Simple Summary
Wolbachia is an intracellular bacterium that occurs in arthropods and in filarial worms. First described nearly a century ago in the reproductive tissues of Culex pipiens mosquitoes, Wolbachia is now known to occur in roughly 50% of insect species, and has been considered the most abundant intracellular bacterium on earth. In insect hosts, Wolbachia modifies reproduction in ways that facilitate spread of the microbe within the host population, but otherwise is relatively benign. In this “gene drive” capacity, Wolbachia provides a tool for manipulating mosquito populations. In mosquitoes, Wolbachia causes cytoplasmic incompatibility, in which the fusion of egg and sperm nuclei is disrupted, and eggs fail to hatch, depending on the presence/absence of Wolbachia in the parent insects. Recent findings demonstrate that Wolbachia from infected insects can be transferred into mosquito species that do not host a natural infection. When transinfected into Aedes aegypti, an important vector of dengue and Zika viruses, Wolbachia causes cytoplasmic incompatibility and, in addition, decreases the mosquito’s ability to transmit viruses to humans. This review addresses the maintenance of Wolbachia in insect cell lines, which provide a tool for high-level production of infectious bacteria. In vitro technologies will improve use of Wolbachia for pest control, and provide the microbiological framework for genetic engineering of this promising biocontrol agent.
Abstract
The obligate intracellular microbe, Wolbachia pipientis (Rickettsiales; Anaplasmataceae), is a Gram-negative member of the alpha proteobacteria that infects arthropods and filarial worms. Although closely related to the genera Anaplasma and Ehrlichia, which include pathogens of humans, Wolbachia is uniquely associated with invertebrate hosts in the clade Ecdysozoa. Originally described in Culex pipiens mosquitoes, Wolbachia is currently represented by 17 supergroups and is believed to occur in half of all insect species. In mosquitoes, Wolbachia acts as a gene drive agent, with the potential to modify vector populations; in filarial worms, Wolbachia functions as a symbiont, and is a target for drug therapy. A small number of Wolbachia strains from supergroups A, B, and F have been maintained in insect cell lines, which are thought to provide a more permissive environment than the natural host. When transferred back to an insect host, Wolbachia produced in cultured cells are infectious and retain reproductive phenotypes. Here, I review applications of insect cell lines in Wolbachia research and describe conditions that facilitate Wolbachia infection and replication in naive host cells. Progress in manipulation of Wolbachia in vitro will enable genetic and biochemical advances that will facilitate eventual genetic engineering of this important biological control agent.
1. Introduction
Wolbachia is an obligate intracellular microbe first described in reproductive tissues of Culex pipiens mosquitoes nearly a century ago [1,2]. Like Escherichia coli, Wolbachia is a Gram-negative bacterium in the phylum Proteobacteria: the purple bacteria and their relatives. Proteobacteria include nine monophyletic classes representing tremendous biodiversity. Among these, the genera Ehrlichia and Anaplasma, which can cause disease in humans, are classified with Wolbachia as members of the alpha-proteobacteria, in the order Rickettsiales, family Anaplasmataceae. Wolbachia is uniquely associated with invertebrates, does not infect vertebrate hosts, and replicates only within a eukaryotic host cell. In contrast, E. coli and many familiar Gram-negative pathogens of humans classified as gamma-proteobacteria can be cultured in liquid medium and plated on solid media as free-living microbes.
Knowledge of well-studied free-living bacteria provides an important framework for investigating the genetics and physiology of Wolbachia, now known to infect a high proportion of insect species, in addition to other arthropods and filarial worms, all members of the Ecdysozoa. Because of its widespread distribution among insects [3,4], Wolbachia provides a model system for exploring biological interactions between an intracellular microbe, the invertebrate host cells in which it resides, and the diversity of reproductive phenotypes with which it is associated [5,6]. In species that harbor Wolbachia, the bacterium is transmitted vertically, from mother to offspring, which retain the infection. In most arthropods, Wolbachia alters reproduction in diverse ways that favor its invasion of naive populations, and is sometimes considered a reproductive parasite. In contrast, Wolbachia is an essential symbiont in filarial worms [7,8,9]. In mosquitoes, Wolbachia causes a reproductive distortion called cytoplasmic incompatibility (CI), which has important applications in vector control [10].
