Simple Summary
Roughly two-thirds of the insect species described from Germany belong to the orders Diptera (flies) or Hymenoptera (wasps, bees, ants and sawflies). However, both orders contain several species-rich families that have received little taxonomic attention until now. This study takes the first step in assessing these “dark taxa” families and provides species estimates for four challenging groups of Diptera (Cecidomyiidae, Chironomidae, Phoridae and Sciaridae). The estimates given in this paper are based on the sequencing results of over 48,000 fly specimens that have been collected in southern Germany via Malaise traps that were operated for one season each. We evaluated the fraction of species in our samples belonging to well-known fly families in order to estimate the species richness of the challenging “dark taxa” (DT families hereafter). Our results suggest a surprisingly high proportion of undetected biodiversity in a supposedly well-investigated country: at least 1800–2200 species await discovery and description in Germany in these four families.
Abstract
Determining the size of the German insect fauna requires better knowledge of several megadiverse families of Diptera and Hymenoptera that are taxonomically challenging. This study takes the first step in assessing these “dark taxa” families and provides species estimates for four challenging groups of Diptera (Cecidomyiidae, Chironomidae, Phoridae, and Sciaridae). These estimates are based on more than 48,000 DNA barcodes (COI) from Diptera collected by Malaise traps that were deployed in southern Germany. We assessed the fraction of German species belonging to 11 fly families with well-studied taxonomy in these samples. The resultant ratios were then used to estimate the species richness of the four “dark taxa” families (DT families hereafter). Our results suggest a surprisingly high proportion of undetected biodiversity in a supposedly well-investigated country: at least 1800–2200 species await discovery in Germany in these four families. As this estimate is based on collections from one region of Germany, the species count will likely increase with expanded geographic sampling.
1. Introduction
Although the Central European insect fauna is considered to be well studied, gaps in knowledge of its taxonomy and biodiversity remain [1]. About 33,300 species of insects are documented from Germany, of which roughly two-thirds of these taxa belong to one of the two orders: Diptera (flies) and Hymenoptera (wasps, bees, ants, and sawflies) [1,2,3,4,5,6,7,8]. However, both orders contain several species-rich families which have received less attention than others in Germany’s long history of taxonomic research [1]. This reflects the confluence of several factors, such as extreme species richness combined with a high rate of cryptic diversity and, most importantly, the limited taxonomic attention directed to small specimens (<2 mm) whose morphological characteristics are difficult to evaluate. Successful identification of species in these groups using morphology is time-consuming and requires taxonomic expertise, the availability of which is decreasing [9,10,11,12,13,14]. This imbalance of few researchers but high species numbers still awaiting documentation is commonly referred to as the taxonomic impediment [9,15,16]. Against the backdrop of a worldwide decline in insect abundance, the taxonomic impediment is an alarming constraint to biodiversity surveys [17,18,19,20,21]. One such constraint is noticeable in the framework of DNA barcoding applications, where species proxies (Barcode Index Numbers, BINs) often lack a linkage to a known species [22]. Page [22] coined the term “dark taxa” for these nameless BINs, and in 2020, Hausmann et al. [1] used it to address species-rich, taxonomically challenging groups of insect families whose diversity remains mostly undescribed. These include certain families of non-brachyceran Diptera (mosquitoes, gnats, midges), some families of Brachycera (flies), and nearly all families of parasitoid Hymenoptera (wasps) which often make up the majority of the insect biodiversity present in environmental and bulk samples [23]. With the shortage of taxonomic specialists, the functional role of “dark taxa” in ecosystems is far too understudied, meaning that they cannot be included in biomonitoring or conservation surveys.
The most recent project in the German Barcode of Life initiative, GBOL III: Dark Taxa, was launched in mid-2020 to tackle these challenging groups. Its two main goals are: (1) to study various DT families using an integrative taxonomic approach which combines morphological and sequence data [1,24], and (2) to expand the DNA barcode reference library established by three earlier initiatives (Barcoding Fauna Bavarica, GBOL I, GBOL II) [24,25,26]. Work conducted by GBOL II generated a reference library for the order Diptera based on 50,963 COI sequences, data that provided barcodes for 5200 BINs [13]. A recent commentary on this study presented a classical dipterist’s perspective on the situation for the better-known families of Diptera [27]. It explored ways to extend the involvement of expert taxonomists in assigning Linnean names to BINs. However, the challenge in implementing similar work on DT families was not addressed, highlighting the need to seek new approaches so these taxa can finally become more accessible to research.
This study begins this effort by considering the German fauna of four DT families of Diptera which lack estimates of their species numbers: Cecidomyiidae (gall midges), Chironomidae (non-biting midges), Phoridae (scuttle flies), and Sciaridae (dark-winged fungus gnats) (Figure 1). To address this goal, we examine the diversity of these DT families in our Malaise trap collections. We employ BIN data resulting from the sequence analysis of samples from southern Germany and use these results to estimate the extent of undocumented biodiversity in these families in Bavaria and Germany. An important backbone to our calculations is species numbers inferred from essential contributions of Germany’s over 200-year-long history of taxonomy [5,6,7,8,28,29,30,31,32,33,34,35,36,37,38].

Figure 1.
Selected representatives of the DT families analyzed in our study: Cecidomyiidae (top left); Phoridae (top right); Sciaridae (bottom left) and Chironomidae (bottom right). Scale bars represent 1 mm.
2. Materials and Methods
2.1. Malaise Tap Sites
In 2012, the Global Malaise Trap Program was launched by the Centre for Biodiversity Genomics (CBG) at the University of Guelph to provide a global overview of arthropod diversity [39]. As part of this project, 14 Malaise traps were deployed at various sites in Germany (Figure 2 and Table 1). In 2012, one trap was operated from May to September in the Bavarian Forest National Park (BFNP), a conifer-dominated montane forest. In 2014, 12 Malaise traps were placed along an altitudinal transect (1036–2160 m) in the Allgäu Alps, ranging from the Oytal to the Schochen and Nebelhorn Mountains. Traps in lower altitudes (Oytal) were deployed in May, whereas those in higher altitudes (Schochen and Koblat) were deployed in June. All traps in the Allgäu Alps were operated until October. Finally, in 2017, one trap was deployed at the Bavarian State Collection of Zoology (ZSM) in Munich, which is situated in a residential neighborhood rich in backyard gardens. This trap was operated from April to December. Altogether, the sampled sites represent a heterogeneous array of habitats typical of southern Germany. The specifics of trap deployment (habitat type, site, orientation, height) strongly influence its catch [40]. Collection dates varied among sites but are detailed in Table A1. Denatured ethanol (80%) was used to preserve specimens.
