Simple Summary
Eriophyoid mites (Eriophyoidea) are strictly phytophagous, consisting of over 5000 extant species, in which many species are pests. The genus Calvittacus includes only four species and is endemic in the Oriental Region. In this study, we combined morphological characters and molecular approaches to delimitate the Calvittacus species and recovered one new species, Calvittacus spectabilus sp. nov. The new species is vagrant on lower leaf surface, causing no apparent symptom to the host plant.
Abstract
Eriophyoid mites (Eriophyoidea) are distributed worldwide and are the largest superfamily in the Acari. After over one and a half centuries of field surveys, regional fauna of eriophyoid mites remains unclear. The genus Calvittacus Xue, Song & Hong 2006 is endemic in the Oriental Region, including four species—C. chenius Xue, Wang, Song & Hong, 2009; C. mollissimus Han, Xue & Hong, 2017; C. regiae Xue, Song & Hong 2006; and C. swidanus Song, Xue & Hong, 2009. In this study, we describe one new species, Calvittacus spectabilus sp. nov., collected on Bougainvillea spectabilis (Nyctaginaceae) from China (the Oriental Region). Phylogenetic analysis based on mitochondrial COI barcode sequences confirmed the C. spectabilus sp. nov., coinciding with the morphological delimitation. We further discussed the potential distribution of the Calvittacus species and underlined the integrative approaches in eriophyoid mite delimitation.
1. Introduction
Eriophyoid mites (Eriophyoidea) comprise over 5000 described species [1], leading to the largest superfamily in the Acari. They are strictly phytophagous and can cause massive economic losses, e.g., Aceria tosichella Keifer and A. guerreronis Keifer [2]. Although eriophyoid mites have a worldwide distribution [3], they are supposed to be distributed mainly in temperate regions [4]. After over one and a half centuries of field surveys of eriophyoid mites, new species continue to be described in recent years. For instance, given more than 1200 eriophyoid mites described in China, an average of 12 new species per year were added in the last three years [5]. Furthermore, the Chinese fauna of eriophyoid mites was suggested, including over 2300 species [6]. It is likely that 1000 species could be recovered in China in future studies.
The genus Calvittacus was established by Xue et al. [7], based on the type species C. regiae Xue, Song & Hong 2006. It is characterized by broad dorsal annuli, forming thickened bands and a furrow; scapular setae ahead of the rear shield margin, in a centrad direction; all coxal setae present; and legs and opisthosoma with the usual setae [7]. To date, only four species have been reported in this genus; all of them infest angiosperms from China.
To understand the diversity of eriophyoid mites in China, especially the Calvittacus species, Xiao-Feng Xue and colleagues conducted a long-term field survey since 2002. In this study, we describe and illustrate Calvittacus spectabilus sp. nov. on Bougainvillea spectabilis (Nyctaginaceae), which was collected from south China. We further discuss the potential distribution of the Calvittacus species and underline the integrative approaches in describing new eriophyoid mite species.
2. Materials and Methods
2.1. Taxa Sampling and Morphological Identification
Samples were collected from Bougainvillea spectabilis (Nyctaginaceae) in the field (Figure 1), using a hand lens (30×), in China. Mite samples were stored in 95–96% ethanol at −20 °C prior to DNA extraction. Mite specimens were also slide-mounted using Keifer’s Booster and a modified Berlese medium [8], but without adding additional fibers as was suggested by de Lillo et al. [9]. The morphological terminology used herein follows Lindquist [10] and Amrine et al. [11], internal female genitalia nomenclature follows Chetverikov [12], and the generic classification is made according to Amrine et al. [11], in combination with descriptions of all the published genera after 2003. Specimens were measured following de Lillo et al. [9]. They were examined with the aid of a Zeiss A2 (Germany) research microscope with phase contrast, and semi-schematic drawings were made. Microphotographs were taken with a Zeiss A2 (microphoto camera AxioCam MRc) research microscope with phase contrast or differential interference, using 10× eyepieces at 100× oil magnification, connected to a computer using Axiovision image analysis software. For each species, the holotype female measurements precede the corresponding range for paratypes (given in parentheses). For males, only ranges are given. If no variation was observed among measurements, it will be indicated with an “*”. All measurements are in micrometres (μm) and represent lengths, when not otherwise specified. The holotype and seven paratypes are deposited in the Arthropod/Mite Collection of the Department of Entomology, Nanjing Agricultural University (NJAU), Jiangsu Province, China [13].

Figure 1.
Host plant and damage symptom. (A) host plant (Bougainvillea spectabilis); (B) damage symptom.
2.2. DNA Extraction and Sequencing
In total, 14 individuals collected from three locations (Baxianshan, Fujian; Wanlu, Hainan; Haikou, Hainan) were selected for molecular analysis. Genomic DNA was extracted using a DNeasy Blood and Tissue Kit (Qiagen) following a modified protocol [14]. PCR was used to amplify the 658 bp barcode region of the mitochondrial COI gene using the primer pairs bcdF01/bcdR04 [14]. PCR reaction, purification, and sequencing followed Yin et al. [6].
2.3. Species Delimitation
Fourteen COI sequences were aligned and trimmed in Geneious 8.1.9 [15]. All sequences were blasted in GenBank and checked for possible contaminants. All the sequences were deposited in GenBank under accession numbers: MZ482549-MZ482557, OM892490-OM892494. In addition, collection data, taxonomic information, sequences, and trace files were submitted to BOLD [16] under number: AEC8688. We constructed a Neighbor-Joining tree that employed the K2P distance metric and used the Taxon ID tree tool on BOLD. Genetic distances were calculated with MEGA 6.0 [17] employing the Kimura 2-parameter (K2P) distance parameter [18].
2.4. Phylogenetic Analysis
The Eriophyoidea includes three families, i.e., Phytoptidae, Eriophyidae, and Diptilomiopidae [11]. To test the phylogenetic position of Calvittacus spectabilus sp. nov. within Eriophyoidea, we constructed an additional data matrix including five species of Phytoptidae (i.e., Trisetacus ehmanni, Trisetacus thujivagrans, Trisetacus juniperinus, Boczekella fabris, Setoptus koraiensis), nine species of Eriophyidae (i.e., Aculus ichnocarpi, Phyllocoptes taishanensis, Surapoda tianlinensis, Aculus populi, Leipothrix sabinae, Tetra zhouzhis, Tegolophus ulmi, Abacarus floridulus, Epitrimerus sabinae), and seven species of Diptilomiopidae (i.e., Apodiptacus rubi, Diptilomiopus nobilus, D. bischofiae, Quadracus cudraniae, Rhyncaphytoptus mori, R. celtis, Trimeroptes luanchuanensis). Osperalycus tenerphagus and Gordialycus sp. from the family Nematalycidae were used to root the tree because they were suggested to be the sister group of Eriophyoidea [19,20]. All COI sequences were aligned using MAFFT v7.2 [21]. The best-fit DNA sequence evolution model for our data was selected using the Bayesian information criterion (BIC) in jModelTest ver. 2.1.1 [22] with the GTR+G+I model selected for COI. Phylogenetic analyses were conducted using maximum likelihood (ML) and Bayesian inference (BI). ML analyses were performed using nucleotide sequences in the RAxML-HPC2 on XSEDE (3.2.3) [23] implemented in CIPRES Science Gateway V. 3.3 [24], using nonparametric bootstrap with 1000 replicates for node support. BI analyses were computed with MrBayes 3.2.6 [25], using two separate data partitions for codons (1st+2nd, 3rd). Two parallel runs of four independent chains were conducted for 10 × 107 generations with samples every 1000 generations. The first 25% of the samples were discarded as burn-in.
