Bacterial Isolates Derived from Nest Soil Affect the Attraction and Digging Behavior of Workers of the Red Imported Fire Ant, Solenopsis invicta Buren
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Collection and Isolation of Bacteria from RIFA Nests
2.2. Identification of Bacterial Isolates
2.3. Isolate Preparations
2.4. Collection and Maintenance of RIFA Colonies
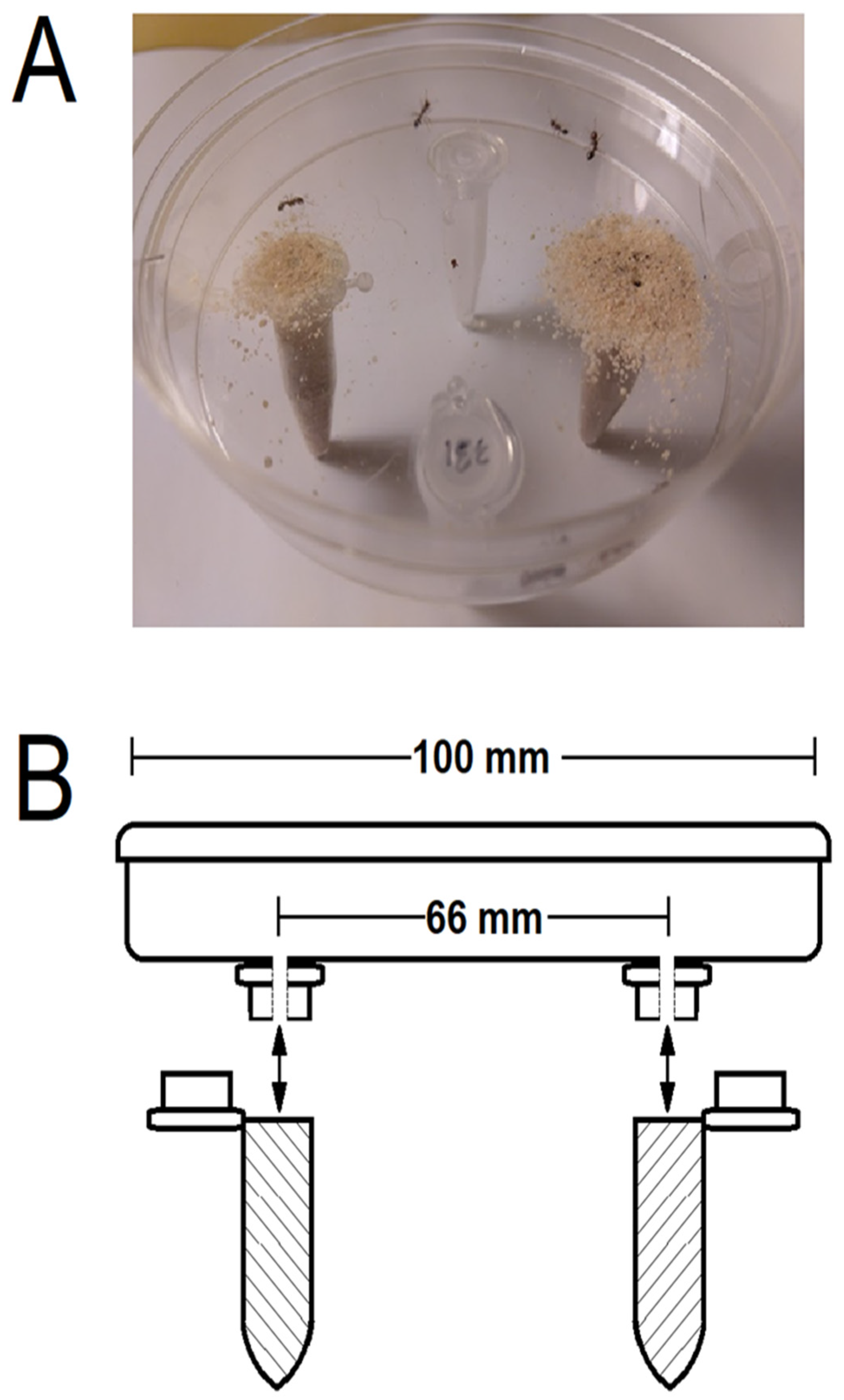
2.5. Bioassay Setup and Validation
2.6. Behavioral Responses of Worker Ants to Bacterial Isolates
2.7. Time-Course Observations
2.8. Statistical Analyses
3. Results
3.1. Bacteria Isolated from Ant Nest Soil
3.2. Behavioral Responses of RIFA Workers to Bacterial Isolates
3.3. Worker Ant Responses to Bacteria Isolates at Lower Cell Densities
3.4. Time-Course of Worker Ant Responses to Bacterial Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Callcott, A.-M.A.; Collins, H.L. Invasion and range expansion of imported fire ants (Hymenoptera: Formicidae) in North America from 1918–1995. Fla. Entomol. 1996, 79, 240–251. [Google Scholar] [CrossRef]
- Ascunce, M.S.; Yang, C.-C.; Oakey, J.; Calcaterra, L.; Wu, W.-J.; Shih, C.-J.; Goudet, J.; Ross, K.G.; Shoemaker, D. Global invasion history of the fire ant Solenopsis invicta. Science 2011, 331, 1066–1068. [Google Scholar] [CrossRef] [PubMed]
- Wetterer, J.K.; Davis, L.R.; White, G.L. Spread in Trinidad of the South American fire ant Solenopsis invicta (Hymenoptera, Formicidae). Fla. Entomol. 2014, 97, 238–241. [Google Scholar] [CrossRef]
- Porter, S.D.; Savignano, D.A. Invasion of polygyne fire ants decimates native ants and disrupts arthropod community. Ecology 1990, 71, 2095–2106. [Google Scholar] [CrossRef]
- Vander Meer, R.; Pereira, R.; Porter, S.; Valles, S.; Oi, D. Area-wide suppression of invasive fire ant Solenopsis spp. populations. In Area-Wide Control of Insect Pests; Springer: New York, NY, USA, 2007; pp. 487–496. [Google Scholar]
- Vinson, S.B. Insect life: Invasion of the red imported fire ant (Hymenoptera: Formicidae). Am. Entomol. 1997, 43, 23–39. [Google Scholar] [CrossRef]
- Eubanks, M.D. Estimates of the direct and indirect effects of red imported fire ants on biological control in field crops. Biol. Control 2001, 21, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Allen, C.R.; Epperson, D.; Garmestani, A. Red imported fire ant impacts on wildlife: A decade of research. Am. Midl. Nat. 2004, 152, 88–103. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wurm, Y.; Nipitwattanaphon, M.; Riba-Grognuz, O.; Huang, Y.-C.; Shoemaker, D.; Keller, L. A Y-like social chromosome causes alternative colony organization in fire ants. Nature 2013, 493, 664–668. [Google Scholar] [CrossRef]
- Byron, D.W.; Hays, S.B. Occurrence and significance of multiple mound utilization by colonies of the red imported fire ant (Hymenoptera: Formicidae). J. Econ. Entomol. 1986, 79, 637–640. [Google Scholar] [CrossRef]