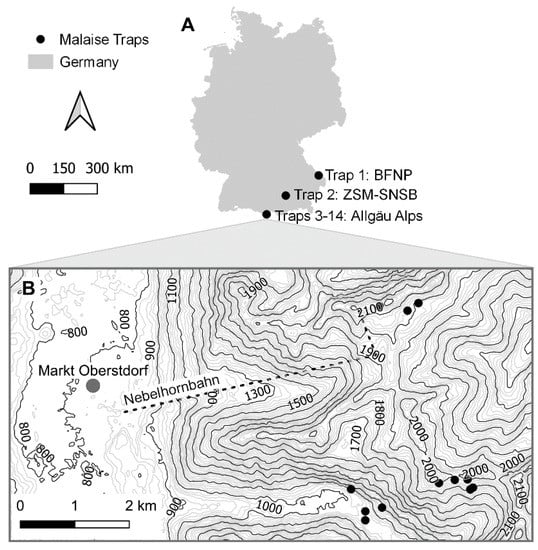
Figure 2.
Malaise trap sites. Locations where the 14 Malaise traps were deployed in 2012, 2014, and 2017 ((A,B) shows enlarged map of Allgäu Alps) as Germany’s contribution to the Global Malaise Trap Program.

Table 1.
Malaise trap information. Trap site, exact location, elevation, and habitat type.
2.2. Processing of Specimens
Samples from two sites (BFNP, ZSM) were sent directly to the CBG for analysis. Due to funding constraints, roughly every second weekly sample from the BFNP and every fourth weekly sample from the ZSM were selected for DNA barcode analysis. Based on the number of specimens in the samples that were processed, the full year of collecting at these sites yielded about 52,000 and 130,000 specimens, respectively. Using morphology, specimens from these locales were sorted to an order prior to sequence analysis and to a family after analysis. In total, tissue samples or whole individuals of 62,073 specimens (29,481 from BFNP; 32,592 from ZSM) were transferred to 96-well microplates for DNA extraction. Samples from the Allgäu Alps were sorted by a dipterist at the ZSM before being dispatched in 96-well microplates to the CBG for sequence analysis. Rough estimates suggest the Allgäu samples included well over a million specimens, but funding was only available to process about 2% of them (20,250 specimens).
At the CBG, specimens were processed using standard protocols for DNA extraction, PCR amplification of the barcode region of COI, and sequencing. Specimens from the BFNP and the Allgäu Alps were Sanger sequenced on an ABI 3730XL [41], while specimens from the ZSM were sequenced on Sequel [42].
2.3. Data Analysis
All specimen metadata and sequence data were uploaded to the Barcode of Life Data System (BOLD), an online workbench and database [32]. These data are publicly available in three datasets: DS-BFNP, DS-ZSMTRAP and DS-ALGALPS. Each sequence ≥ 300 base pairs (bp) was automatically assigned to a Barcode Index Number (BIN) already in BOLD if sequence similarity based on the (RESL-) BIN algorithm was fulfilled [43]. Sequences ≥ 500 bp which did not find a match served as founders of new BINs. All data were downloaded on 8 February 2021 for further analysis. Therefore, the present results correspond to BINs assigned at that time (BIN assignments can change as new sequences are added to BOLD).
Employing BINs as a proxy for species, we employed Chao1 [44] to estimate species counts for the dipteran families selected for analysis. We then calculated the ratio between the observed number of BINs in our samples to the estimate of species richness generated by Chao1 to ascertain the proportion of species at the sampling sites that have not been captured by our Malaise traps and that await analysis. We also generated continuous diversity profiles that illustrated variation in three standard metrics of biodiversity, which are quantified by Hill numbers (q): species richness (q = 0), Shannon diversity (q = 1), and Simpson diversity (q = 2) [34]. Hill numbers are a mathematically consolidated group of diversity indices which include relative species abundances in order to quantify biodiversity [45]. All calculations were performed in R version 3.3.6 with the Chao1 estimates calculated using the SpadeR package [46].
2.4. Extrapolating Species Numbers
We selected, more or less randomly, 11 dipteran families whose taxonomy and fauna have been intensively studied to date in order to assess the fractions of the Bavarian and German faunas represented in our samples (Table 2). By comparing the known species counts for these 11 families with the species recovered from our Malaise traps, we could estimate the percentage of these taxa that were recovered, providing a basis for estimating the completeness of our sampling. These values could then be used to estimate species diversity for our four DT families: Cecidomyiidae—gall midges; Chironomidae—non-biting midges; Phoridae—scuttle flies, and Sciaridae—dark-winged fungus gnats.

Table 2.
Species numbers for 15 families of Diptera. Species numbers for the Bavarian and German faunas are shown for 11 families of Diptera with well-established taxonomy and for four families with limited knowledge (Cecidomyiidae, Chironomidae, Phoridae, Sciaridae). *—estimated at 80% of German fauna.
Species numbers for Germany and for Bavaria were obtained from extensive literature (Table 2). For each family where a species count for Bavaria was unavailable, we adopted a count equal to 0.80 of the species number for Germany. This value was conservative because where species lists were available for both Bavaria and Germany, the ratio often exceeded 0.80 (Table 2). Moreover, this proportion corresponds to past evidence that Bavaria hosts 80–85% of the German fauna in well-studied invertebrate groups, both terrestrial and limnic [2,47].
We estimated species numbers for the DT families through the following steps:
- We calculated a Recovery Ratio by dividing the number of BINs detected through sequencing by the species count for each of the 15 families and for all Diptera (BIN/species ratio). This approach generated a ratio for each well-known family, for each DT family, and for all Diptera.
- We estimated the maximum number of species for each “dark taxon” for both Germany and Bavaria by dividing its BIN count by the average BIN/species ratio of all 11 well-known families.