2. A Brief History of Wolbachia and Cytoplasmic Incompatibility in Mosquitoes
The species name for Wolbachia, Wolbachia pipientis, reflects its discovery in reproductive tissues of the mosquito Cx. pipiens [1,2]. Wolbachia was described during the historical period when arthropod-borne intracellular bacteria were first appreciated as pathogens that cause disease in humans. Notable discoveries during that time included those of Howard Ricketts, who observed that a tick-borne bacterium, now known as Rickettsia rickettsii, was the causative agent of Rocky Mountain Spotted Fever [11], and Henrique da Rocha Lima, who described Rickettsia prowazekii as the cause of epidemic typhus [12]. Simeon Burt Wolbach was involved in identification of the louse as the vector of typhus [13], and it is not surprising that he noticed similarities between arthropod-borne pathogens and the intracellular bacteria now known as Wolbachia. Within the Rickettsiales, members of the genus Rickettsia are now assigned to the family Rickettsiaceae (short rods or coccobacilli), and Wolbachia, to the family Anaplasmataceae (small pleomorphic cocci); these families have been distinguished based on genetic analyses [14]. For many years, Wolbachia was thought to be unique to Cx. pipiens mosquitoes because it appeared to be restricted to reproductive tissues, whereas other microbial symbionts in arthropods were more widely distributed among host tissues [1,2]. In retrospect, however, it should be noted that methods for distinguishing species of intracellular microbes in insects were poorly developed at that time.
The best-studied effect of Wolbachia is the reproductive distortion known as cytoplasmic incompatibility (CI). Discovery of CI was a fortuitous result of the fact that Cx. pipiens mosquitoes mate in small cages and establish breeding populations under laboratory conditions. In studies unrelated to the microbiology of Wolbachia, crosses between Cx. pipiens from independent laboratory colonies representing diverse geographic regions sometimes exhibited the peculiar, maternally inherited mating distortion called CI. CI was manifested when eggs from crosses between male and female mosquitoes originating from different regions failed to hatch, in a pattern eventually defined by 17 distinct cytotypes [15,16,17,18]. Even before the connection between CI and Wolbachia was established, maternal inheritance of CI was recognized as a gene drive mechanism potentially useful for population replacement of vector mosquitoes [19,20]. The association between Wolbachia and CI was established when Yen and Barr demonstrated reversal of the CI phenotype in Cx. pipiens after treatment with antibiotics, which eliminated the bacterium [21,22]. The molecular biology and biochemistry of CI and the genes associated with this phenotype in Drosophila and mosquitoes have been reviewed, and will not be discussed in detail here [17,18].
3. Contemporary Wolbachia Research
Development of genomic sequencing and related molecular technologies, and the discovery of Wolbachia sequences in DNA samples from Drosophila melanogaster and other insects, contributed substantially to our understanding of the biology, distribution, and diversity of Wolbachia. Advancement of Wolbachia research through the use of D. melanogaster mutants is only beginning and, among mosquitoes, the presence of Wolbachia in Aedes albopictus, and its absence in important vectors such as Aedes aegypti and Anopheles gambiae, has provided an important incentive for Wolbachia research. Wolbachia also occurs in agricultural pests, including planthoppers that feed on rice, and lepidopteran pests of food crops.
Publication of the first Wolbachia genome from strain wMel infecting D. melanogaster revealed a streamlined, AT-rich genome of 1.27 Mbp encoding 1144 proteins (NC_002978.6), compared to the E. coli genome of 4.64 Mbp, 4242 proteins for K-12 sub-strain MG1655 (NC_000913.3). The wMel genome contains a putative prophage/s and an unusual abundance of mobile elements, relative to genomes from other intracellular bacteria [23]. Comparative analysis of Wolbachia genomes has provided important insights into the correlations between gene loss, symbiosis, Wolbachia’s dependence on its host for essential nutrients, and its genetic capabilities; complete or nearly-complete sequence annotation for several representative genomes is available on the NCBI website (ncbi.nih.nlm.gov).
As noted above, a Wolbachia isolate is typically called a strain, and is usually named for its host species; for example, Wolbachia from D. melanogaster is called wMel; that from Cx. pipiens is known as wPip. Individual strains are further subdivided into 17 monophyletic clusters of diversity, or supergroups, designated by letters [24,25]. As more strains are described, nomenclature based on species names of hosts is becoming cumbersome. For example, wCtub could refer to a supergroup J Wolbachia from a filarial worm [26] or a supergroup F Wolbachia from a termite [27].