3. Results
3.1. Taxonomy
- Family Eriophyidae Nalepa
- Subfamily Phyllocoptinae Nalepa
- Tribe Phyllocoptini Nalepa
- Genus Calvittacus Xue, Song & Hong
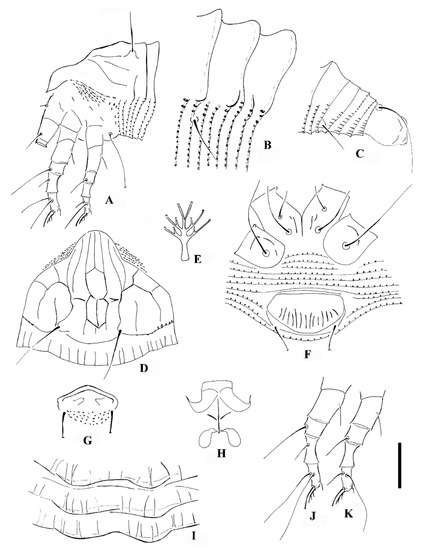
Figure 2.
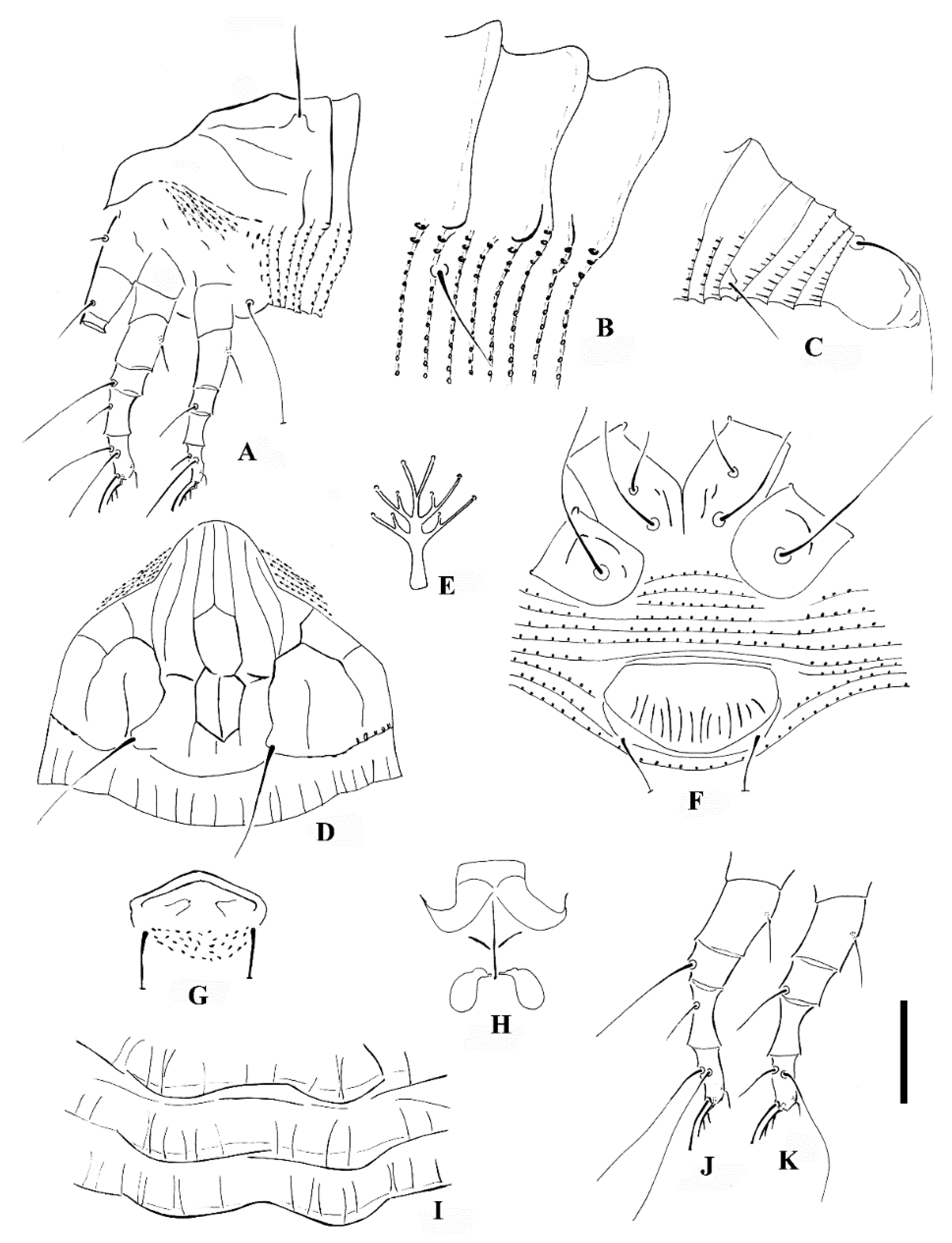
Calvittacus spectabilus sp. nov. (A) lateral view of anterior part of body; (B) lateral microtubercles; (C) lateral view of telosoma; (D) prodorsal shield; (E) empodium; (F) female coxigenital area; (G) male external genitalia; (H) female internal genitalia; (I) dorsal annuli; (J) leg I; (K) leg II. Scale bar: 18 μm for (A,C); 15 μm for (D,F–H,J,K); 4 μm for (B,E,I).
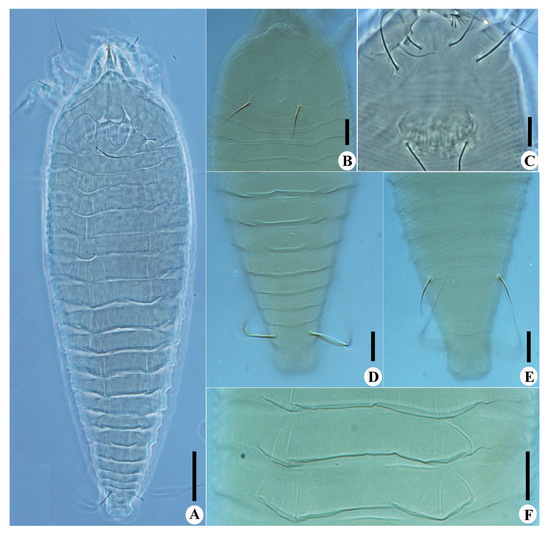
Figure 3.
