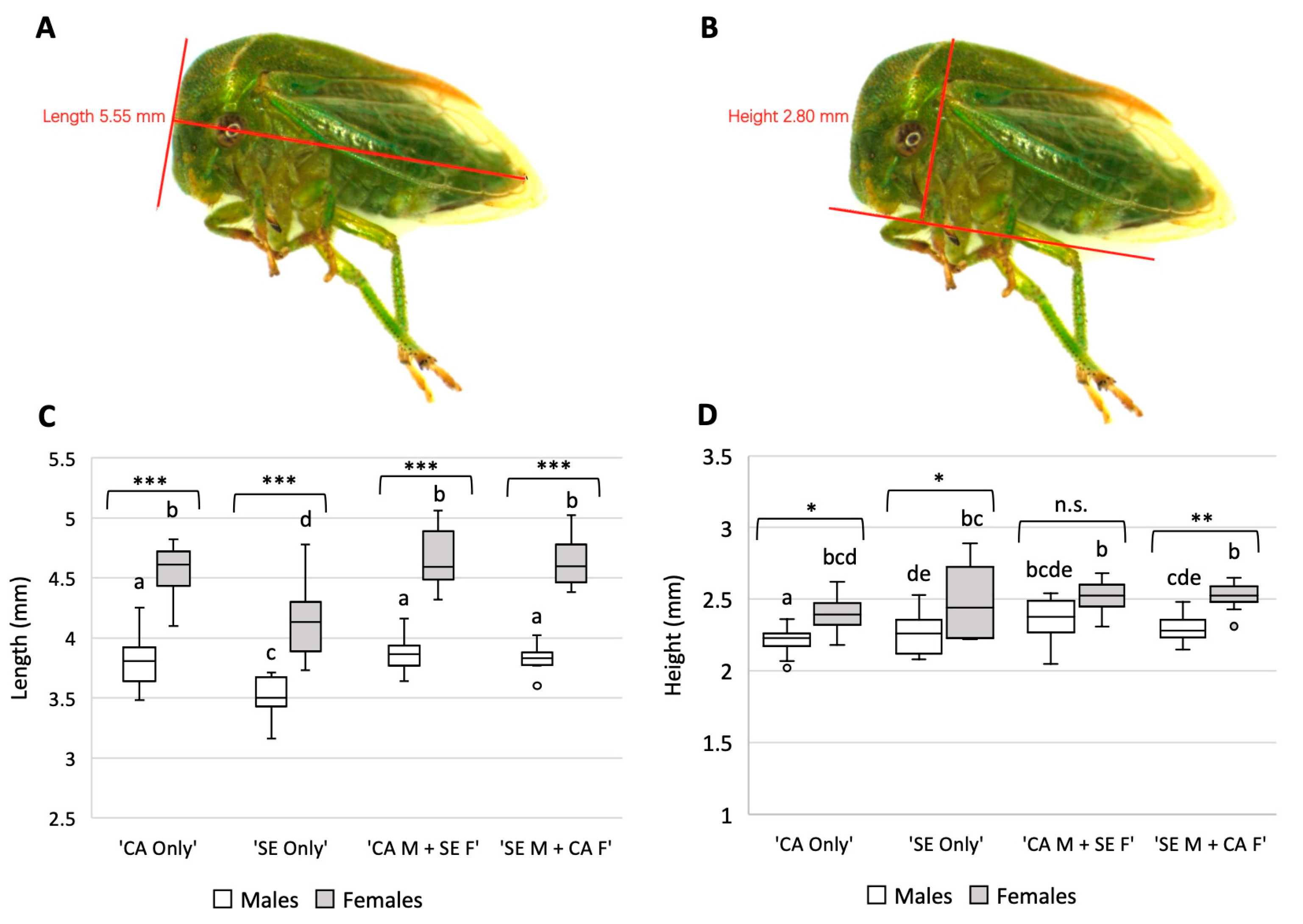
2.4. Morphological Assessment of F1 Progeny of S. festinus Mixed Mating Pairs
2.5. Genotype Determination of F1 Progeny of S. festinus Mixed Mating Pairs
2.6. GRBV Inoculation of P. vulgaris
2.7. Acquisition of GRBV by S. festinus Populations
2.8. Transmission Assays of GRBV by Both S. festinus Genotypes
2.9. Nucleic acid Extraction from Plant and S. festinus Tissues
2.10. GRBV Detection and Quantification
2.11. Statistics
3. Results
3.1. Production of S. festinus Mating Pairs
3.2. The CA and SE S. festinus Genotypes Are Capable of Mating
3.3. S. festinus F1 Generations Vary Phenotypically
3.4. Mixed Mating Pairs Are Capable of Acquiring GRBV
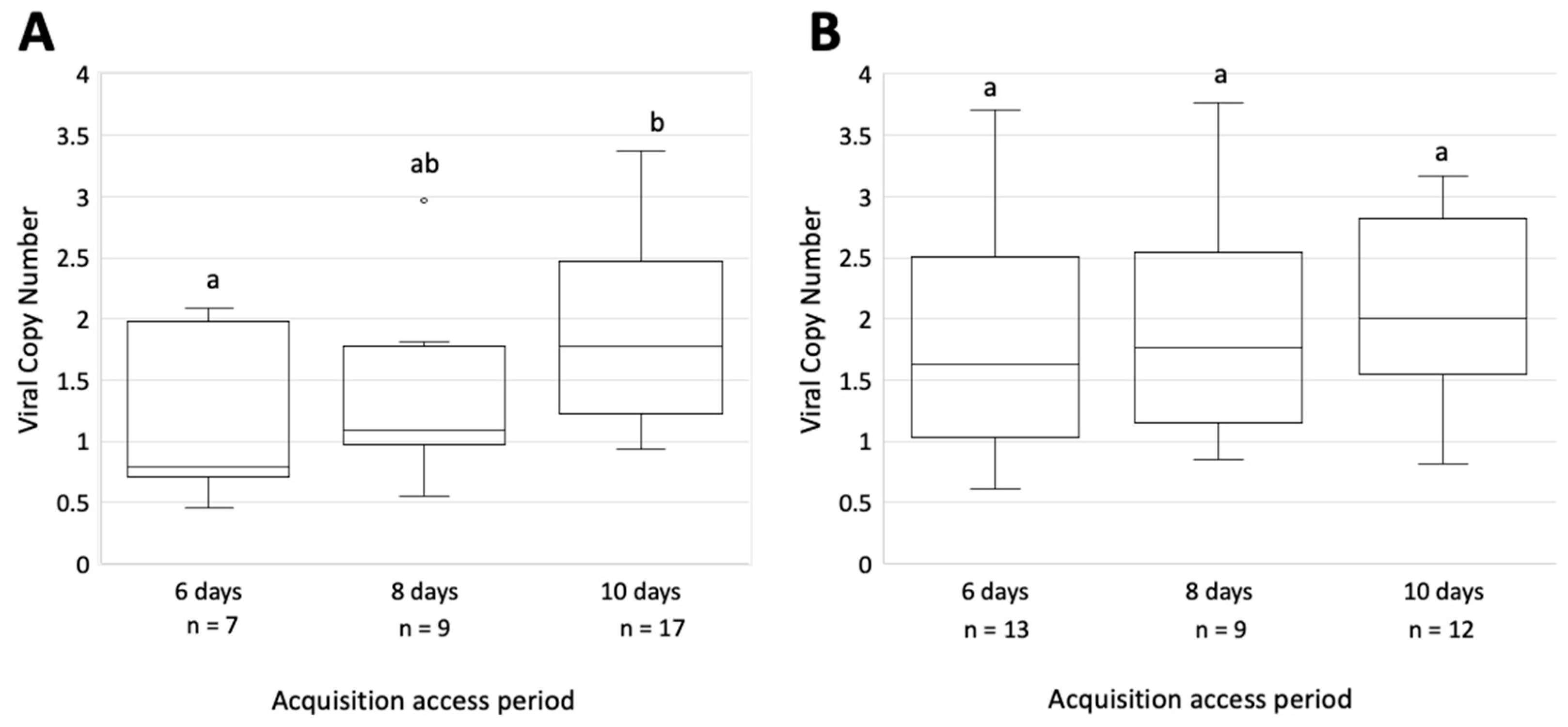
3.5. The Rate of GRBV Acquisition Is Similar between the CA and SE S. festinus Genotypes
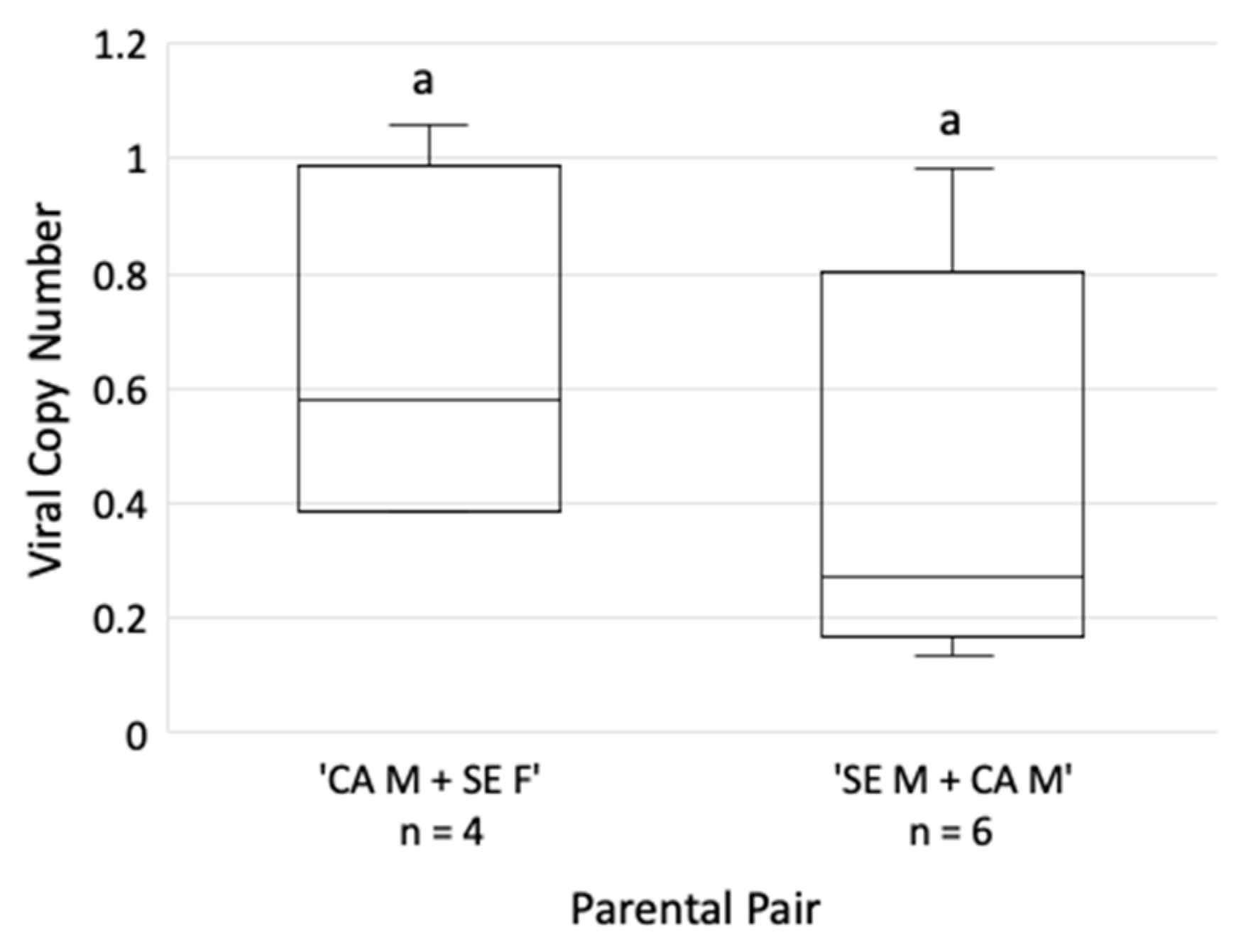
3.6. GRBV Titer in the Salivary Glands Varies with the S. festinus Genotype
3.7. S. festinus of the SE Genotype Transmit GRBV at a Higher Rate Than the CA Genotype
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beyer, B.A.; Srinivasan, R.; Roberts, P.M.; Abney, M.R. Biology and management of the three-cornered alfalfa hopper (hemiptera: Membracidae) in alfalfa, soybean, and peanut. J. Integr. Pest Manag. 2017, 8, 10. [Google Scholar] [CrossRef]
- Flasco, M.T.; Hoyle, V.; Cieniewicz, E.J.; Loeb, G.; McLane, H.; Perry, K.; Fuchs, M.F. The three-cornered alfalfa hopper, Spissistilus festinus, is a vector of grapevine red blotch virus in vineyards. Viruses 2023, 15, 927. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera Bustamante, R.F.; Zerbini, F.M.; Adkins, S.; et al. World management of geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Roumagnac, P.; Fuchs, M.; Navas-Castillo, J.; Moriones, E.; Idris, A.; Briddon, R.W.; Rivera-Bustamante, R.; Murilo Zerbini, F.; Martin, D.P. Capulavirus and Grablovirus: Two new genera in the family Geminiviridae. Arch. Virol. 