- We estimated the minimum species number for each “dark taxon” by dividing all Diptera BINs by all Diptera species (i.e., 9544). Because this calculation includes numerous families with cryptic diversity, the resultant values underestimate the diversity of the DT families.
In the same fashion, we extrapolated species numbers employing the Chao1 values for the four DT families.
3. Results
3.1. Sequencing Results
COI sequences were recovered from 85.4% of the insects (70,293/82,323) that were analyzed (Table 3) and success was even higher for Diptera (91%). Diptera comprised nearly two thirds of the specimens that were analyzed and more than half of the resultant BINs. When results for Diptera from the three collection sites were pooled, the resulting 48,230 COI sequences were assigned to 4863 BINs and included species from 85 families. Across all sites, roughly 20% of the BINs were new to BOLD and almost 70% of them were Diptera with representatives from 56 families. Almost half of all dipteran BINs (2146; 44.1%) and 55% of the new dipteran BINs belonged to the four DT families.

Table 3.
Sequence results for the three sampling sites. Total sample size, number of processed specimens, sequences recovered, BINs, BINs new to BOLD, Diptera specimens, and Diptera BINs.
3.2. Estimation of Taxon Diversity Using BIN/Species Ratios
The 11 well-known families of Diptera displayed BIN/species ratios that ranged from 0.19–0.60 (ø 0.33 ± 0.9) for Bavaria and from 0.15–0.48 (ø 0.27 ± 0. 7) for Germany (Table 4, Figure A1a). Dividing all Diptera BINs by all known Diptera species produced a ratio of 0.64 for Bavaria and 0.51 for Germany. While one DT family (Chironomidae) possessed a ratio (0.38, Germany) that overlapped the upper end of the values for the 11 well-known families, the other three had far higher ratios. In fact, the BIN count for Phoridae and Sciaridae nearly matched the known species count for Germany, while the count for Cecidomyiidae exceeded it.

Table 4.
Fifteen families of Diptera, 11 with well-developed taxonomy and four that are less well known. The number of BINs recovered in this study is followed by the known species count for Bavaria and Germany, the ratio of species counts for Bavaria and Germany, and BIN/Species ratios for Bavaria and Germany.
3.3. Estimation of Taxon Diversity Using Chao1/Species Ratios
Chao1 estimates of species richness were obtained for the 15 families of Diptera (Table 5). BIN/Chao1 ratios averaged 0.76 for the 11 well-known families. The diversity profiles for 10 of these families showed overlap between the species richness in our samples and that estimated to occur at the sites sampled by our Malaise traps (Hill number q = 0, Figure 3). Muscidae was the sole exception as its predicted diversity was considerably higher than currently recognized. Chao1/species ratios ranged from 0.21–0.82 (0.46 ± 0.2) for Bavaria and from 0.16–0.66 (0.37 ± 0.2) for Germany (Table 5).

Table 5.
Proportion of undocumented Diptera biodiversity for Bavaria and Germany based on Chao1 estimates for 15 families.
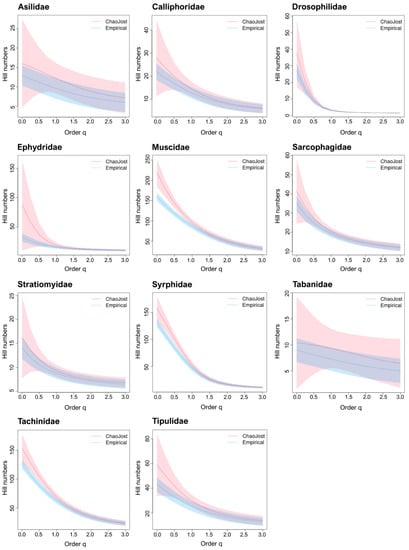
Figure 3.
Diversity profiles for 11 well-known taxa. The empirical (BIN counts; dotted blue) and estimated (Chao1; red) diversity profiles for communities where Malaise traps were deployed, as quantified by Hill numbers for each of the 11 well-known families for values of the diversity order (q) from 0–3 with 95% confidence intervals (shaded areas based on bootstrap analysis of 100 permutations). Species richness is depicted by q = 0; Shannon diversity by q = 1; and Simpson diversity by q = 2.
The BIN/Chao1 ratios for the DT families were similar to those for the well-known families, ranging from 0.60–0.81 (ø 0.69 ± 0.8). The diversity profiles for all four families (Figure 4) showed no overlap between observed and estimated species richness (i.e., Hill number q = 0). Chao1/species ratios indicated coverages of 0.83–5.91 for Bavaria and 0.61–2.25 for Germany (Table 5). Excluding Chironomidae, all DT families possessed ratios well above 1. Considering all Diptera, our samples recovered about 70% of the species estimated to occur at the study sites, meaning that as many as 6927 BINs of Diptera could have been collected during sampling. Chao1/species ratios were 0.91 for Bavaria and 0.73 for Germany.
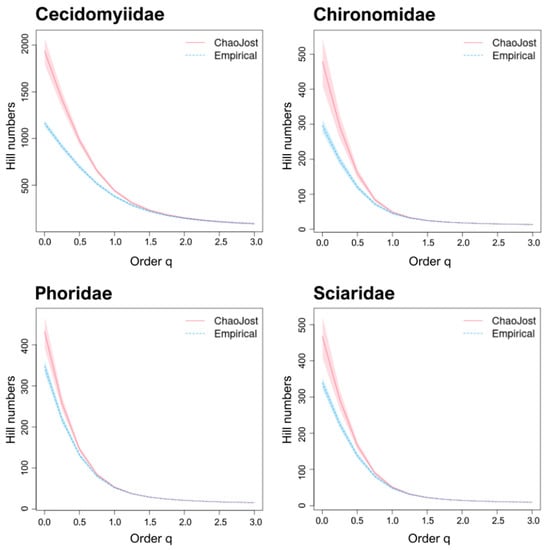
Figure 4.