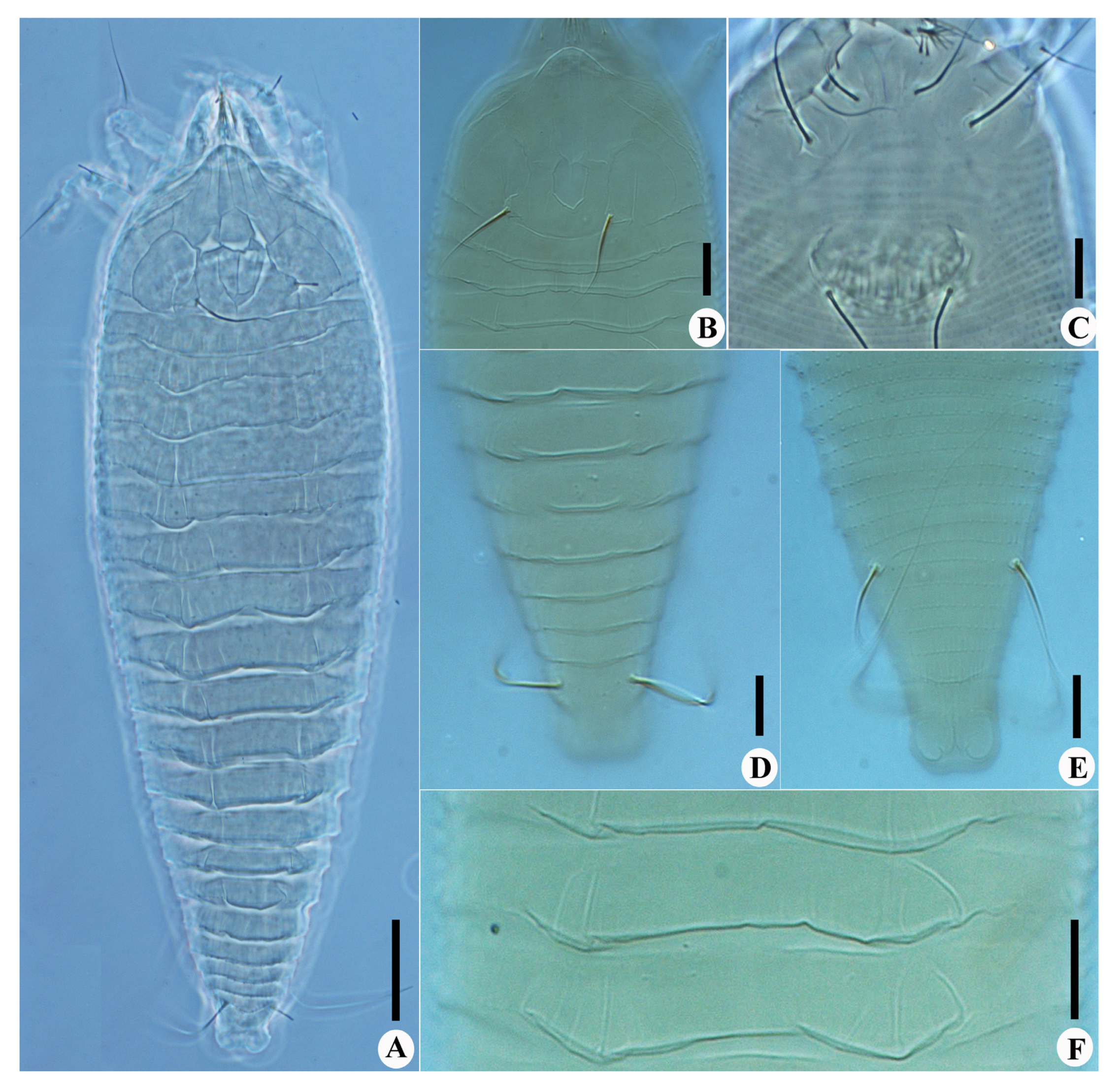
Calvittacus spectabilus sp. nov. (A) dorsal view; (B) prodorsal shield; (C) female coxigenital area; (D) dorsal view of telosoma; (E) ventral view of telosoma; (F) dorsal annuli. Scale bar: 20 μm for A; 10 μm for (B–F).
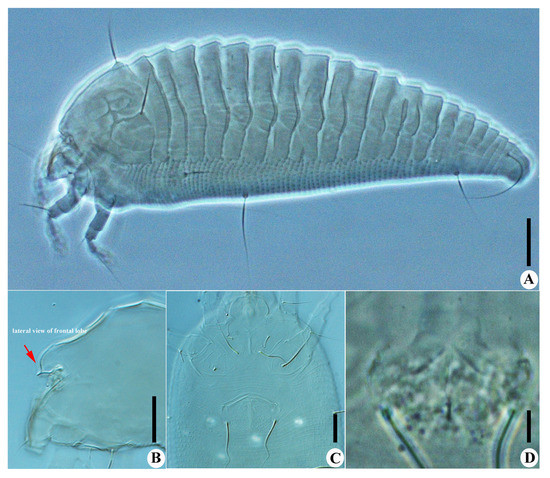
Figure 4.
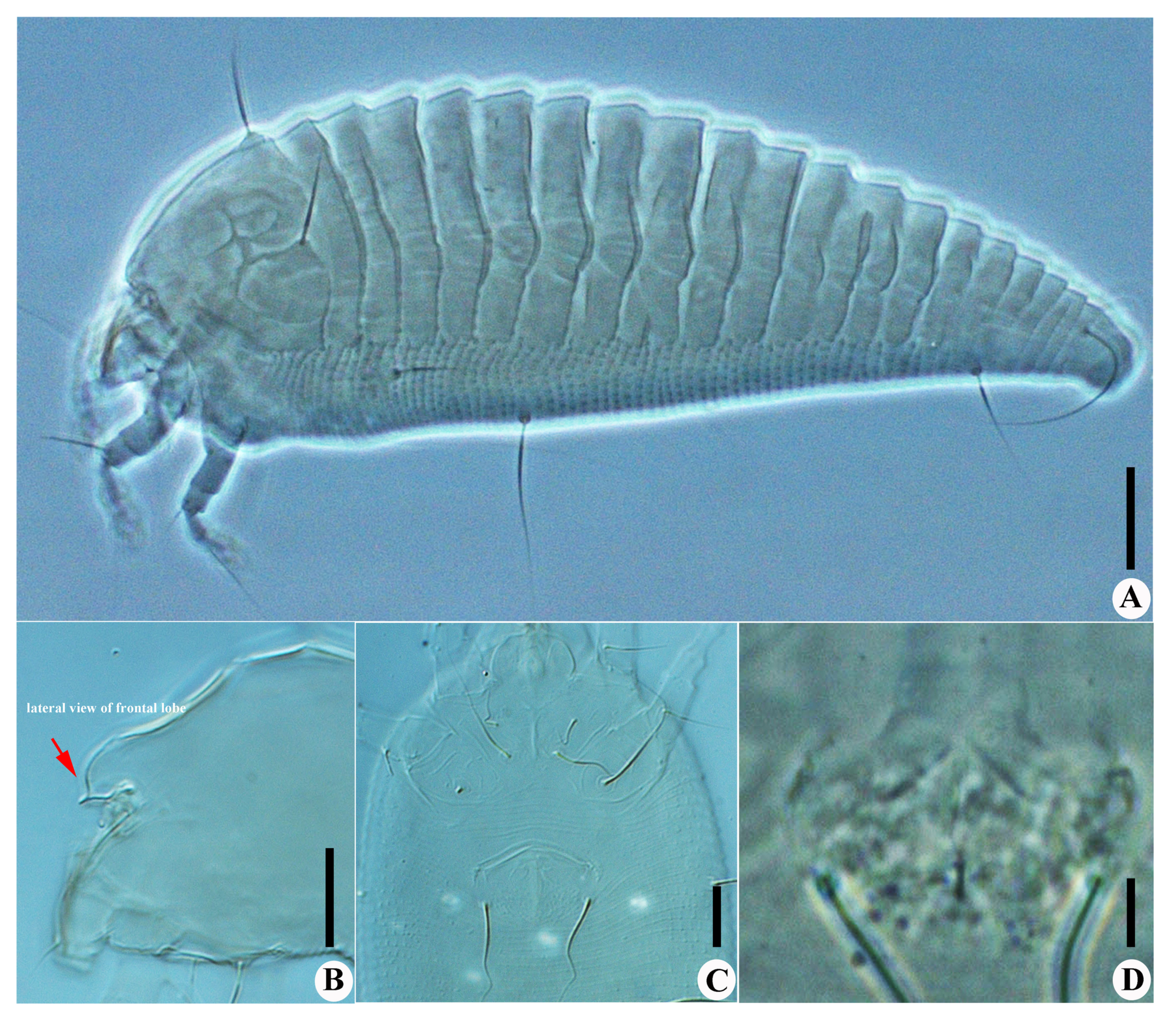
Calvittacus spectabilus sp. nov. (A) lateral view; (B) lateral view of gnathosoma; (C) male coxigenital area; (D) female internal genitalia. Scale bar: 20 μm for A; 10 μm for (B,C); 5 μm for (D).