- Tschinkel, W.R.; King, J.R. Ant community and habitat limit colony establishment by the fire ant, Solenopsis invicta. Funct. Ecol. 2017, 31, 955–964. [Google Scholar] [CrossRef] [Green Version]
- Stiles, J.H.; Jones, R.H. Distribution of the red imported ant, Solenopsis invicta, in road and powerline habitats. Landsc. Ecol. 1998, 335, 335–346. [Google Scholar] [CrossRef]
- Zettler, J.A.; Taylor, M.D.; Allen, C.R.; Spira, T.P. Consequences of forest clear-cuts for native and nonindigenous ants (Hymenoptera: Formicidae). Ann. Entomol. Soc. Am. 2004, 97, 513–518. [Google Scholar] [CrossRef]
- Apperson, C.S.; Powell, E.E. Foraging activity of ants (Hymenoptera: Formicidae) in a pasture inhabited by the red imported fire ant. Fla. Entomol. 1984, 67, 383. [Google Scholar] [CrossRef]
- Morrison, L.W.; Porter, S.D.; Daniels, E.; Korzukhin, M.D. Potential global range expansion of the invasive fire ant, Solenopsis invicta. Biol. Invasions 2004, 6, 183–191. [Google Scholar] [CrossRef]
- Tschinkel, W.R. The Fire Ants; Harvard University Press: Cambrdige, MA, USA, 2006. [Google Scholar]
- Sternberg, T.; Perry, G.; Britton, C. Grass repellency to the red imported fire ant. Rangel. Ecol. Manag. 2006, 59, 330–333. [Google Scholar] [CrossRef]
- Schmid-Hempel, P. Parasites in Social Insects; Princeton University Press: Princeton, NJ, USA, 1998; Volume 60. [Google Scholar]
- Oi, D.H.; Williams, D.F.; Pereira, R.M.; Horton, P.M.; Davis, T.S.; Hyder, A.H.; Bolton, H.T.; Zeichner, B.C.; Porter, S.D.; Hoch, L.A. Combining biological and chemical controls for the management of red imported fire ants (Hymenoptera: Formicidae). Am. Entomol. 2008, 54, 46–55. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Lu, L.; Feng, X.; Chen, H.; Zhou, X.; Zhang, D.; Huang, H.; He, Y. Advance in biological control of red imported fire ant, Solenopsis invicta (Hymenoptera: Formicidae). Guangdong Agric. Sci. 2006, 5, 18–23. [Google Scholar]
- Li, J.; Guo, Q.; Lin, M.; Jiang, L.; Ye, J.; Chen, D.; Li, Z.; Dai, J.; Han, S. Evaluation of a new entomopathogenic strain of Beauveria bassiana and a new field delivery method against Solenopsis invicta. PLoS ONE 2016, 11, e0158325. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.E.; Buren, W.F. Microsporidan and fungal diseases of Solenopsis invicta Buren in Brazil. J. N. Y. Entomol. Soc. 1974, 82, 125–130. [Google Scholar]
- Woolfolk, S.W. Bacteria and Fungi Associated with Red Imported Fire Ants Solenopsis Invicta Buren (Hymenoptera: Formicidae) and Mounds in Mississippi, and Their Potential Use as Biological Control Agents; Mississippi State University: Oxford, MS, USA, 2020. [Google Scholar]
- Nickle, W.; Jouvenaz, D. Tetradonema solenopsis n. sp.(Nematoda: Tetradonematidae) parasitic on the red imported fire ant Solenopsis invicta Buren from Brazil. J. Nematol. 1987, 19, 311. [Google Scholar]
- Stimac, J.L.; Pereira, R.M.; Alves, S.B.; Wood, L.A. Beauveria bassiana (Balsamo) Vuillemin (Deuteromycetes) applied to laboratory colonies of Solenopsis invicta Buren (Hymenoptera: Formicidae) in soil. J. Econ. Entomol. 1993, 86, 348–352. [Google Scholar] [CrossRef]