2017, 162, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Yepes, L.M.; Cieniewicz, E.; Krenz, B.; McLane, H.; Thompson, J.R.; Perry, K.L.; Fuchs, M. Causative role of grapevine red blotch virus in red blotch disease. Phytopathology 2018, 108, 902–909. [Google Scholar] [CrossRef]
- Calvi, B.L. Effects of Red-Leaf Disease on Cabernet Sauvignon at the Oakville Experimental Vineyard and Mitigation by Harvest Delay and Crop Adjustment. Available online: https://www.proquest.com/openview/1fd04fad88855d648357391969b4d9b2/1?pq-origsite=gscholar&cbl=18750&diss=y (accessed on 8 June 2020).
- Sudarshana, M.R.; Perry, K.L.; Fuchs, M.F. Grapevine red blotch-associated virus, an emerging threat to the grapevine industry. Phytopathology 2015, 105, 1026–1032. [Google Scholar] [CrossRef]
- Cieniewicz, E.J.; Pethybridge, S.J.; Loeb, G.; Perry, K.; Fuchs, M. Insights into the ecology of grapevine red blotch virus in a diseased vineyard. Phytopathology 2018, 108, 94–102. [Google Scholar] [CrossRef]
- Rumbaugh, A.C.; Girardello, R.C.; Cooper, M.L.; Plank, C.; Kurtural, S.K.; Oberholster, A. Impact of rootstock and season on red blotch disease expression in cabernet sauvignon (V. vinifera). Plants 2021, 10, 1583. [Google Scholar] [CrossRef]
- Blanco-Ulate, B.; Hopfer, H.; Figueroa-Balderas, R.; Ye, Z.; Rivero, R.M.; Albacete, A.; Pérez-Alfocea, F.; Koyama, R.; Anderson, M.M.; Smith, R.J.; et al. Red blotch disease alters grape berry development and metabolism by interfering with the transcriptional and hormonal regulation of ripening. J. Exp. Bot. 2017, 68, 1225–1238. [Google Scholar] [CrossRef]
- Cauduro Girardello, R.; Rich, V.; Smith, R.J.; Brenneman, C.; Heymann, H.; Oberholster, A. The impact of grapevine red blotch disease on Vitis vinifera L. Chardonnay grape and wine composition and sensory attributes over three seasons. J. Sci. Food Agric. 2020, 100, 1436–1447. [Google Scholar] [CrossRef]