Description. Female (n = 29): Body fusiform, 191 (182–205), 64 (58–65) wide, 65 (63–67) thick; light yellow in colour. Gnathosoma 16 (16–17), projecting downwards, cheliceral stylets 14 (14–15), pedipalp coxal seta (ep) 3 (2–3), dorsal pedipalp genual setae (d) 8 (7–8), palp tarsal ventral setae (v) absent. Prodorsal shield 41 (37–45), including the frontal lobe, 56 (52–56) wide, frontal lobe broad (Figure 2D and Figure 3B), with a spine in lateral view (Figure 4B); median line discontinuous, present at anterior 1/3 and posterior 1/3, admedian lines connected with median line by short transverse lines, forming a large cell at center and two small cells at posterior, submedian lines complete. Scapular tubercles ahead of rear shield margin, setae sc 22 (21–22), 21 (20–21) apart, projecting centered. Coxigenital region with 6* semiannuli between coxae and genitalia, smooth; coxal plates with granules, anterolateral setae on coxisternum I (1b) 7 (7–8), 13 (13–14) apart; proximal setae on coxisternum I (1a) 15 (15–18), 9 (8–9) apart; proximal setae on coxisternum II (2a) 32 (32–38), 24 (22–24) apart. Prosternal apodeme present, 9*. Leg I 28 (28–30), femur 10 (9–10), basiventral femoral setae (bv) 10 (10–11); genu 4 (4–5), antaxial genual setae (l″) 21 (20–21); tibia 6*, paraxial tibial setae (l′) 5 (5–6), located at basal 1/3; tarsus 6*, paraxial fastigial tarsal setae ft′ 15 (15–17), antaxial fastigial tarsal setae ft″ 18 (18–19), setae u′ 4 (4–5); tarsal empodium (em) 6 (6–7), simple, 3-rayed, tarsal solenidion (ω) 7*, rod-like. Leg II 25 (25–27), femur 8 (8–10), basiventral femoral setae (bv) 10 (10–12); genu 4 (4–5), antaxial genual setae (l″) 8 (6–8); tibia 6*; tarsus 5*, paraxial fastigial tarsal setae ft′9 (8–9), antaxial fastigial tarsal setae ft″ 21 (19–21), setae u′ 4*; tarsal empodium (em) 6*, simple, 3-rayed, tarsal solenidion (ω) 7*, rod-like. Opisthosoma dorsally with 18 (18–19) semiannuli, with elliptical granules on ridges, with three ridges, middorsal ridge ended in a broad furrow; ventrally with 71 (69–71) semiannuli, with elliptical to linear microtubercles. Setae c2 16 (16–17), on ventral semiannulus 14 (14–15), 61 (59–61) apart; setae d 31 (29–31), on ventral semiannulus 26 (26–27), 40 (40–42) apart; setae e 14 (14–20), on ventral semiannulus 43 (41–43), 20 (19–20) apart; setae f 18 (18–24), 20 (20–21) apart, on 5th ventral semiannulus from rear; setae h1 absent, setae h2 50 (45–50). Female genitalia 13 (13–14), 25 (22–25) wide, coverflap with 12 to 14 ridges, setae 3a 40 (39–40), 18* apart. Internal genitalia: spermathecae ovoid, oriented posterolaterad; spermathecal tubes relatively short; transverse genital apodeme trapezoidal.
Male (n = 9): Body fusiform, 153–181, 56–58 wide; light yellow in colour. Gnathosoma 14–15, projecting downwards, cheliceral stylets 13*, pedipalp coxal seta (ep) 3*, dorsal pedipalp genual setae (d) 6*, palp tarsal ventral setae (v) absent. Prodorsal shield 42–45, including the frontal lobe, 50–52 wide. Scapular tubercles ahead of rear shield margin, setae sc 18–20, 18–20 apart, projecting centrad. Coxigenital region with 4 semiannuli between coxae and genitalia, smooth; coxal plates with short lines, anterolateral setae on coxisternum I (1b) 5–6, 12* apart; proximal setae on coxisternum I (1a) 11*, 8–9 apart; proximal setae on coxisternum II (2a) 22–25, 21–22 apart. Prosternal apodeme present, 8*. Leg I 26–28, femur 9*, basiventral femoral setae (bv) 10–13; genu 4–5, antaxial genual setae (l″) 15–20; tibia 6*, paraxial tibial setae (l′) 5*, located at basal 1/3; tarsus 5*, paraxial fastigial tarsal setae ft′ 15*, antaxial fastigial tarsal setae ft″ 17–19, setae u′ 4*; tarsal empodium (em) 5–6, simple, 3-rayed, tarsal solenidion (ω) 6–7, rod-like. Leg II 25–28, femur 8*, basiventral femoral setae (bv) 10–11; genu 5*, antaxial genual setae (l″) 6–7; tibia 5–6; tarsus 5*, paraxial fastigial tarsal setae ft′ 6–7, antaxial fastigial tarsal setae ft″ 16–19, setae u′ 4*; tarsal empodium (em) 5*, simple, 3-rayed, tarsal solenidion (ω) 6*, rod-like. Opisthosoma dorsally with 17–18 semiannuli, smooth, with three ridges, middorsal ridge ended in a broad furrow; ventrally with 65–70 semiannuli, with elliptical to linear microtubercles. Setae c2 15–17, on ventral semiannulus 14*, 52–53 apart; setae d 26–28, on ventral semiannulus 26*, 33* apart; setae e 14–15, on ventral semiannulus 41–42, 16–17 apart; setae f 18–20, 17* apart, on 5th ventral semiannulus from rear; setae h1 absent, setae h2 38–45. Male genitalia 11*, 19* wide, setae 3a 28–30, 15* apart.