- Williams, D.F.; Oi, D.H.; Knue, G.J. Infection of red imported fire ant (Hymenoptera: Formicidae) colonies with the entomopathogen Thelohania solenopsae (Microsporidia: Thelohaniidae). J. Econ. Entomol. 1999, 92, 830–836. [Google Scholar] [CrossRef]
- Leroy, P.D.; Sabri, A.; Verheggen, F.J.; Francis, F.; Thonart, P.; Haubruge, E. The semiochemically mediated interactions between bacteria and insects. Chemoecology 2011, 21, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Vander Meer, R. Ant interactions with soil organisms and associated semiochemicals. J. Chem. Ecol. 2012, 38, 728–745. [Google Scholar] [CrossRef] [PubMed]
- Wielkopolan, B.; Obrępalska-Stęplowska, A. Three-way interaction among plants, bacteria, and coleopteran insects. Planta 2016, 244, 313–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engl, T.; Kaltenpoth, M. Influence of microbial symbionts on insect pheromones. Nat. Prod. Rep. 2018, 35, 386–397. [Google Scholar] [CrossRef]
- Huang, H.; Ren, L.; Li, H.; Schmidt, A.; Gershenzon, J.; Lu, Y.; Cheng, D. The nesting preference of an invasive ant is associated with the cues produced by actinobacteria in soil. PLoS Pathog. 2020, 16, e1008800. [Google Scholar] [CrossRef]
- Travanty, N.V.; Vargo, E.L.; Apperson, C.S.; Ponnusamy, L. Colonization by the red imported fire ant, Solenopsis invicta, modifies soil bacterial communities. Microb. Ecol. 2021, 9, 1–17. [Google Scholar] [CrossRef]
- Valles, S.M.; Porter, S.D. Identification of polygyne and monogyne fire ant colonies (Solenopsis invicta) by multiplex PCR of Gp-9 alleles. Insectes Soc. 2003, 50, 199–200. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Ponnusamy, L.; Xu, N.; Nojima, S.; Wesson, D.M.; Schal, C.; Apperson, C.S. Identification of bacteria and bacteria-associated chemical cues that mediate oviposition site preferences by Aedes aegypti. Proc. Natl. Acad. Sci. USA 2008, 105, 9262–9267. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.; McGarrell, D.M.; Marsh, T.; Garrity, G.M. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Allen, M. Significance of digging behavior to mortality of red imported fire ant workers, Solenopsis invicta, in fipronil-treated sand. J. Econ. Entomol. 2006, 99, 476–482. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, G. Effect of gland extracts on digging and residing preferences of red imported fire ant workers (Hymenoptera: Formicidae). Insect Sci. 2013, 20, 456–466. [Google Scholar] [CrossRef]
- Woolfolk, S.; Stokes, C.E.; Watson, C.; Brown, R.; Baird, R. Bacteria associated with red imported fire ants (Solenopsis invicta) from mounds in Mississippi. Microb. Ecol. 2016, 15, 83–101. [Google Scholar] [CrossRef]
- Stahly, D.P.; Andrews, R.E.; Yousten, A.A. The genus Bacillus—insect pathogens. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 563–608. [Google Scholar] [CrossRef]
- Matheson, A. World Bee Health Report. Bee World 1993, 74, 176–212. [Google Scholar] [CrossRef]