Diversity profiles for the four DT families. The empirical (BIN counts; dotted blue) and estimated (Chao1; red) diversity profiles for communities where Malaise traps were deployed, as quantified by Hill numbers for each of the four “dark taxa” families for values of the diversity order (q) from 0–3 with 95% confidence intervals (shaded areas based on bootstrap analysis of 100 permutations). Species richness is depicted by q = 0; Shannon diversity by q = 1; and Simpson diversity by q = 2.
3.4. Extrapolating Species Numbers
We employed the two ratios to estimate the number of species in the DT families. First, we used BIN/species ratios to extrapolate species numbers based on the number of observed BINs. Second, we used the Chao1/species ratios to estimate species numbers based on the estimated BIN diversity. The first approach generates more conservative values than the second. We divided the number of observed BINs by the (BIN or Chao1)/species ratio for all Diptera to calculate minimum species numbers. To obtain an upper limit, we divided the number of observed BINs for each family by the average (BIN or Chao1)/species ratio for all well-known families. The following calculation is presented below (e.g., Sciaridae).
As 339 Sciaridae BINs were recovered, the minimum species estimate for Bavaria was 530 (339/0.64), while the upper estimate was 1027 (339/0.33). Similarly, the number of species in Germany could be estimated as ranging from 665 (339/0.51) to 1255 (339/0.27) species. By making similar calculations for each DT family, an overall estimate for total species numbers in Bavaria and Germany was obtained (Table 6). The number of species that await discovery in each region can then be obtained by subtracting the number of known species from these estimates.

Table 6.
BINs and calculated estimates. Total number of BINs recovered for each family from all traps, our calculated estimates, number of recorded species, and potential amplitude of new records for Bavaria and Germany.
In total, we recovered 2146 BINs for the DT families which is 22% of the total count of dipteran species known from Germany. Our conservative estimate suggested that just the DT families comprise about 3300–6500 species in Bavaria versus 4200–7900 in Germany. Based on the current species count for Diptera in Bavaria (7635) and Germany (9544), and our estimate of new record, this implies an increase of 25–66% and by 19–59% respectively.
By comparison, the Chao1 analysis suggested that 3316 BINs of the DT families occurred at our sampling sites, a 54% increase from current estimates. Based on this approach, there about 2200–5800 species in Bavaria and 2200–6600 in Germany that may still await documentation. Hence, this approach raises the species count for Diptera by 29–75% for Bavaria and by 22–69% for Germany.
4. Discussion
Although members of the order Diptera comprise almost a third of Germany’s insect fauna, the true diversity of the four highly diverse families [1] examined in this study is likely much higher than previously assumed [13,38]. By assessing the number of BINs sequenced from our collections and extrapolating species numbers, we obtained an initial estimate of their species numbers. Our results suggest that at least 1900–2200 dipteran species await discovery in Bavaria versus 1800–2200 in Germany. Although our species estimates were only based on sequencing Bavarian specimens, they are likely a good approximation of diversity in Germany as 80–85% of the invertebrate species found in Germany occur in Bavaria [2,36]. While Bavaria does have some habitats (e.g., alpine) that are not found in other regions of Germany, other habitats (e.g., coastal marshes) are absent [2], meaning that species specialized in the latter habitats will not occur in the state.
4.1. DNA Barcoding: Using BIN Numbers as Proxies for Species Numbers
Prior studies [49] have demonstrated that DNA barcoding is not only effective for specimen identification, but is also valuable for estimating species numbers [50,51,52,53]. Although there is strong correspondence between BIN counts and species numbers [49,54], several factors can lead to differences [54]. For example, COI numts can lead to the overestimation of species numbers if they are preferentially amplified in some specimens [55,56,57,58]. Conversely, the introgression of mitochondrial DNA (mtDNA), incomplete lineage sorting, and recent speciation can lead to underestimation of species numbers [59,60,61]. Other factors that challenge COI-based species identifications include heteroplasmy [62] and the homogenization of mtDNA haplotypes due to the maternally inherited endosymbiont Wolbachia [63,64]. These underlying molecular factors can lead the BIN algorithm on BOLD to assign members of a single species to several BINs or to assign several species to a single BIN. In groups with well-developed taxonomic systems, the BIN algorithm typically underestimates the true species count by about 10% as it was designed to deliver a conservative value for species diversity [65]. In addition to this internal constraint, two operational factors may have led our study to substantially underestimate actual species numbers:
- Limited geographic sampling as our data originates from few sites in Bavaria only, covering a tiny fraction of habitat types otherwise present.
- Limited funding constrained analysis to just 5% of the 1.2 million specimens that were collected.
4.2. BIN & Chao1/Species Ratios: Well-Known Families versus DT Families
We assessed the completeness of the species coverage provided by our Malaise trap samples in two ways. First, we calculated the ratio of the BINs recovered for each family and its known species count for Bavaria and Germany. We then made the same calculation employing Chao1 estimates, which, in contrast to the first approach, includes species that were present at our sampling sites but not caught nor sequenced. Thus, it is important to note that our first approach generates more conservative values than the second. By calculating the BIN/Chao1 ratios for each taxon, we were able to make the proportion of diversity that was not captured tangible.
Overall, the resulting (BIN or Chao1)/species ratios were much higher for the DT families than for the well-known ones (Table 4 and Table 5). Average ratios among the well-known families were well under 1 (ranging from 0.33–0.46 for Bavaria and 0.27–0.37 for Germany), indicating that our collections only included a fraction of the known diversity from Bavaria and Germany. This was expected because we only sampled few sites and only processed a fraction of our dipteran specimens. The much higher ratios for the DT families (average ranging from 1.67–2.55 for Bavaria and 0.91–1.34 for Germany) strongly suggest the presence of undescribed, unknown species. The Cecidomyiidae were the most dramatic case as we detected 1163 BINs, a value 35% higher than the species count for this family in Germany [8]. In fact, a quarter of all Diptera BINs belonged to this family, reinforcing conclusions from earlier studies indicating that this is the most diverse family of flies [13,49]. For example, extensive sampling at sites across Canada [49] revealed more than 10,000 BINs, a result which suggested that the Cecidomyiidae may include two million species worldwide. The Bavarian fauna has received little taxonomic attention as only 328 species are recorded versus a likely count of 687 species based on the presumption that 80% of the German fauna occurs there. By contrast, our analysis of 7148 specimens revealed 1163 BINs, a count for Bavaria which is threefold higher than the number of recorded species. Chironomidae was an exception among our DT families, as we obtained ratios that were consistent with those of the well-known families (Table 5). Although Chironomidae is a dark taxon, extensive research concerning the systematics, taxonomy, and nomenclature of European and Neotropical species has and is being conducted at the Bavarian State Collection of Zoology (ZSM) by the late Ernst Fittkau (former director of the ZSM) and his students including Martin Spies, the current editor of the Chironomid Home Page [66]. We therefore expect that the chironomid fauna of Bavaria and Germany is well documented and that, in contrast to the other DT families, a much lower amplitude of new species will be discovered in the following years of GBOL III. Among the well-known families, the Muscidae displayed the highest BIN/species indicating that the current species count considerably underrepresents its actual diversity. As a result, the Muscidae should also be recognized as a DT family.