Type material. Holotype, female (slide number NJAUFJ2.1; marked Holotype), found on Bougainvillea spectabilis Willd. (Nyctaginaceae), Baxianshan Park, Jinjiang, Quanzhou city, Fujian province, China, 24°30′48″ N, 118°24’37” E, elevation 251 m, 15 August 2015, coll. Yan Dong, deposited as slide-mounted specimen in the Arthropod/Mite Collection of the Department of Entomology, NJAU. Paratypes, five females on five slides and two males on two slides (slide number NJAUFJ2.2–NJAUFJ2.8; marked Paratypes), from Bougainvillea spectabilis Willd. (Nyctaginaceae), same details as holotype, deposited as slide-mounted specimens in the Arthropod/Mite Collection of the Department of Entomology, NJAU.
Other material. Eight females on Eight slides and three males on three slides (slide number NJAUQ314.1–NJAUQ314.11; marked Paratypes), from Bougainvillea spectabilis Willd. (Nyctaginaceae), Wanlu Park, Haikou city, Hainan province, China, 20°02’48” N, 110°18’47” E, elevation 10 m, 27 May 2019, coll. Yue Yin and Liang-Fei Yao, deposited as slide-mounted specimens in the Arthropod/Mite Collection of the Department of Entomology, NJAU; fifteen females on fifteen slides and four males on four slides (slide number NJAUQ330.1–NJAUQ330.19; marked Paratypes), from Bougainvillea spectabilis Willd. (Nyctaginaceae), Volcano Geological Park, Haikou city, Hainan province, China, 19°55’54” N, 110°13’06” E, elevation 140 m, 29 May 2019, coll. Yue Yin and Liang-Fei Yao.
Deposited as slide-mounted specimens in the Arthropod/Mite Collection of the Department of Entomology, NJAU.
GenBank accession numbers. MZ482551 (NJAUFJ2); MZ482550, MZ482553-MZ482555 (NJAUQ314); MZ482554, MZ482552, MZ482549, OM892490-OM892494 (NJAUQ330).
BOLD number. AEC8688.
Relation to the plant host. Vagrant on lower leaf surface. No apparent symptom to the host plant was observed. We suppose that the bronze in colour for the upper leaf surface of young leaves was normal for the host plant, while not induced by mites (Figure 1B).
Etymology. The specific designation spectabilus is derived from the species name of the host plant, spectabilis, changing postfix -is to -us; masculine in gender.
Differential diagnosis. The new species is morphologically similar to four Calvittacus species (Table 1), but can be differentiated by median, admedian and submedian lines present on the prodorsal shield (simple prodorsal shield design with few lines in the other four Calvittacus species), 18 dorsal annuli (11 to 13 annuli in C. chenius, C. mollissimus, and C. regiae; 23 annuli in C. swidanus), and empodium 3-rayed (empodium 5-rayed in the other four species).

Table 1.
List of Calvittacus species.
Key to species of Calvittacus
1. Coxal plates smooth………………………………………………………………………2
-. Coxal plates with granules and short lines……………………………………………..3
2. Dorsal opisthosoma with 11 annuli, the first 7 annuli form large bounds……………
……………………………………………….. C. chenius Xue, Wang, Song & Hong, 2009