- Evans, J.D. Transcriptional immune responses by honey bee larvae during invasion by the bacterial pathogen, Paenibacillus larvae. J. Invertebr. Pathol. 2004, 85, 105–111. [Google Scholar] [CrossRef]
- Bulla, L.A., Jr.; Candas, M. Formicidae (Ant) Control Using Bacillus thuringiensis toxin. U.S. Patent 6,797,490, 28 September 2004. [Google Scholar]
- Funke, G.; Hutson, R.A.; Bernard, K.A.; Pfyffer, G.E.; Wauters, G.; Collins, M.D. Isolation of Arthrobacter spp. from clinical specimens and description of Arthrobacter cumminsii sp. nov. and Arthrobacter woluwensis sp. nov. J. Clin. Microbiol. 1996, 34, 2356–2363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.; Keddie, R.M. The genus Arthrobacter. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 945–960. [Google Scholar] [CrossRef]
- Ishak, H.D.; Plowes, R.; Sen, R.; Kellner, K.; Meyer, E.; Estrada, D.A.; Dowd, S.E.; Mueller, U.G. Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb. Ecol. 2011, 61, 821–831. [Google Scholar] [CrossRef]
- Powell, C.M.; Hanson, J.D.; Bextine, B.R. Bacterial community survey of Solenopsis invicta Buren (red imported fire ant) colonies in the presence and absence of Solenopsis invicta virus (SINV). Curr. Microbiol. 2014, 69, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Zahner, V.; Lucarotti, C.J.; McIntosh, D. Application of 16S rDNA-DGGE and plate culture to characterization of bacterial communities associated with the sawfly, Acantholyda erythrocephala (Hymenoptera, Pamphiliidae). Curr. Microbiol. 2008, 57, 564–569. [Google Scholar] [CrossRef]
- Danismazoglu, M.; Demir, İ.; Sevim, A.; Demirbag, Z.; Nalcacioglu, R. An investigation on the bacterial flora of Agriotes lineatus (Coleoptera: Elateridae) and pathogenicity of the flora members. Crop. Prot. 2012, 40, 1–7. [Google Scholar] [CrossRef]
- González-Dominici, L.I.; Saati-Santamaría, Z.; García-Fraile, P. Genome analysis and genomic comparison of the novel species Arthrobacter ipsi reveal its potential protective role in its bark beetle host. Microb. Ecol. 2020, 81, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Munaganti, R.K.; Muvva, V.; Konda, S.; Naragani, K.; Mangamuri, U.K.; Dorigondla, K.R.; Akkewar, D.M. Antimicrobial profile of Arthrobacter kerguelensis VL-RK_09 isolated from Mango orchards. Braz. J. Microbiol. 2016, 47, 1030–1038. [Google Scholar] [CrossRef] [Green Version]
- Ponnusamy, L.; Schal, C.; Wesson, D.M.; Arellano, C.; Apperson, C.S. Oviposition responses of Aedes mosquitoes to bacterial isolates from attractive bamboo infusions. Paras. Vectors 2015, 8, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada-Katsumata, A.; Zurek, L.; Nalyanya, G.; Roelofs, W.L.; Zhang, A.; Schal, C. Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. USA 2015, 112, 15678–15683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goelen, T.; Vuts, J.; Sobhy, I.S.; Wäckers, F.; Caulfield, J.C.; Birkett, M.A.; Rediers, H.; Jacquemyn, H.; Lievens, B. Identification and application of bacterial volatiles to attract a generalist aphid parasitoid: From laboratory to greenhouse assays. Pest Manag. Sci. 2020, 77, 930–938. [Google Scholar] [CrossRef]