4.3. Discrepancies in Taxa Coverage in Our Malaise Traps
Our estimated species counts for the DT families are based on the presumption that recovery success for the 11 families with strong taxonomy is a useful predictor of recovery success for the DT families. Our results did reveal threefold differences in recovery success among the well-known families, being lowest for Asilidae and Tabanidae and highest for the Muscidae. In our study, we used Malaise traps as a source of insect material, because they enable sampling of high numbers of flying insects, especially Diptera [67,68,69]. However, a bias favoring the sampling of some taxa over others is always present, meaning that the community captured with such traps does not depict the true insect community of a sampled site [67]. Furthermore, the setup of a Malaise trap in terms of site choice, orientation, and above-ground-level is another source of bias, and these factors strongly influence sampling results [40]. To incorporate such variations, we used different approaches for extrapolating species numbers including Chao1 estimate calculations, which consider the unsampled taxa present at the sampling sites. The resulting Chao1 values indicated that we only recovered about 70% of the dipteran species present at the sites. In this manner, we obtained BIN estimates for each family that consider recovery success and unsampled taxa. Our results indicate that more than 3316 more BINs await detection, a total that would raise the number of Dipteran species in Germany by a third.
5. Conclusions
In this study, we aimed at estimating the number of species in the Bavarian and German faunas for four families of Diptera that are prime examples of “dark taxa”. Our estimates were inferred from the analysis of sequence data, reproducible genetic patterns, rather than on speculations. The confidence intervals on these estimates are broad (Table 5), reflecting the various factors that influence any effort to gauge species diversity. Despite our limited geographic sampling effort, our results strongly suggest that a surprisingly high proportion of Germany’s biodiversity is yet to be discovered.
Author Contributions
Substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data for the work: A.H. (Axel Hausmann), C.C., V.B., D.D., S.S.; drafting the work or revising it critically for important intellectual content: A.H. (Axel Hausmann), C.C., V.B., P.D.N.H., S.S., G.H., M.J., A.H. (Amelie Höcherl), J.H., M.J.R., R.A.; final approval of the version to be published: A.H. (Axel Hausmann), C.C., S.S.; agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: A.H. (Axel Hausmann), C.C., V.B., P.D.N.H., S.S., G.H., M.J., A.H. (Amelie Höcherl), J.H., M.J.R., R.A. All authors have read and agreed to the published version of the manuscript.
Funding
GBOL projects were supported by grants from the German Federal Ministry of Education and Research (GBOL I: 01LI1101; GBOL II: 01LI1501, GBOL III: 16LI1901B). The Barcoding Fauna Bavarica project was funded by the Bayerisches Staatsministerium für Wissenschaft und Kunst. Sequence analyses were partially financed by funding from the government of Canada through Genome Canada and the Ontario Genomics Institute in support of the International Barcode of Life project.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The datasets containing all sequence data are publicly available in three datasets on the Barcode of Life Data System: DS-BFNP, DS-ZSMTRAP and DS-ALGALPS.
Acknowledgments
The GBOL III project is generously supported by a grant from the German Federal Ministry of Education and Research (FKZ 16LI1901B). We express our sincere thanks to the team at the Zoological Research Museum Alexander Koenig (ZMFK) for leading the GBOL III consortium and extensive project coordination. In particular, we would like to thank Marion Kotrba for valuable scientific advice and support.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Collection events for each Malaise trap.
Table A1.
Collection events for each Malaise trap.
| Site | Trap | Processed Collection Events |
|---|---|---|
| BFNP 2012 | 1 | 8 May; 22 May; 8 June; 20 June; 4 July; 25 July; 12 August; 3 September; 22 September 2012. |
| ZSM-SNSB | 2 | 10 April; 8 May; 5 June; 3 July; 31 July; 28 August; 25 September; 23 October; 20 November; 29 December 2017. |
| Allgäu Alps: Oytal | 3 | 4 May; 17 May; 1 June; 16 June; 5 July: 20 July; 7 August; 29 August; 2 October; 27 October 2014. |
| Allgäu Alps: Oytal | 4 | 4 May; 17 May; 1 June; 16 June; 5 July: 20 July; 7 August; 29 August; 2 October; 27 October 2014. |
| Allgäu Alps: Oytal | 5 | 4 May; 17 May; 1 June; 16 June; 5 July: 20 July; 7 August; 29 August; 2 October; 27 October 2014. |
| Allgäu Alps: Oytal | 6 | 4 May; 17 May; 1 June; 16 June; 5 July: 20 July; 7 August; 29 August; 2 October; 27 October 2014. |
| Allgäu Alps: Schochen | 7 | 21 June; 4 July; 17 July; 6 August; 4 September; 29 September; 19 October 2014. |
| Allgäu Alps: Schochen | 8 | 21 June; 4 July; 17 July; 6 August; 4 September; 29 September; 19 October 2014. |
| Allgäu Alps: Schochen | 9 | 21 June; 4 July; 17 July; 6 August; 4 September; 29 September; 19 October 2014. |
| Allgäu Alps: Schochen | 10 | 21 June; 4 July; 17 July; 6 August; 4 September; 29 September; 19 October 2014. |
| Allgäu Alps: Schochen | 11 | 21 June; 4 July; 17 July; 6 August; 4 September; 29 September; 19 October 2014. |
| Allgäu Alps: Koblat | 12 | 23 June, 4 July, 17 July; 8 August; 8 September; 5 September, 27 September; 20 October 2014. |
| Allgäu Alps: Koblat | 13 | 23 June, 4 July, 17 July; 8 August; 8 September; 5 September, 27 September; 20 October 2014. |
| Allgäu Alps: Koblat | 14 | 23 June, 4 July, 17 July; 8 August; 8 September; 5 September, 27 September; 20 October 2014. |
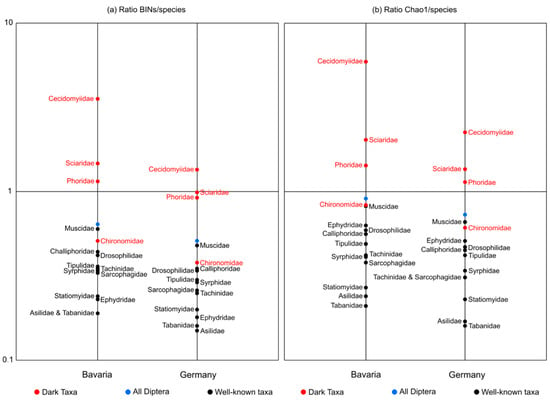
Figure A1.