-. Dorsal opisthosoma with 23 annuli, the first 19 annuli form large bounds…………
………………………………………………………C. swidanus Song, Xue & Hong, 2009
3. Dorsal opisthosoma with 11 to 13 annuli, median or admedian lines present………4
-. Dorsal opisthosoma with 18 annuli, median, admedian and submedian lines present
……………………………………………………………………..C. spectabilus sp. nov.
4. Ventral annuli with round microtubercles, dorsal annuli smooth……………………
…………………………………………………… C. mollissimus Han, Xue & Hong, 2017
-. Ventral annuli with spiny microtubercles, dorsal annuli with filamentous micro-
tubercles………………………………………………C. regiae Xue, Song & Hong 2006.
3.2. Genetic Distance and Phylogenetic Analysis
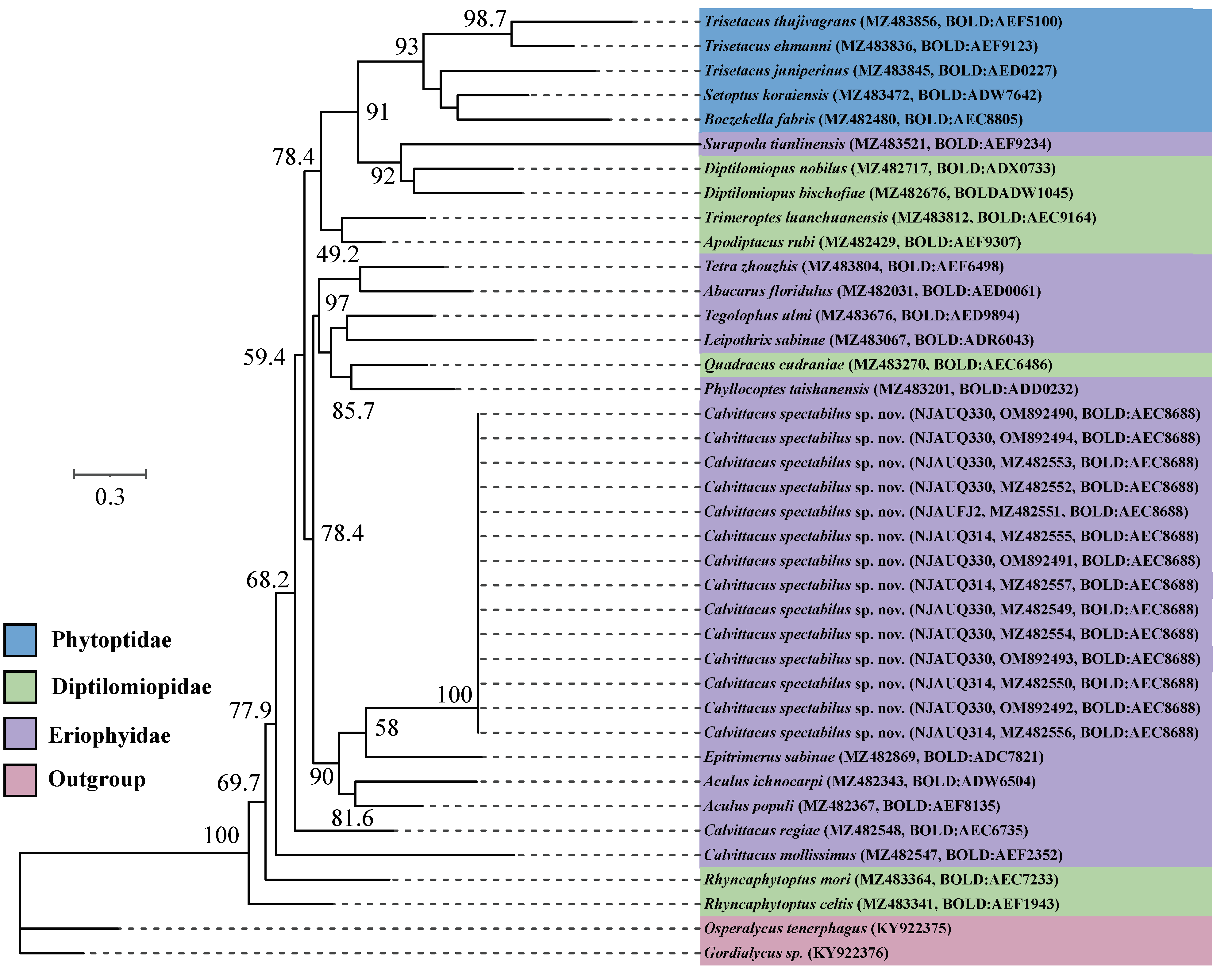
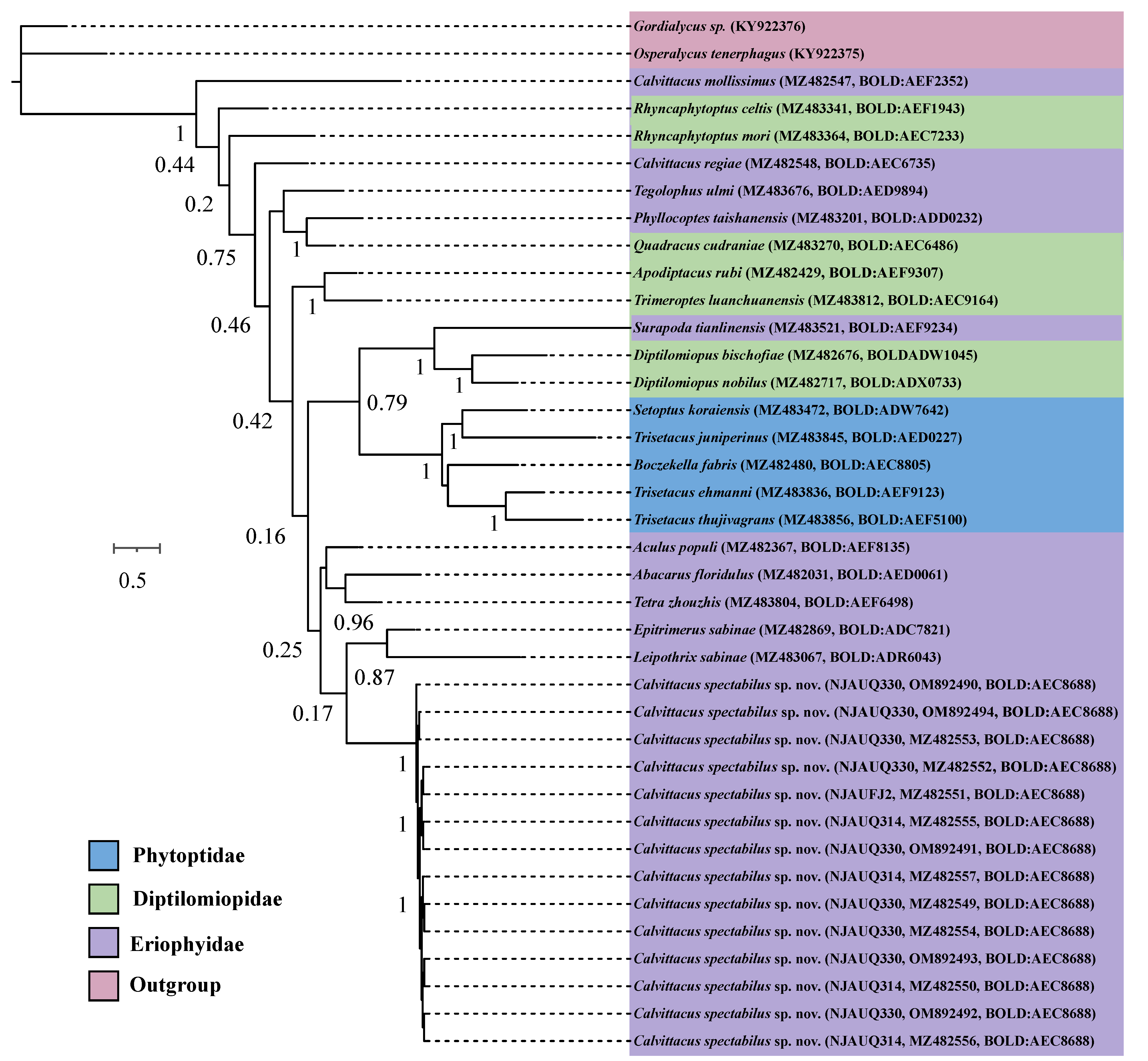
Fourteen COI sequences of C. spectabilus sp. nov. from three populations were nearly identical in composition, with an intraspecific distance (K2P) ranging from 0.000 to 0.001. We tried to sequence the COI sequences of C. chenius and C. swidanus, but failed, possibly due to DNA degradation of samples, which were collected for over 10 years and were kept in 75% ethanol at room temperature. We therefore included three Calvittacus species in the phylogenetic analyses.
The ML and BI analyses showed that C. spectabilus sp. nov. was nested within the Eriophyidae (Figure 5 and Figure 6), which is consistent with the morphological assignment.
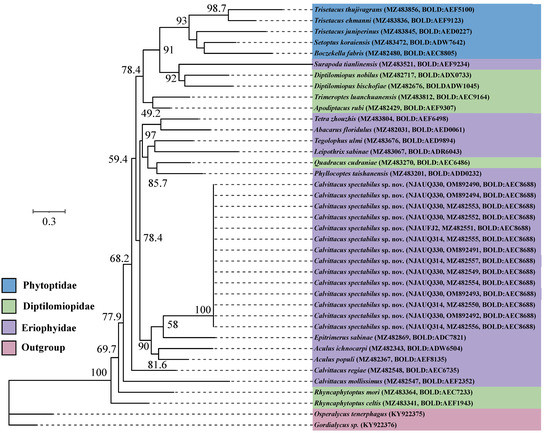
Figure 5.
Maximum likelihood (ML) tree for Calvittacus species based on mitochondrial COI barcode nucleotide sequences.
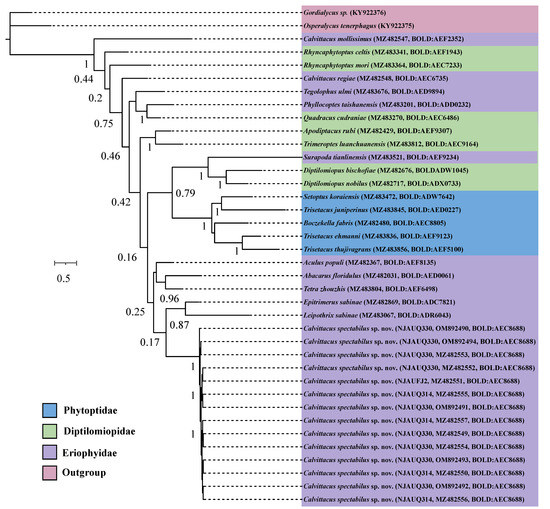
Figure 6.
Bayesian inference (BI) tree for Calvittacus species based on mitochondrial COI barcode nucleotide sequences by codons (1st+2nd, 3rd).
4. Discussion
In this study, we described and illustrated one new eriophyoid mite species belonging to the genus Calvittacus, based on the distinct morphological characters of dorsal annuli with thickened bands, forming a furrow [7]. Eriophyoid mites are characterized by ringed opisthosoma, in which dorsal annuli vary in number (ranged from 10 to 80) and shape (forming ridges, furrows, or evenly rounded) [11]. However, dorsal annuli formed by thickened bands were occasionally found in genus Achaetocoptes [29], Calvittacus [7], Johnella [30], Neovittacus [26], and Vittacus [31]; all these genera hold few species (less than ca. 10). Furthermore, this morphological character has not been phylogenetically tested at the generic level to determine whether it is a synapomorphy. Although four species have been described in the Calvittacus, all generic assignments were based on morphological characters. Our phylogenetic results showed a non-monophyly of Calvittacus; however, this result was based on a fragment of mitochondrial COI gene sequences. Multiple genes from mitochondria and nuclear, especially genome sequences, should be determined in future analyses to account for the position of C. spectabilus sp. nov. and to unveil the monophyly of Calvittacus.