- Martínez-Lüscher, J.; Plank, C.M.; Brillante, L.; Cooper, M.L.; Smith, R.J.; Al-Rwahnih, M.; Yu, R.; Oberholster, A.; Girardello, R.; Kurtural, S.K. Grapevine red blotch virus may reduce carbon translocation leading to impaired grape berry ripening. J. Agric. Food Chem. 2019, 67, 2437–2448. [Google Scholar] [CrossRef] [PubMed]
- Cieniewicz, E.; Thompson, J.R.; McLane, H.; Perry, K.L.; Dangl, G.S.; Corbett, Q.; Martinson, T.; Wise, A.; Wallis, A.; O’Connell, J.; et al. Prevalence and genetic diversity of grabloviruses in free-living Vitis spp. Plant Dis. 2018, 102, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- KC, A.N.; DeShields, J.B.; Levin, A.D.; Hilton, R.; Rijal, J. Epidemiology of grapevine red blotch disease progression in southern Oregon vineyards. Am. J. Enol. Vitic. 2022, 73, 116–124. [Google Scholar] [CrossRef]
- Krenz, B.; Thompson, J.R.; McLane, H.L.; Fuchs, M.; Perry, K.L. Grapevine red blotch-associated virus is widespread in the United States. Phytopathology 2014, 104, 1232–1240. [Google Scholar] [CrossRef]
- Perry, K.; Mclane, H.; Hyder, M.; Dangl, G.; Thompson, J.; Fuchs, M. Grapevine red blotch-associated virus is present in free-living Vitis spp. proximal to cultivated grapevines. Phytopathology 2016, 106, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Flasco, M.; Hoyle, V.; Cieniewicz, E.; Roy, B.; McLane, H.; Perry, K.L.; Loeb, G.M.; Nault, B.; Cilia, M.; Fuchs, M. Grapevine red blotch virus is transmitted by the three-cornered alfalfa hopper in a circulative, nonpropagative mode with unique attributes. Phytopathology 2021, 111, 1851–1861. [Google Scholar] [CrossRef] [PubMed]
- Cieniewicz, E.; Poplaski, V.; Brunelli, M.; Dombroskie, J.; Fuchs, M. Two distinct genotypes of Spissistilus festinus (Say, 1830) (Hemiptera, Membracidae) in the United States revealed by phylogenetic and morphological analyses. Insects 2020, 11, 80. [Google Scholar] [CrossRef]