Ratio of BIN or Chao1 counts versus recorded species counts ratios for each family on a logarithmic scale. (a) BINs/species and (b) Chao1/species for well-known families, problematic families, and for all Diptera for Bavaria and Germany.
References
- Hausmann, A.; Krogmann, L.; Peters, R.; Rduch, V.; Schmidt, S. GBOL III: DARK TAXA. iBOL Barcode Bull. 2020, 10, 4. [Google Scholar] [CrossRef]
- Dathe, H.H.; Taeger, A.; Blank, S.M. Entomofauna Germanica Band 4 (Hymenoptera). Verzeichnis der Hautflügler Deutschlands. Entomol. Nachr. Ber. 2001, Beiheft 7, 1–180. [Google Scholar]
- Dathe, H.H.; Blank, S.M. Nachträge zum Verzeichnis der Hautflügler Deutschlands, Entomofauna Germanica Band 4 (Hymenoptera) (1). Entomol. Nachr. Ber. 2004, 48, 179–183. [Google Scholar]
- Völkl, W.; Blick, T.; Kornacher, P.M.; Martens, H. Quantitativer Überblick über die rezente Fauna von Deutschland. Nat. Landsch. 2004, 79, 293–295. [Google Scholar]
- Schumann, H. Checkliste der Dipteren Deutschlands. In Entomofauna Germanica 2. Studia Dipterologica Supplement; Ampyx-Verlag: Halle/Saale, Germany, 1999; Volume 2, p. 354. [Google Scholar]
- Schumann, H. Erster Nachtrag zur “Checkliste der Dipteren Deutschlands”. Studia Dipterol. 2002, 9, 437–445. [Google Scholar]
- Schumann, H. Zweiter Nachtrag zur “Checkliste der Dipteren Deutschlands”. Studia Dipterol. 2004, 11, 619–630. [Google Scholar]
- Schumann, H. Dritter Nachtrag zur “Checkliste der Dipteren Deutschlands”. Studia Dipterol. 2010, 16, 17–27. [Google Scholar]
- de Carvalho, M.R. Revisiting the taxonomic impediment. Science 2005, 307, 353b. [Google Scholar] [CrossRef]
- Mandelik, Y.; Roll, U.; Fleischer, A. Cost-efficiency of biodiversity indicators for Mediterranean ecosystems and the effects of socio-economic factors. J. Appl. Ecol. 2010, 47, 1179–1188. [Google Scholar] [CrossRef]
- Chan, A.; Chiang, L.-P.; Hapuarachchi, H.C.; Tan, C.-H.; Pang, S.-C.; Lee, R.; Lee, K.-S.; Ng, L.-C.; Lam-Phua, S.-G. DNA Barcoding: Complementing morphological identification of mosquito species in Singapore. Parasites Vectors 2014, 7, 569. [Google Scholar] [CrossRef]
- Porter, T.M.; Gibson, J.F.; Shokralla, S.; Baird, D.J.; Golding, G.B.; Hajibabaei, M. Rapid and accurate taxonomic classification of insect (class Insecta) cytochrome c oxidase subunit 1 (COI) DNA barcode sequences using a naïve Bayesian classifier. Mol. Ecol. Resour. 2014, 14, 929–942. [Google Scholar] [CrossRef] [Green Version]
- Morinière, J.; Balke, M.; Doczkal, D.; Geiger, M.F.; Hardulak, L.A.; Haszprunar, G.; Hausmann, A.; Hendrich, L.; Regalado, L.; Rulik, B.; et al. A DNA barcode library for 5,200 German flies and midges (Insecta: Diptera) and its implications for metabarcoding-based biomonitoring. Mol. Ecol. Resour. 2019, 19, 900–928. [Google Scholar] [CrossRef] [Green Version]
- Brown, B.V. Sampling methods for adult flies (Diptera). In Measuring Arthropod Biodiversity: A Handbook of Sampling Methods; Santos, J.C., Fernandes, G.W., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 187–204. [Google Scholar] [CrossRef]
- Giangrande, A. Biodiversity, conservation, and the ‘taxonomic impediment’. Aquat. Conserv. Mar. Freshw. Ecosyst. 2003, 13, 451–459. [Google Scholar] [CrossRef]
- de Carvalho, M.R.; Bockmann, F.A.; Amorim, D.S.; Brandão, C.R.F.; de Vivo, M.; de Figueiredo, J.L.; Britski, H.A.; de Pinna, M.C.C.; Menezes, N.A.; Marques, F.P.L.; et al. Taxonomic impediment or impediment to taxonomy? A commentary on systematics and the cybertaxonomic-automation paradigm. Evol. Biol. 2007, 34, 140–143. [Google Scholar] [CrossRef]
- Wenzel, M.; Schmitt, T.; Weitzel, M.; Seitz, A. The severe decline of butterflies on western German calcareous grasslands during the last 30 years: A conservation problem. Biol. Conserv. 2006, 4, 542–552. [Google Scholar] [CrossRef]
- Sodhi, N.S.; Brook, B.W.; Bradshaw, C.J.A. Causes and consequences of species extinctions. In The Princeton Guide to Ecology; Levin, S.A., Ed.; Princton University Press: Princeton, NJ, USA, 2009; pp. 514–520. [Google Scholar]
- Coleman, C.O. Taxonomy in times of the taxonomic impediment—Examples from the community of experts on amphipod crustaceans. J. Crustacean Biol. 2015, 35, 729–740. [Google Scholar] [CrossRef] [Green Version]
- Hallmann, C.A.; Sorg, M.; Jongejans, E.; Siepel, H.; Hofland, N.; Schwan, H.; Stenmans, W.; Müller, A.; Sumser, H.; Hörren, T.; et al. More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PLoS ONE 2017, 12, e0185809. [Google Scholar] [CrossRef] [Green Version]
- Goulson, D. The insect apocalypse, and why it matters. Curr. Biol. 2019, 29, R967–R971. [Google Scholar] [CrossRef]
- Page, R.D.M. DNA barcoding and taxonomy: Dark taxa and dark texts. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150334. [Google Scholar] [CrossRef] [Green Version]
- GBOL III: Dark Taxa—Dark Taxa. Available online: https://bolgermany.de/home/gbol3/ (accessed on 16 December 2021).