Consistent with the wide diversity of its hosts, Wolbachia infections are associated with phenotypes other than CI, including parthenogenesis, male killing, and feminization of genetic males, which are not necessarily uniform among members of a supergroup. Like CI, these phenotypes have a net effect of increasing the abundance of Wolbachia in host populations [5,6,28] and, in many cases, the arthropod host can be cured of the Wolbachia infection, with loss of associated phenotypes, by treatment with rifampicin and/or tetracycline [29]. Filarial worms, and a few arthropods including the bedbug Cimex lectularius (Hemiptera), the wasp Asobara tabida (Hymenoptera), and the springtail Folsomia candida (Collembola), in which Wolbachia has become a symbiont, are refractory to antibiotic cure. In humans, antibiotics have therapeutic value for reducing filarial infection [9].
Although reproductive phenotypes can be validated by comparing infected relative to antibiotic-cured populations, it has been difficult to document Wolbachia’s more general effects on the biology and ecology of its hosts [30]. Evaluation of host fitness under laboratory conditions is subject to complex variables [31], and although both positive and negative effects have been described, the narrative has been confounded by undefined contributions of the host’s genetic background [31,32,33]. Similarly, unpublished comparisons between infected and tetracycline-cured Cx. pipiens adults in the author’s laboratory have thus far yielded inconsistent results that may reflect, at least in part, variability in larval rearing conditions. Wolbachia’s individual associations with diverse hosts may preclude broad generalizations, and the complexity of Wolbachia’s interactions with host tissues provides a rich field for further investigation. As described below, fitness issues have become particularly important in efforts to establish novel Wolbachia associations for control of mosquito vectors.
Present day interest in Wolbachia as an environmentally-friendly approach to vector control builds upon early successful demonstrations that its associated gene drive can be harnessed for vector replacement [19,20,34], coupled with more recent methods for rapid detection of Wolbachia infections; successful introduction (transinfection) into non-host species such as the dengue vector, Ae aegypti, and the malaria vector, Anopheles gambiae [10]; retention of the CI phenotype and maternal transmission after transinfection; and, in mosquitoes, the unanticipated suppression of pathogen transmission [10,35,36,37]. Additional biological factors, including fecundity, hatch rate, diapause survival, longevity, Wolbachia density, and tissue distribution, and virus-blocking capacity, are similarly important for successful Wolbachia-based control applications. Several ongoing studies address optimization of Wolbachia to control disease transmission, including combination with sterile insect techniques [38]. Although such applications are underway, much remains to be learned about the basic biology of Wolbachia, and the potential for its genetic modification using technologies that have become routine for E. coli.
4. Wolbachia in Insect Cell Lines
Wolbachia’s obligate intracellular lifestyle complicates the biochemical and genetic analyses that could advance pest control and anti-filarial applications. Even with hosts amenable to laboratory rearing, maintenance of colonies, dissection of infected tissues, and embryonic microinjection are labor-intensive and time-consuming. Moreover, many existing laboratory colonies are highly inbred, complicating cage studies that address fitness. The utility of Wolbachia in control applications would be enhanced if the microbe could be experimentally manipulated by genetic engineering to express selectable markers, which in turn will be advanced by improving manipulation of Wolbachia in cell lines and expanding the diversity of Wolbachia strains that can be investigated in culture. A modest advance would be adaptation of a filarial strain of Wolbachia to a cell line; at present, Wolbachia-infected insect cell lines are used as a surrogate to identify new drugs that target Wolbachia for treatment of filarial diseases [39,40,41].
The author’s research focuses on systematic exploration of Wolbachia propagation in cultured cells as a substitute for the differentiated host tissues, such as ovaries and testes, in which Wolbachia is most abundant. Cell lines used to propagate Wolbachia are listed in Table 1, wherein supergroup designations are noted after the strain name; for example, wPip_B indicates that wPip is classified in supergroup B. With the exception of a single member of supergroup F from the cat flea [42], only members of supergroups A and B, sometimes called the “pandemic” supergroups, have been maintained in insect cell lines. The reader should note that, in some cases, an infected cell line may have been sub-cultured only a limited number of times and/or has a very long doubling time, and that the same cell line may have been infected with the same strain of Wolbachia by different investigators, and given a different name. An important incentive for employing cell lines was the possibility that preadaption to cultured cells might improve the likelihood that Wolbachia would establish in novel hosts infected by embryonic microinjection, and towards this end, a few lines have been maintained for several years [43]. In other cases, which are not reviewed in detail here, infected cell lines have been used to test effects of Wolbachia on viral replication in efforts that generally validate the anti-pathogen responses seen in transinfected mosquitoes. Finally, as with Wolbachia itself, a uniform descriptive label for infected cell lines remains to be developed.