- Caldwell, J.S. A generic revision of the treehoppers of the tribe Ceresini in America north of Mexico, based on a study of the male genitalia. Proc. U. S. Natl. Mus. 1949, 98, 491–521, plates 18–23. [Google Scholar] [CrossRef]
- Jones, T.; Nita, M. A survey of Virginia vineyards revealed high incidences of grapevine rupestris stem pitting-associated virus, grapevine red blotch virus, and two mealybug species. Plant Health Prog. 2019, 20, 207–214. [Google Scholar] [CrossRef]
- Hoffmann, M.; Talton, W.; Nita, M.; Jones, T.; Al Rwahnih, M.; Sudarshana, M.R.; Almeyda, C. First report of grapevine red blotch virus, the causal agent of grapevine red blotch disease, in Vitis vinifera in North Carolina. Plant Dis. 2020, 104, 1266. [Google Scholar] [CrossRef]
- Soltani, N.; Hu, R.; Hensley, D.D.; Lockwood, D.L.; Perry, K.L.; Hajimorad, M.R. A survey for nine major viruses of grapevines in Tennessee vineyards. Plant Health Prog. 2020, 21, 157–161. [Google Scholar] [CrossRef]
- Ouro-Djobo, A.; Stevens, K.A.; Scheiner, J.; Tsolova, V.M.; Pontasch, F.M.; McBride, S.; Appel, D.N.; Al Rwahnih, M.; Alabi, O.J. Molecular characterization of divergent isolates of grapevine red blotch virus from Blanc Du Soleil, an interspecific hybrid white grapevine cultivar. PhytoFrontiers 2023, 3, 290–295. [Google Scholar] [CrossRef]
- Brannen, P.M.; Deom, C.M.; Alabi, O.J.; Naidu, R.A. Prevalence of viruses in commercial wine grape vineyards in Georgia. Plant Health Prog. 2018, 19, 342–346. [Google Scholar] [CrossRef]
- Seiter, J.; Cieniewicz, E.J. Vector ecology of grapevine red blotch virus in a southeastern U.S. vineyard. Presented at the Plant Health: Pittsburgh, PA, USA, 24 June 2021. [Google Scholar]
- Preto, C.R.; Sudarshana, M.R.; Bollinger, M.L.; Zalom, F.G. Vitis vinifera (Vitales: Vitaceae) as a reproductive host of Spissistilus festinus (Hemiptera: Membracidae). J. Insect Sci. 2018, 18, 20. [Google Scholar] [CrossRef]