- Davis, T.S.; Crippen, T.L.; Hofstetter, R.W.; Tomberlin, J.K. Microbial volatile emissions as insect semiochemicals. J. Chem. Ecol. 2013, 39, 840–859. [Google Scholar] [PubMed]
- Morel, L.; Meer, R.K.V.; Lofgren, C.S. Comparison of nestmate recognition between monogyne and polygyne populations of Solenopsis invicta (Hymenoptera: Formicidae). Ann. Entomol. Soc. Am. 1990, 83, 642–647. [Google Scholar] [CrossRef]
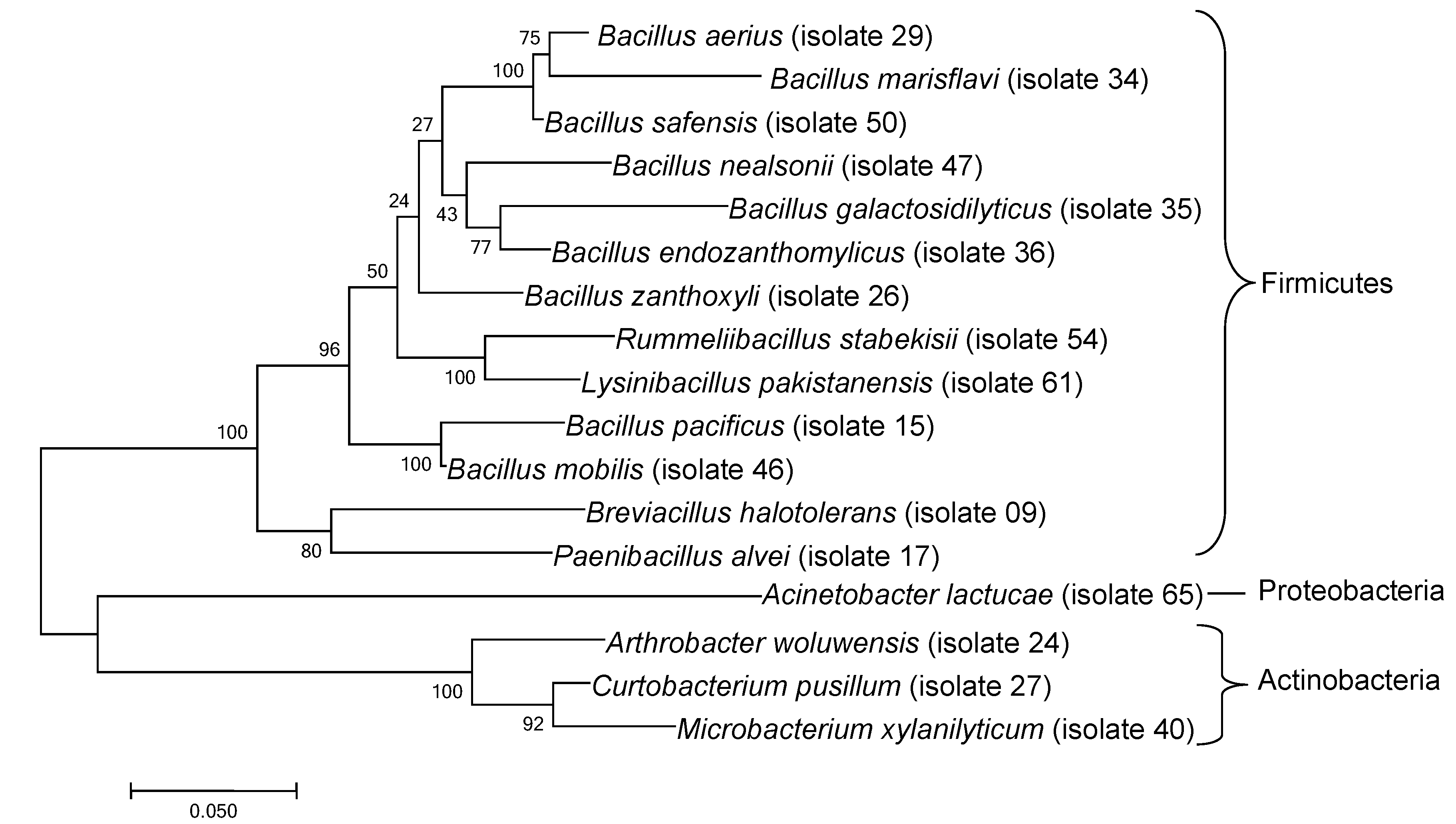


| Closest Cultured Bacteria/Sequence from NCBI (Strain) | Classification (Phylum) | Similarity (%) | Closest Match NCBI Accession Number | Deposited in NCBI with Accession Number |
|---|---|---|---|---|
| Brevibacillus halotolerans (LAM0312) | Firmicutes | 99.80 | NR156834 | MW255490 |
| Bacillus pacificus (MCCC 1A06182) | Firmicutes | 99.80 | NR157733 | MW255491 |
| Paenibacillus alvei (NBRC 3343) | Firmicutes | 99.20 | NR113577 | MW255492 |
| Arthrobacter woluwensis (1551) | Actinobacteria | 98.30 | NR044894 | MW255493 |
| Bacillus zanthoxyli (1433) | Firmicutes | 99.80 | NR164882 | MW255494 |
| Curtobacterium pusillum (DSM 20527) | Actinobacteria | 98.20 | NR042315 | MW255495 |
| Bacillus aerius (24K) | Firmicutes | 99.80 | NR118439 | MW255496 |
| Bacillus marisflavi (TF-11) | Firmicutes | 99.80 | NR118437 | MW255497 |
| Bacillus galactosidilyticus (LMG 17892) | Firmicutes | 96.70 | NR025580 | MW255498 |
| Bacillus endozanthoxylicus (1404) | Firmicutes | 96.20 | NR158107 | MW255499 |
| Microbacterium xylanilyticum (S3-E) | Actinobacteria | 98.20 | NR042350 | MW255500 |
| Bacillus mobilis (MCCC 1A05942) | Firmicutes | 99.80 | NR157731 | MW255501 |
| Bacillus nealsonii (DSM 15077) | Firmicutes | 98.50 | NR044546 | MW255502 |
| Bacillus safensis (FO-36b) | Firmicutes | 100 | NR041794 | MW255503 |
| Rummeliibacillus stabekisii (NBRC 104870) | Firmicutes | 99.80 | NR114270 | MW255504 |
| Lysinibacillus pakistanensis (NCCP-54) | Firmicutes | 99.80 | NR113166 | MW255505 |
| Acinetobacter lactucae (NRRL B-41902) | Proteobacteria | 99.30 | NR152004 | MW255506 |
| Bacterial Isolate | Number of Ants (±SE) | % Ants Responding (±SE) | t-Test on Ant Choice t-Value (P > t) | DF | |
|---|---|---|---|---|---|
| Treatment | Control | ||||
| B. halotolerans | 8.1 (0.6) | 8.5 (0.8) | 82.6 (3.5) | −0.29 (0.387) | 15 |
| B. pacificus | 3.4 (1.1) | 13.8 (1.2) | 84.2 (2.9) | −4.70 (<0.001) | 15 |
| P. alvei | 3.1 (1.4) | 13.1 (1.7) | 80.6 (6.1) | −3.45 (0.002) | 15 |
| A. woluwensis | 9.3 (0.9) | 5.0 (0.9) | 71.0 (4.2) | 2.63 (0.010) | 15 |
| B. zanthoxyli | 1.6 (0.6) | 13.4 (1.1) | 74.5 (5.6) | −8.98 (<0.001) | 15 |
| C. pusillum | 8.8 (1.3) | 6.5 (1.2) | 75.8 (5.0) | 0.97 (0.174) | 15 |
| B. aerius | 3.3 (0.7) | 8.33 (1.5) | 58.9 (7.6) | −2.85 (0.006) | 15 |
| B. marisflavi | 1.9 (0.8) | 14.4 (1.0) | 81.5 (3.5) | −7.54 (<0.001) | 15 |