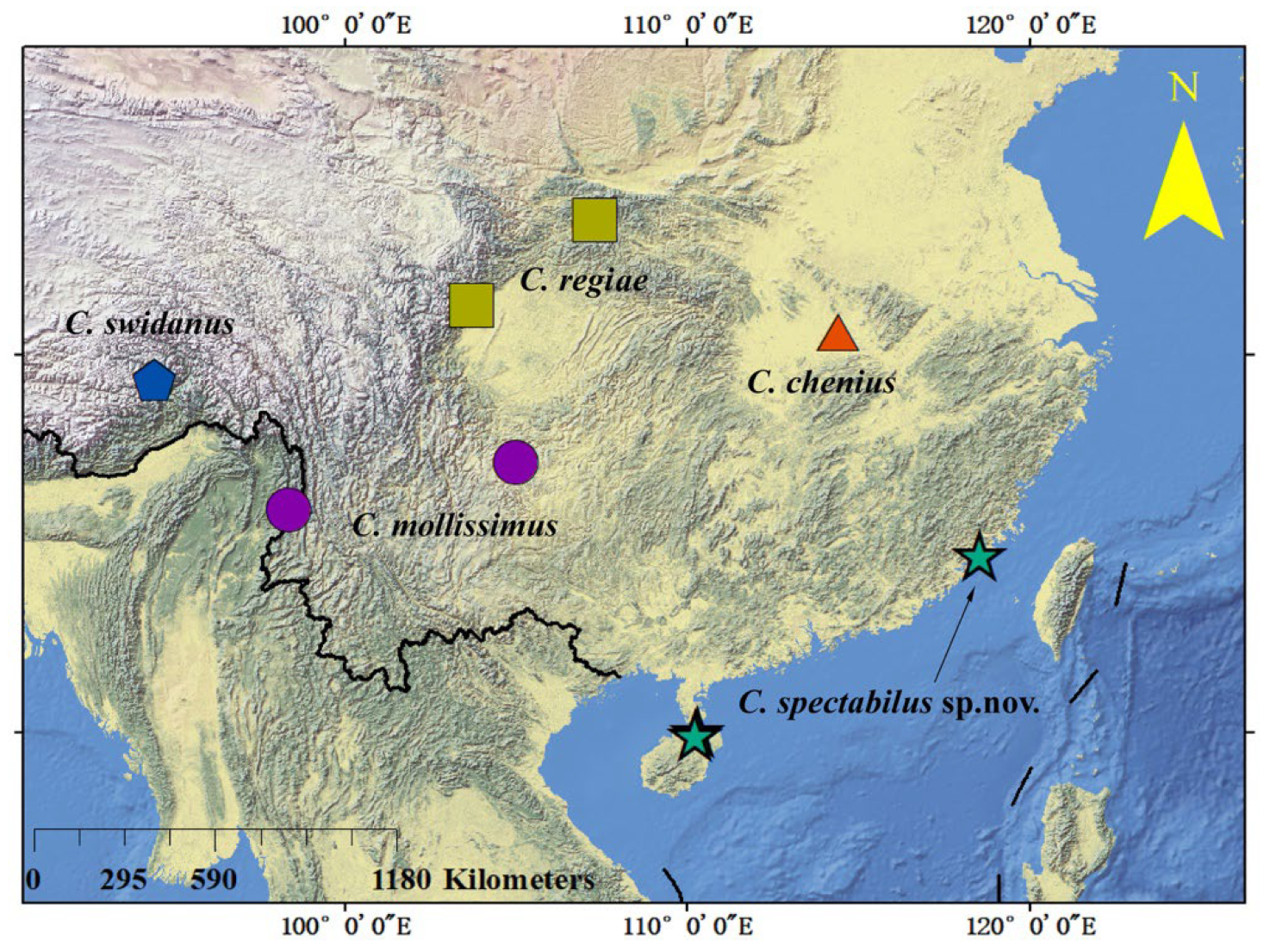
The genus Calvittacus was established by Xue et al. [7] based on the type species C. regiae Xue, Song & Hong 2006, which was collected from the Oriental Region (Figure 7). After that, three more species, collected from the Oriental Region, were assigned to this genus (Figure 7). Herein, the fifth species, C. spectabilus sp. nov., were also collected from the Oriental Region. Under these contexts, it is likely that the genus Calvittacus was endemic in the Oriental Region. However, more field surveys are warranted in future studies to understand the origin and biogeographical distribution of the Calvittacus species.
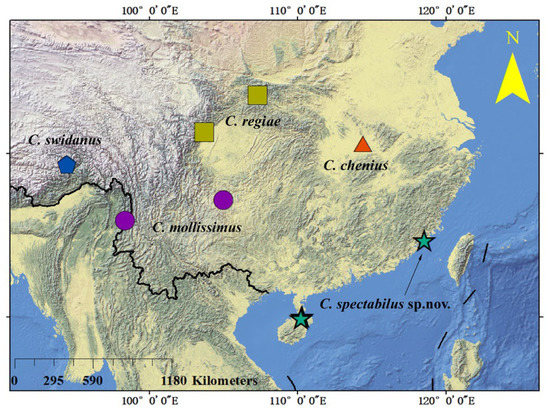
Figure 7.
The distribution of Calvittacus species.
COI barcodes were recently explored in eriophyoid mite delimitation, showing a clear gap between interspecific divergence and intraspecific divergence, and thus enabling a discrimination of 99% of the eriophyoid mite species [6]. We provided 14 COI barcode sequences of C. spectabilus sp. nov., which were collected from three locations. Sequence analysis showed no intraspecific divergence. After comparing with the eriophyoid mite sequences in the Barcode of Life Data Systems database (DS-ERIYYDNA Barcodes for Eriophyoidea, available at doi: dx.doi.org/10.5883/DS-ERIYY.), all 14 sequences were assigned into one Barcode Index Number (BOLD: AEC8688), which reflects a single species; this molecular delimitation is in line with our morphological studies, confirming a new eriophyoid mite species of C. spectabilus sp. nov.
Simply relying upon few morphological characters to delimitate eriophyoid mites has its limitations. Inaccurate species delimitation could result from (1) species complex (e.g., Abacarus hystrix complex [6,32]), (2) cryptic diversity (e.g., Diptilomiopus species [33]), and (3) two morphological forms (protogyne and deutogyne [34]). We therefore highlight the integrative methods that combined molecular sequences (even genome sequences) and morphological characters in the description of new eriophyoid mite species.
Author Contributions
Y.Y. and X.-F.X. conceived and designed the study. Y.Y., Y.-W.L. and X.-Y.L. performed the research. Y.Y., Y.-W.L., X.-Y.L. and X.-F.X. participated in writing, reviewing, and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32161143014).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All data is available in this paper. All sequences were deposited in the GenBank under accession numbers of MZ482549-MZ482557, OM892490-OM892494.