- Preto, C.R.; Sudarshana, M.R.; Zalom, F.G. Feeding and reproductive hosts of Spissistilus festinus (Say) (Hemiptera: Membracidae) found in Californian vineyards. J. Econ. Entomol. 2018, 111, 2531–2535. [Google Scholar] [CrossRef]
- Preto, C.R.; Bahder, B.W.; Bick, E.N.; Sudarshana, M.R.; Zalom, F.G. Seasonal dynamics of Spissistilus festinus (Hemiptera: Membracidae) in a Californian vineyard. J. Econ. Entomol. 2019, 112, 1138–1144. [Google Scholar] [CrossRef]
- Wilson, H.; Yazdani, A.S.; Daane, K.M. Influence of riparian habitat and ground covers on three-cornered alfalfa hopper (Hemiptera: Membracidae) populations in vineyards. J. Econ. Entomol. 2020, 113, 2354–2361. [Google Scholar] [CrossRef]
- Hoyle, V.; Flasco, M.T.; Choi, J.; Cieniewicz, E.J.; McLane, H.; Perry, K.; Dangl, G.; Rwahnih, M.A.; Heck, M.; Loeb, G.; et al. Transmission of grapevine red blotch virus by Spissistilus festinus [Say, 1830] (Hemiptera: Membracidae) between free-living vines and Vitis vinifera ‘Cabernet franc’. Viruses 2022, 14, 1156. [Google Scholar] [CrossRef]
- Setiono, F.J.; Chatterjee, D.; Fuchs, M.; Perry, K.L.; Thompson, J.R. The distribution and detection of grapevine red blotch virus in its host depend on time of sampling and tissue type. Plant Dis. 2018, 102, 2187–2193. [Google Scholar] [CrossRef]
- De Barro, P.J.; Hart, P.J. Mating interactions between two biotypes of the whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) in Australia. Bull. Entomol. Res. 2000, 90, 103–112. [Google Scholar] [CrossRef]
- Omondi, B.A.; Sseruwagi, P.; Obeng-Ofori, D.; Danquah, E.Y.; Kyerematen, R.A. Mating interactions between okra and cassava biotypes of Bemisia tabaci (Homoptera: Aleyrodidae) on eggplant. Int. J. Trop. Insect Sci. 2005, 25, 159–167. [Google Scholar] [CrossRef]