| B. galactosidilyticus | 6.3 (1.0) | 8.6 (1.0) | 74.2 (3.7) | −1.24 (0.116) | 15 |
| B. endozanthoxylicus | 3.9 (1.3) | 11.8 (1.4) | 78.4 (3.4) | −3.07 (0.004) | 15 |
| M. xylanilyticum | 7.9 (0.9) | 8.3 (1.1) | 80.6 (3.0) | −0.20 (0.424) | 15 |
| B. mobilis | 2.3 (0.7) | 12.1 (1.2) | 73.0 (4.3) | −5.88 (<0.001) | 15 |
| B. nealsonii | 5.8 (1.1) | 10.2 (1.3) | 79.4 (4.2) | −1.95 (0.035) | 15 |
| B. safensis | 7.4 (1.2) | 7.2 (1.1) | 73.2 (5.3) | 0.09 (0.464) | 15 |
| R. stabekisii | 4.2 (1.2) | 11.2 (1.6) | 77.1 (5.2) | −2.63 (0.010) | 15 |
| L. pakistanensis | 7.3 (1.8) | 8.4 (1.8) | 78.4 (3.1) | −0.30 (0.385) | 15 |
| A. lactucae | 3.7 (0.7) | 10.1 (1.0) | 73.3 (4.2) | −4.83 (<0.001) | 15 |
| Bacterial Isolate | Sand Removed (mg) (±SE) | t-Test on Sand Removed t-Value (P > t) | DF | |
|---|---|---|---|---|
| Treatment | Control | |||
| B. halotolerans | 775.3 (62.0) | 998.4 (66.7) | −4.09 (<0.001) | 15 |
| B. pacificus | 123.4 (48.2) | 430.9 (82.0) | −4.56 (<0.001) | 15 |
| P. alvei | 144.4 (56.3) | 332.8 (65.9) | −1.97 (0.034) | 15 |
| A. woluwensis | 531.0 (86.6) | 351.0 (53.2) | 2.36 (0.016) | 15 |
| B. zanthoxyli | 205.4 (71.5) | 667.4 (57.7) | −9.04 (<0.001) | 15 |
| C. pusillum | 634.7 (69.1) | 783.6 (99.0) | −1.19 (0.127) | 15 |
| B. aerius | 71.8 (17.6) | 184.1 (47.9) | −2.24 (0.021) | 15 |
| B. marisflavi | 136.8 (49.3) | 347.0 (61.8) | −3.65 (0.001) | 15 |
| B. galactosidilyticus | 376.7 (65.4) | 501.6 (66.7) | −2.32 (0.017) | 15 |
| B. endozanthoxylicus | 253.1 (69.5) | 472.9 (83.1) | −2.95 (0.005) | 15 |
| M. xylanilyticum | 577.0 (68.5) | 556.4 (58.8) | 0.40 (0.346) | 15 |
| B. mobilis | 267.6 (77.0) | 509.6 (70.0) | −4.26 (<0.001) | 15 |
| B. nealsonii | 415.8 (92.7) | 559.8 (76.9) | −2.17 (0.023) | 15 |
| B. safensis | 956.2 (71.2) | 1124.4 (61.6) | −2.77 (0.007) | 15 |
| R. stabekisii | 351.3 (63.2) | 615.6 (66.9) | −2.23 (0.021) | 15 |
| L. pakistanensis | 312.6 (59.7) | 880.3 (65.6) | −8.63 (<0.001) | 15 |
| A. lactucae | 337.6 (65.6) | 535.4 (80.5) | −2.70 (0.008) | 15 |
| Bacterial Isolate | Average DPI (±SE) | Tukey’s Test to Identify Significantly Different Means | |||
|---|---|---|---|---|---|
| A. woluwensis | 0.21 (0.11) | A | |||
| M. xylanilyticum | 0.01 (0.05) | A | B | ||
| C. pusillum | −0.05 (0.11) | A | B | C | |
| B. safensis | −0.09 (0.03) | A | B | C | |
| B. halotolerans | −0.13 (0.03) | A | B | C | |
| B. galactosidilyticus | −0.20 (0.08) | A | B | C | |
| B. mobilis | −0.22 (0.12) | A | B | C | D |
| R. stabekisii | −0.27 (0.14) | A | B | C | D |
| B. aerius | −0.28 (0.13) | A | B | C | D |
| A. lactucae | −0.28 (0.10) | A | B | C | D |
| B. endozanthoxylicus | −0.34 (0.14) | B | C | D | |
| P. alvei | −0.44 (0.19) | B | C | D | |
| B. mobilis | −0.45 (0.10) | B | C | D | |
| B. marisflavi | −0.52 (0.12) | C | D | ||
| L. pakistanensis | −0.55 (0.07) | C | D | ||
| B. zanthoxyli | −0.66 (0.08) | D | |||
| B. pacificus | −0.68 (0.09) | D | |||