- German Barcode of Life. Available online: https://bolgermany.de (accessed on 8 October 2021).
- Hausmann, A.; Haszprunar, G.; Hebert, P.D.N. DNA barcoding the geometrid fauna of Bavaria (Lepidoptera): Successes, surprises, and questions. PLoS ONE 2011, 6, e17134. [Google Scholar] [CrossRef]
- Hendrich, L.; Morinière, J.; Haszprunar, G.; Hebert, P.D.N.; Hausmann, A.; Köhler, F.; Balke, M. A comprehensive DNA barcode database for Central European beetles with a focus on Germany: Adding more than 3500 identified species to BOLD. Mol. Ecol. Resour. 2015, 15, 795–818. [Google Scholar] [CrossRef] [PubMed]
- Kotrba, M. The DNA barcoding project on German Diptera: An appreciative and critical analysis with four suggestions for improving the development and reliability of DNA-based identification. Eur. J. Entomol. 2020, 117, 315–327. [Google Scholar] [CrossRef]
- Schacht, W. Katalog der Zweiflügler (Mücken und Fliegen) Bayerns Familienliste—Artenliste—Literaturverzeichnis—Bestimmungsliteratur (Insecta: Diptera). Available online: http://www.zsm.mwn.de/docs_zsm/htdocs/dip/DiptBayKat.pdf (accessed on 8 October 2021).
- Wolff, D.; Gebel, M.; Geller-Grimm, F. Die Raubfliegen Deutschlands; Quelle & Meyer: Wiebelsheim, Germany, 2018. [Google Scholar]
- Jentzsch, M. Erster Entwurf einer Bibliographie zur Waffenfliegen-Fauna Deutschlands (Diptera: Stratiomyidae). Studia Dipterol. 2015, 22, 187–197. [Google Scholar]
- Doczkal, D.; Claußen, C.; Ssymank, A. Erster Nachtrag und Korrekturen zur Checkliste der Schwebfliegen Deutschlands (Diptera, Syrphidae). Volucella 2002, 6, 167–173. [Google Scholar]
- Heiss, R.; Fahldieck, M.; Stuke, J.-H. Kommentierte Checkliste der Schnaken der Bundesländer Niedersachsen und Bremen (Diptera, Tipulidae). Entomol. Nachr. Ber. 2019, 63, 245–259. [Google Scholar]
- Heiss, R.; Merkel-Wallner, G. Ein Beitrag zur Schnaken-Fauna Bayerns—Neue und wenig bekannte Arten aus Malaisefallen-Fängen 2007 bis 2009. Beiträge Bayer. Entomofaunist. 2013, 12, 17–30. [Google Scholar]
- Menzel, F. Die Trauermücken—Fauna der Bundesrepublik Deutschland (Diptera: Sciaridae). Beiträge Entomol. 2000, 50, 317–355. [Google Scholar] [CrossRef]
- Drees, M. Die Fleischfliegen des Hagener Raumes (Diptera: Sarcophagidae). Entomol. Z. 2014, 124, 3. [Google Scholar]
- Stuke, J.-H. Eine Kritische Liste der aus Deutschland nachgewiesenen Ephydridae mit der Beschreibung einer neuen Art (Diptera). Entomol. Z. 2011, 121, 115–126. [Google Scholar]
- Stuke, J.-H. Die Fruchtfliegen Niedersachsens und Bremens (Diptera, Drosophilidae). Linz. Biol. Beiträge 2020, 52, 475–499. [Google Scholar]
- Skuhravá, M.; Skuhravý, V.; Meyer, H. Gall midges (Diptera: Cecidomyiidae: Cecidomyiinae) of Germany—Faunistics, ecology and zoogeography. Faun.-Ökol. Mitt. 2014, Suppl. 38, 1–200. [Google Scholar]
- Global Malaise Program. Available online: https://biodiversitygenomics.net/projects/gmp/ (accessed on 8 October 2021).
- Chan-Canché, R.; Ballina-Gómez, H.; Leirana-Alcocer, J.; Bordera, S.; González-Moreno, A. Sampling of parasitoid Hymenoptera: Influence of the height on the ground. J. Hymenopt. Res. 2020, 78, 19–31. [Google Scholar] [CrossRef]
- Resources of CCDB. Available online: https://ccdb.ca/resources/ (accessed on 8 October 2021).