- Blanc, S.; Drucker, M.; Uzest, M. Localizing viruses in their insect vectors. Annu. Rev. Phytopathol. 2014, 52, 403–425. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Pan, L.-L.; Liu, S.-S.; Navas-Castillo, J. Transmission of begomoviruses and other whitefly-borne viruses: Dependence on the vector species. Phytopathology 2020, 110, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Blanc, S. Insect transmission of plant single-stranded DNA viruses. Annu. Rev. Entomol. 2021, 66, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479–480, 278–289. [Google Scholar] [CrossRef]
- Shi, X.; Tang, X.; Zhang, X.; Zhang, D.; Li, F.; Yan, F.; Zhang, Y.; Zhou, X.; Liu, Y. Transmission efficiency, preference and behavior of Bemisia tabaci MEAM1 and MED under the influence of tomato chlorosis virus. Front. Plant Sci. 2018, 8, 2271. [Google Scholar] [CrossRef]
- Chowda-Reddy, R.; Kirankumar, M.; Seal, S.E.; Muniyappa, V.; Valand, G.B.; Govindappa, M.; Colvin, J. Bemisia Tabaci phylogenetic groups in India and the relative transmission efficacy of tomato leaf curl Bangalore virus by an indigenous and an exotic population. J. Integr. Agric. 2012, 11, 235–248. [Google Scholar] [CrossRef]
- Buchner, P. Endosymbiosis of Animals with Plant Microorganisms; Wiley: New York, NY, USA, 1965; ISBN 978-0-470-11517-6. [Google Scholar]
- Luan, J.-B.; Shan, H.-W.; Isermann, P.; Huang, J.-H.; Lammerding, J.; Liu, S.-S.; Douglas, A.E. Cellular and molecular remodelling of a host cell for vertical transmission of bacterial symbionts. Proc. Biol. Sci. 2016, 283, 20160580. [Google Scholar] [CrossRef]
- Xu, X.-R.; Li, N.-N.; Bao, X.-Y.; Douglas, A.E.; Luan, J.-B. Patterns of host cell inheritance in the bacterial symbiosis of whiteflies. Insect Sci. 2020, 27, 938–946. [Google Scholar] [CrossRef]