| Bacterial Isolate | Ant Social Form † | Number of Ants (±SE) | Mean Amount of Sand (mg) Excavated (±SE) | Average DPI (±SE) ‡ | t-Test on Ant Choice t-Value (P) § | t-test on Ant Digging Activity t-Value (P > t) § | DF | ||
|---|---|---|---|---|---|---|---|---|---|
| Treatment | Control | Treatment | Control | ||||||
| A. woluwensis | M | 16.1 (1.1) | 3.0 (0.9) | 477.3 (92.9) | 209.0 (70.3) | 0.53 (0.13) a | 6.86 (<0.001) | 3.56 (0.005) | 7 |
| P | 11.8 (1.8) | 5.4 (1.3) | 293.9 (95.5) | 145.8 (32.1) | 0.13 (0.19) a,b | 2.14 (0.035) | 1.82 (0.056) | 7 | |
| M. xylanilyticum | M | 8.9 (2.3) | 8.5 (2.1) | 744.0 (166.5) | 798.8 (103.1) | −0.10 (0.18) a,b,c | 0.09 (0.467) | −0.22 (0.415) | 7 |
| P | 7.5 (1.6) | 8.6 (1.8) | 532.0 (146.5) | 589.5 (69.9) | −0.19 (0.18) b,c | −0.37 (0.363) | −0.45 (0.334) | 7 | |
| C. pusillum | M | 1.9 (0.8) | 14.3 (1.7) | 561.5 (108.2) | 1084.8 (67.8) | −0.24 (0.11) b,c | −4.90 (<0.001) | −4.05 (0.002) | 7 |
| P | 4.1 (1.1) | 8.0 (0.5) | 485.8 (90.3) | 713.1 (81.0) | −0.36 (0.11) b,c | −2.83 (0.013) | −3.28 (0.007) | 7 | |
| L. pakistanensis | M | 7.4 (2.0) | 8.6 (1.4) | 103.7 (78.4) | 149.0 (55.4) | −0.37 (0.17) b,c | −0.32 (0.365) | −0.91 (0.198) | 6 |
| P | 6.0 (1.3) | 10.0 (0.9) | 115.9 (36.0) | 207.4 (45.8) | −0.37 (0.07) b,c | −1.90 (0.050) | −3.46 (0.005) | 7 | |
| B. zanthoxyli | M | 4.5 (2.2) | 12.0 (2.0) | 553.8 (96.6) | 1120.5 (49.6) | −0.38 (0.15) b,c | −1.81 (0.057) | −6.98 (<0.001) | 7 |
| P | 3.6 (1.5) | 10.5 (2.1) | 328.3 (115.0) | 595.1 (91.1) | −0.39 (0.11) b,c | −2.08 (0.038) | −1.78 (0.059) | 7 | |
| B. pacificus | M | 3.0 (0.6) | 13.9 (1.3) | 480.3 (140.8) | 904.3 (113.4) | −0.46 (0.15) b,c | −5.89 (<0.001) | −2.97 (0.010) | 7 |
| P | 1.1 (0.4) | 15.0 (1.0) | 56.9 (17.2) | 562.9 (138.0) | −0.65 (0.18) c | −11.84 (<0.001) | −3.84 (0.003) | 7 | |
| Bacterial Isolate | Ant Social Form † | Number of Ants (±SE) | Mean Amount of Sand (mg) Excavated (±SE) | Average DPI (±SE) ‡ | t-test on Ant Choice t-Value (P) § | t-Test on ant Digging Activity t-Value (P > t) § | DF | ||
|---|---|---|---|---|---|---|---|---|---|
| Treatment | Control | Treatment | Control | ||||||
| A. woluwensis | M | 8.9 (2.2) | 9.3 (2.6) | 535.5 (93.5) | 560.9 (135.5) | 0.29 (0.19) | −0.08 (0.470) | −0.22 (0.418) | 7 |
| P | 12.6 (1.2) | 4.4 (1.4) | 349.5 (105.0) | 252.4 (82.6) | 0.11 (0.12) | 3.51 (0.005) | 0.85 (0.212) | 7 | |
| M. xylanilyticum | M | 4.4 (1.1) | 10.5 (1.6) | 77.4 (14.2) | 144.3 (35.7) | 0.06 (0.06) | −2.34 (0.026) | −1.53 (0.085) | 7 |
| P | 1.8 (0.6) | 3.6 (0.7) | 12.9 (1.7) | 56.4 (22.6) | 0.02 (0.12) | −1.64 (0.072) | −1.94 (0.047) | 7 | |
| C. pusillum | M | 5.3 (1.0) | 9.6 (0.9) | 626.4 (115.9) | 658.9 (145.4) | −0.01 (0.10) | −2.54 (0.019) | −0.35 (0.368) | 7 |
| P | 8.6 (0.7) | 3.5 (1.0) | 371.1 (31.9) | 342.4 (49.1) | −0.14 (0.12) | 3.52 (0.005) | 0.7 (0.252) | 7 | |
| L. pakistanensis | M | 7.0 (0.8) | 9.3 (0.6) | 900.9 (56.6) | 810.6 (138.0) | −0.19 (0.33) | −1.76 (0.061) | 0.62 (0.279) | 7 |
| P | 3.9 (1.2) | 9.0 (1.1) | 247.3 (50.7) | 303.4 (37.7) | −0.21 (0.21) | −2.42 (0.023) | 0.84 (0.214) | 7 | |