Acknowledgments
We would like to thank Liangfei Yao (Nanjing Agricultural University, China) for his help with collecting mite specimens.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, Z.Q. Eriophyoidea and allies: Where do they belong? Syst. Appl. Acarol. 2017, 22, 1091–1095. [Google Scholar] [CrossRef]
- de Lillo, E.; Pozzebon, A.; Valenzano, D.; Duso, C. An intimate relationship between eriophyoid mites and their host plants. Front. Plant Sci. 2018, 9, 1786. [Google Scholar] [CrossRef] [PubMed]
- Amrine, J.W., Jr.; Stasny, T.A. Catalog of the Eriophyoidea (Acarina: Prostigmata) of the World; Indira Publishing House: West Bloomfield, MI, USA, 1994; pp. ix, 798. [Google Scholar]
- de Lillo, E.; Skoracka, A. What’s “cool” on eriophyoid mites? Exp. Appl. Acarol. 2010, 51, 3–30. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.F.; Lu, Y.W. Two new species of eriophyoid mites from China (Acari: Eriophyoidea). Syst. Appl. Acarol. 2022, 27, 660–669. [Google Scholar] [CrossRef]
- Yin, Y.; Yao, L.F.; Hu, Y.; Shao, Z.K.; Hong, X.Y.; Hebert, P.D.N.; Xue, X.F. DNA barcoding uncovers cryptic diversity in minute herbivorous mites (Acari, Eriophyoidea). Mol. Ecol. Resour. 2022. [Google Scholar] [CrossRef]
- Xue, X.F.; Song, Z.W.; Hong, X.Y. A new genus and three new species of Phyllocoptini (Acari: Eriophyidae: Phyllocoptinae) from the Qinling Mountains, Shaanxi Province, northwestern China. Zootaxa 2006, 1275, 31–41. [Google Scholar] [CrossRef]
- Amrine, J.W., Jr.; Manson, D.C.M. Preparation, mounting and descriptive study of eriophyoid mites. In World Crop Pests, Eriophyoid Mites: Their Biology, Natural Enemies and Control; Lindquist, E.E., Sabelis, M.W., Bruin, J., Eds.; Elsevier: Amsterdam, The Netherlands, 1996; Volume 6, pp. 383–396. [Google Scholar] [CrossRef]
- de Lillo, E.; Craemer, C.; Amrine, J.W., Jr.; Nuzzaci, E.G. Recommended procedures and techniques for morphological studies of Eriophyoidea (Acari: Prostigmata). Exp. Appl. Acarol. 2010, 51, 283–307. [Google Scholar] [CrossRef]
- Lindquist, E.E. External anatomy and notation of structures. In World Crop Pests, Eriophyoid Mites: Their Biology, Natural Enemies and Control; Lindquist, E.E., Sabelis, M.W., Bruin, J., Eds.; Elsevier: Amsterdam, The Netherlands, 1996; Volume 6, pp. 3–31. [Google Scholar] [CrossRef]
- Amrine, J.W., Jr.; Stasny, T.A.H.; Flechtmann, C.H.W. Revised Keys to World Genera of Eriophyoidea (Acari: Prostigmata); Indira Publishing House: West Bloomfield, MI, USA, 2003; pp. iv, 244. [Google Scholar]
- Chetverikov, P.E. Comparative confocal microscopy of internal genitalia of phytoptine mites (Eriophyoidea, Phytoptidae): New generic diagnoses reflecting host-plant associations. Exp. Appl. Acarol. 2014, 62, 129–160. [Google Scholar] [CrossRef]
- Zhang, Z.Q. Repositories for mite and tick specimens: Acronyms and their nomenclature. Syst. Appl. Acarol. 2018, 23, 2432–2466. [Google Scholar] [CrossRef]
- Dabert, J.; Ehrnsberger, R.; Dabert, M. Glaucalges tytonis sp. n. (Analgoidea, Xolalgidae) from the barn owl Tyto alba (Strigiformes, Tytonidae): Compiling morphology with DNA barcode data for taxon descriptions in mites (Acari). Zootaxa 2008, 1719, 41–52. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Bolton, S.J.; Chetverikov, P.E.; Klompen, H. Morphological support for a clade comprising two vermiform mite lineages: Eriophyoidea (Acariformes) and Nematalycidae (Acariformes). Syst. Appl. Acarol. 2017, 22, 1096–1131. [Google Scholar] [CrossRef]
- Klimov, P.B.; Oconnor, B.M.; Chetverikov, P.E.; Bolton, S.J.; Pepato, A.R.; Mortazavi, A.L.; Tolstikov, A.V.; Bauchan, G.R.; Ochoa, R. Comprehensive phylogeny of acariform mites (Acariformes) provides insights on the origin of the four-legged mites (Eriophyoidea), a long branch. Mol. Phylogenetetics Evol. 2018, 119, 105–117. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Xue, X.F.; Wang, Z.; Song, Z.W.; Hong, X.Y. Eriophyoid mites on Fagaceae with descriptions of seven new genera and eleven new species (Acari: Eriophyoidea). Zootaxa 2009, 22, 1–95. [Google Scholar] [CrossRef]
- Han, X.; Xue, X.F.; Hong, X.Y. Eriophyoid mites from Hengduan Mountains, southwestern China with descriptions of nine new species (Acari, Eriophyoidea). Syst. Appl. Acarol. 2017, 22, 1132–1166. [Google Scholar] [CrossRef][Green Version]
- Song, Z.W.; Xue, X.F.; Hong, X.Y. Seven new species of Phyllocoptini from Tibet Autonomous Region, China (Acari: Eriophyidae: Phyllocoptinae). Zootaxa 2009, 2291, 1–23. [Google Scholar] [CrossRef]
- Farkas, H.K. Uber die Eriophyiden (Acarina) Ungarns II. Acta Zool. Acad. Sci. Hung. 1961, 7, 73–76. [Google Scholar]
- Keifer, H.H. New eriophyid mites. Ann. Entomol. Soc. Am. 1959, 52, 649–657. [Google Scholar] [CrossRef]
- Keifer, H.H. Eriophyid Studies C-2; U.S. Department of Agriculture, Agricultural Research Service: Washington, DC, USA, 1969; p. 24.
- Laska, A.; Majer, A.; Szydlo, W.; Karpicka-Ignatowska, K.; Hornyak, M.; Labrzycka, A.; Skoracka, A. Cryptic diversity within grass-associated Abacarus species complex (Acariformes: Eriophyidae), with the description of a new species, Abacarus plumiger n. sp. Exp. Appl. Acarol. 2018, 76, 1–28. [Google Scholar] [CrossRef]
- Liu, Q.; Yuan, Y.M.; Lai, Y.; Wang, G.Q.; Xue, X.F. Unravelling the phylogeny, cryptic diversity and morphological evolution of Diptilomiopus mites (Acari: Eriophyoidea). Exp. Appl. Acarol. 2019, 79, 323–344. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yao, L.-F.; Zhang, Q.; Hebert, P.D.N.; Xue, X.-F. Using multiple lines of evidence to delimit protogynes and deutogynes of four-legged mites: A case study on Epitrimerus sabinae s.l. (Acari: Eriophyidae). Invertebr. Syst. 2020, 34, 757–768. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).