- Cieniewicz, E.J.; Pethybridge, S.J.; Gorny, A.; Madden, L.V.; McLane, H.; Perry, K.L.; Fuchs, M. Spatiotemporal spread of grapevine red blotch-associated virus in a California vineyard. Virus Res. 2017, 241, 156–162. [Google Scholar] [CrossRef]
- Cieniewicz, E.; Flasco, M.; Brunelli, M.; Onwumelu, A.; Wise, A.; Fuchs, M.F. Differential spread of grapevine red blotch virus in California and New York vineyards. Phytobiomes J. 2019, 3, 203–211. [Google Scholar] [CrossRef]
- Dalton, D.T.; Hilton, R.J.; Kaiser, C.; Daane, K.M.; Sudarshana, M.R.; Vo, J.; Zalom, F.G.; Buser, J.Z.; Walton, V.M. Spatial associations of vines infected with grapevine red blotch virus in Oregon vineyards. Plant Dis. 2019, 103, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Cieniewicz, E.J.; Qiu, W.; Saldarelli, P.; Fuchs, M. Believing is seeing: Lessons from emerging viruses in grapevine. J. Plant Pathol. 2020, 102, 619–632. [Google Scholar] [CrossRef]
- Fall, M.L.; Xu, D.; Lemoyne, P.; Moussa, I.E.B.; Beaulieu, C.; Carisse, O. A diverse virome of leafroll-infected grapevine unveiled by dsRNA sequencing. Viruses 2020, 12, 1142. [Google Scholar] [CrossRef] [PubMed]
- LaFond, H.F.; Volenberg, D.S.; Schoelz, J.E.; Finke, D.L. Identification of potential grapevine red blotch virus vector in Missouri vineyards. Am. J. Enol. Vitic. 2022, 73, 247–255. [Google Scholar] [CrossRef]
- Wilson, H.; Hogg, B.N.; Blaisdell, G.K.; Andersen, J.C.; Yazdani, A.S.; Billings, A.C.; Ooi, K.M.; Soltani, N.; Almeida, R.P.P.; Cooper, M.L.; et al. Survey of vineyard insects and plants to identify potential insect vectors and noncrop reservoirs of grapevine red blotch virus. PhytoFrontiers 2022, 2, 66–73. [Google Scholar] [CrossRef]

| Parent Pair a | Replicate b | Observed c | Average d |
|---|---|---|---|
| CA Only | 1 | 94 | 50.6 |
| 2 | 35 | ||
| 3 | 58 | ||
| 4 | 43 | ||
| 5 | 23 | ||
| SE Only | 1 | 1 | 54.2 |
| 2 | 70 | ||
| 3 | 63 | ||
| 4 | 82 | ||
| 5 | 55 | ||
| CA Male + SE Female | 1 | 55 | 52 |
| 2 | 1 | ||