| B. zanthoxyli | M | 4.3 (1.6) | 10.4 (2.3) | 372.8 (130.0) | 559.5 (132.9) | −0.25 (0.11) | −1.61 (0.076) | −2.65 (0.016) | 7 |
| P | 5.8 (2.3) | 10.0 (2.5) | 124.6 (62.6) | 152.8 (44.0) | −0.27 (0.19) | −0.92 (0.195) | −0.31 (0.383) | 7 | |
| B. pacificus | M | 2.8 (1) | 10.0 (2.4) | 63.4 (44.8) | 216.1 (85.5) | −0.37 (0.27) | −2.27 (0.029) | −2.38 (0.025) | 7 |
| P | 9.5 (3.2) | 6.9 (2.8) | 194.3 (106.6) | 245.3 (84.8) | −0.42 (0.15) | 0.47 (0.327) | −0.74 (0.242) | 7 | |
| RIFA Social Form | Bacterial Isolate | Sand Removed (±SE) mg | t-Test on Amount of Sand Removed t-Value (P > t) | DF | DPI (±SE) | |
|---|---|---|---|---|---|---|
| Treatment | Control | |||||
| Monogyne | B. pacificus | 21.3 (5.5) | 41.4 (12.2) | −1.563 (0.081) | 7 | −0.197 (0.173) |
| A. woluwensis | 169.7 (62.2) | 65.7 (29.7) | 2.126 (0.043) | 6 | 0.377 (0.194) | |
| B. zanthoxyli | 34.1 (21.6) | 107.6 (38.7) | −2.735 (0.015) | 7 | −0.342 (0.232) | |
| C. pusillum | 211.5 (77.4) | 82 (32.6) | 1.432 (0.098) | 7 | 0.137 (0.267) | |
| M. xylanilyticum | 18.6 (7.6) | 86.4 (32.1) | −2.366 (0.025) | 7 | −0.526 (0.139) | |
| L. pakistanensis | 22.8 (11.9) | 75.9 (37.6) | −1.214 (0.132) | 7 | −0.27 (0.288) | |
| Polygyne | B. pacificus | 6.9 (1.8) | 8 (1.3) | −0.471 (0.327) | 6 | −0.136 (0.181) |
| A. woluwensis | 49.7 (36) | 16.6 (2.7) | 0.893 (0.203) | 6 | −0.09 (0.276) | |
| B. zanthoxyli | 40.1 (21.5) | 90.7 (62.6) | −1.193 (0.139) | 6 | −0.133 (0.239) | |
| C. pusillum | 23 (5.4) | 10 (3) | 2.024 (0.042) | 7 | 0.321 (0.174) | |
| M. xylanilyticum | 9.4 (1.9) | 30.5 (24.1) | −0.838 (0.218) | 7 | 0.063 (0.194) | |
| L. pakistanensis | 9.3 (3.3) | 20.1 (5.9) | −1.748 (0.066) | 6 | −0.406 (0.123) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Travanty, N.V.; Vargo, E.L.; Schal, C.; Apperson, C.S.; Ponnusamy, L. Bacterial Isolates Derived from Nest Soil Affect the Attraction and Digging Behavior of Workers of the Red Imported Fire Ant, Solenopsis invicta Buren. Insects 2022, 13, 444. https://doi.org/10.3390/insects13050444
Travanty NV, Vargo EL, Schal C, Apperson CS, Ponnusamy L. Bacterial Isolates Derived from Nest Soil Affect the Attraction and Digging Behavior of Workers of the Red Imported Fire Ant, Solenopsis invicta Buren. Insects. 2022; 13(5):444. https://doi.org/10.3390/insects13050444
Chicago/Turabian StyleTravanty, Nicholas V., Edward L. Vargo, Coby Schal, Charles S. Apperson, and Loganathan Ponnusamy. 2022. "Bacterial Isolates Derived from Nest Soil Affect the Attraction and Digging Behavior of Workers of the Red Imported Fire Ant, Solenopsis invicta Buren" Insects 13, no. 5: 444. https://doi.org/10.3390/insects13050444
APA StyleTravanty, N. V., Vargo, E. L., Schal, C., Apperson, C. S., & Ponnusamy, L. (2022). Bacterial Isolates Derived from Nest Soil Affect the Attraction and Digging Behavior of Workers of the Red Imported Fire Ant, Solenopsis invicta Buren. Insects, 13(5), 444. https://doi.org/10.3390/insects13050444