- Hebert, P.D.N.; Braukmann, T.W.A.; Prosser, S.W.J.; Ratnasingham, S.; deWaard, J.R.; Ivanova, N.V.; Janzen, D.H.; Hallwachs, W.; Naik, S.; Sones, J.E.; et al. A sequel to Sanger: Amplicon sequencing that scales. BMC Genom. 2018, 19, 219. [Google Scholar] [CrossRef] [Green Version]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-based registry for all animal species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [Green Version]
- Chao, A.; Chiu, C.-H. Estimation of species richness and shared species richness. In Methods and Applications of Statistics in the Atmospheric and Earth Sciences; Balakrishnan, N., Ed.; Wiley: New York, NY, USA, 2012; pp. 76–111. [Google Scholar]
- Chao, A.; Gotelli, N.J.; Hsieh, T.C.; Sander, E.L.; Ma, K.H.; Colwell, R.K.; Ellison, A.M. Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies. Ecol. Monogr. 2014, 84, 45–67. [Google Scholar] [CrossRef] [Green Version]
- Chao, A.; Ma, K.H.; Chiu, T.C.H. Spade R: Species-richness prediction and diversity estimation with R. Available online: https://cran.r-project.org/web/packages/SpadeR/SpadeR.pdf (accessed on 16 December 2021).
- Haszprunar, G. Barcoding Fauna Bavarica—Eine Chance für die Entomologie. Nachr. Bayer. Entomol. 2009, 58, 4. [Google Scholar]
- Büro für Freilandökologie Dr. Jürgen Esser. Available online: https://freilandoekologie-esser.de/ (accessed on 16 December 2021).
- Hebert, P.D.N.; Ratnasingham, S.; Zakharov, E.V.; Telfer, A.C.; Levesque-Beaudin, V.; Milton, M.A.; Pedersen, S.; Jannetta, P.; deWaard, J.R. Counting animal species with DNA barcodes: Canadian insects. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150333. [Google Scholar] [CrossRef] [Green Version]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. Available online: http://www.barcodinglife.org (accessed on 16 December 2021). [CrossRef] [Green Version]
- Hebert, P.D.N.; deWaard, J.R.; Landry, J.-F. DNA barcodes for 1/1000 of the animal kingdom. Biol. Lett. 2010, 6, 359–362. [Google Scholar] [CrossRef]
- Reid, B.N.; Le, M.; McCord, W.P.; Iverson, J.B.; Georges, A.; Bergmann, T.; Amato, G.; Desalle, R.; Naro-Maciel, E. Comparing and combining distance-based and character-based approaches for barcoding turtles. Mol. Ecol. Resour. 2011, 11, 956–967. [Google Scholar] [CrossRef] [Green Version]
- Chimeno, C.; Morinière, J.; Podhorna, J.; Hardulak, L.; Hausmann, A.; Reckel, F.; Grunwald, J.E.; Penning, R.; Haszprunar, G. DNA barcoding in forensic entomology—Establishing a DNA reference library of potentially forensic relevant arthropod species. J. Forensic Sci. 2019, 64, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Raupach, M.J.; Astrin, J.J.; Hannig, K.; Peters, M.K.; Stoeckle, M.Y.; Wägele, J.-W. Molecular species identification of Central European ground beetles (Coleoptera: Carabidae) using nuclear rDNA expansion segments and DNA barcodes. Front. Zool. 2010, 7, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pamilo, P.; Viljakainen, L.; Vihavainen, A. Exceptionally high density of NUMTs in the honeybee genome. Mol. Biol. Evol. 2007, 24, 1340–1346. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Buhay, J.E.; Whiting, M.F.; Crandall, K.A. Many Species in One: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc. Natl. Acad. Sci. USA 2008, 105, 13486–13491. [Google Scholar] [CrossRef] [Green Version]
- Buhay, J.E. “COI-like” sequences are becoming problematic in molecular systematic and DNA barcoding studies. J. Crustacean Biol. 2009, 29, 96–110. [Google Scholar] [CrossRef]
- Hazkani-Covo, E.; Zeller, R.M.; Martin, W. Molecular poltergeists: Mitochondrial DNA copies (Numts) in sequenced nuclear genomes. PLoS Genet. 2010, 6, e1000834. [Google Scholar] [CrossRef]
- Nesi, N.; Nakouné, E.; Cruaud, C.; Hassanin, A. DNA barcoding of African fruit bats (Mammalia, Pteropodidae). the mitochondrial genome does not provide a reliable discrimination between Epomophorus gambianus and Micropteropus pusillus. Comptes Rendus Biol. 2011, 334, 544–554. [Google Scholar] [CrossRef]
- Ermakov, O.A.; Simonov, E.; Surin, V.L.; Titov, S.V.; Brandler, O.V.; Ivanova, N.V.; Borisenko, A.V. Implications of hybridization, Numts, and overlooked diversity for DNA barcoding of Eurasian ground squirrels. PLoS ONE 2015, 10, e0117201. [Google Scholar] [CrossRef]
- Dufresnes, C.; Berroneau, M.; Dubey, S.; Litvinchuk, S.N.; Perrin, N. The effect of phylogeographic history on species boundaries: A comparative framework in Hyla tree frogs. Sci. Rep. 2020, 10, 5502. [Google Scholar] [CrossRef] [Green Version]
- Kmiec, B.; Woloszynska, M.; Janska, H. Heteroplasmy as a Common State of Mitochondrial Genetic Information in Plants and Animals. Curr. Genet. 2006, 50, 149–159. [Google Scholar] [CrossRef]
- Dobson, S.L. Evolution of Wolbachia cytoplasmic incompatibility. Evolution 2007, 58, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Bouchon, D.; Boutin, S.; Bellamy, L.; Zhou, L.; Engelstädter, J.; Hurst, G.D. the diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, A.; Godfray, H.C.J.; Huemer, P.; Mutanen, M.; Rougerie, R.; van Nieukerken, E.J.; Ratnasingham, S.; Hebert, P.D.N. Genetic patterns in European geometrid moths revealed by the Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e84518. [Google Scholar] [CrossRef]
- The Chironomid Home Page. Available online: www.chironomidae.net (accessed on 16 December 2021).
- Karlsson, D.; Forshage, M.; Holston, K.; Ronquist, F. The data of the Swedish Malaise Trap Project, a countrywide inventory of Sweden’s insect fauna. Biodivers. Data J. 2020, 8, e56286. [Google Scholar] [CrossRef] [PubMed]
- Moeed, A.; Meads, M.J. Seasonality of arthropods caught in a malaise trap in mixed lowland forest of the Orongorongo Valley, New Zealand. N. Z. J. Zool. 1987, 14, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Matthews, R.; Matthews, J. The Malaise trap: Its utility and potential for sampling insect populations. Great Lakes Entomol. 2017, 4, 4. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).