| 3 | 52 | ||
| 4 | 80 | ||
| 5 | 31 | ||
| 6 | 93 | ||
| SE Male + CA Female | 1 | 3 | 46.8 |
| 2 | 88 | ||
| 3 | 42 | ||
| 4 | 32 | ||
| 5 | 69 |
| Parental Pair a | Total b | AverageT c | Males | AverageM d | Females | AverageF e |
|---|---|---|---|---|---|---|
| CA Male + SE Female | 106 | 17.7 | 59 | 9.8 | 47 | 7.8 |
| SE Male + CA Female | 51 | 10.2 | 30 | 12 | 21 | 4.2 |
| Genotype a | |||
|---|---|---|---|
| Acquisition Access Period b | CA c | SE c | Total d |
| 6 | 23/34 (68%) | 28/44 (64%) | 51/78 (65%) |
| 8 | 15/33 (45%) | 19/31 (61%) | 34/64 (53%) |
| 10 | 23/38 (61%) | 18/31 (58%) | 41/69 (59%) |
| Total e | 61/105 (58%) | 65/106 (61%) | |
| Genotype a | |||
|---|---|---|---|
| Acquisition Access Period b | CA c | SE c | Total d |
| 6 | 3/5 (60%) | 5/5 (100%) | 8/10 (80%) |
| 8 | 3/5 (60%) | 5/7 (71%) | 8/12 (67%) |
| 10 | 2/6 (33%) | 6/6 (100%) | 8/12 (67%) |
| Total e | 8/16 (50%) | 16/18 (89%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flasco, M.T.; Fuchs, M.F. Two Distinct Genotypes of Spissistilus festinus (Say, 1830) Reproduce and Differentially Transmit Grapevine Red Blotch Virus. Insects 2023, 14, 831. https://doi.org/10.3390/insects14100831
Flasco MT, Fuchs MF. Two Distinct Genotypes of Spissistilus festinus (Say, 1830) Reproduce and Differentially Transmit Grapevine Red Blotch Virus. Insects. 2023; 14(10):831. https://doi.org/10.3390/insects14100831
Chicago/Turabian StyleFlasco, Madison T., and Marc F. Fuchs. 2023. "Two Distinct Genotypes of Spissistilus festinus (Say, 1830) Reproduce and Differentially Transmit Grapevine Red Blotch Virus" Insects 14, no. 10: 831. https://doi.org/10.3390/insects14100831
APA StyleFlasco, M. T., & Fuchs, M. F. (2023). Two Distinct Genotypes of Spissistilus festinus (Say, 1830) Reproduce and Differentially Transmit Grapevine Red Blotch Virus. Insects, 14(10), 831. https://doi.org/10.3390/insects14100831