Table 1.
Cell lines in which Wolbachia strains have been propagated.
5. Cell Lines Derived from Infected Insects
In Table 1, two types of Wolbachia-infected cell line are noted: those established directly from infected tissues, which retain Wolbachia from the host, and lines established from uninfected cells into which Wolbachia has been artificially introduced, either from infected tissues or from a previously infected cell line. O’Neill and coworkers were the first to describe a line with a host-derived Wolbachia infection, which they named Aa23 [44]. Aa23 cells are persistently infected with wAlbB, one of two strains that co-infected the host Ae. albopictus mosquitoes. Curiously, at least in some geographic areas, when a single Wolbachia strain is found in Ae. albopictus, it types to strain wAlbA [70]. Given that Aedes albopictus hosts a natural infection, one might wonder why Wolbachia is absent from older Ae. albopictus lines dating back to the late 1960s, before Wolbachia was known to be widespread in insects [63]. The possibility exists that early in their history, such lines may have been infected, followed by loss of Wolbachia over decades of use in multiple laboratories.
Other cell lines developed directly from infected hosts include JW-18, which carries wMelPop from D. melanogaster [41]. In addition, a Wolbachia strain in a primary culture called Dm2008Wb1, from which the Wolbachia was subsequently transferred to Drosophila S2 cells, has been developed in Russia [65]. Although it can take considerable time to establish primary cultures, which require additional time to become established, it will be of interest to learn whether primary cultures established directly from carefully chosen Drosophila mutants will provide insights into Wolbachia’s interactions with host cells.
Extrapolation of Wolbachia’s behavior in host tissues to cultured cells is not straightforward; similarly, it is not known how cultured cells should be manipulated to favor successful establishment of a persistent infection. For example, studies with Ae. albopictus eggs suggest that the highest Wolbachia densities occur during embryogenesis, and that replication is dependent on host cell division [71], whereas wMelPop replicates best in postmitotic cells of adult flies [72]. How established infections coevolve with host cells and potentially modify the host cell cycle are yet to be explored. Methods for distinguishing live Wolbachia from metabolically active and/or infectious Wolbachia are not yet available, and will be needed to determine whether maintenance of Wolbachia in one cell line facilitates its establishment in a second cell line, or whether the initial cell line simply provides a higher multiplicity of infectious particles. When examined by fluorescence microscopy, a single Cx. pipiens ovary appears to contain innumerable Wolbachia, but it is not known whether all are equally infectious, whether the proportion of microbes capable of initiating infection changes during the mosquito reproductive cycle, and what factors define the small proportion of Wolbachia that establish in the embryonic germline. Finally, it will be of interest to learn whether Wolbachia (or other endogenous infectious agents such as insect-specific flaviviruses) influence how easily a permanent cell line can be established from a host species. Compared to those from Ae. albopictus and Ae. aegypti mosquitoes, relatively few established cell lines are available from Cx pipiens [63,73].
6. Transfers between Cell Lines
As with infected insects, wAlbB can be removed from Aa23 cells by treatment with tetracycline, and these cured Aa23(T) cells have been a popular host for establishment of new infections [43,45,46]. Indeed, the majority of cell lines into which Wolbachia has been introduced derive from Ae. albopictus, and the possibility that these successes are influenced by host factors that coevolved with the natural Wolbachia infection in ancestral mosquitoes has not been addressed. Note, for example, that the RML-12 cell line, originally used to “preadapt” Wolbachia for transfer to Ae. aegypti, is now known to have originated from Ae. albopictus [43,45]. Aa23(T) cells host the single example of wPip in culture [46]. Despite the importance of wPip as one of the best-understood models for CI [74,75,76], this author has been unable to establish wPip in culture, and is not aware of a cell line that maintains a long-term persistent infection with wPip.
One of the most interesting Wolbachia strains introduced into Aa23(T) cells is wMelPop, an unusually virulent strain isolated from D. melanogaster. Briefly, to investigate brain degeneration in Drosophila, Min and Benzer crossed an X-chromosome deficiency strain with reduced life span into the white mutant w1118, which had a normal life span [72]. Resulting progeny expressed the short life span, which was associated with a maternally-transmitted bacterium identified as the “popcorn” strain of Wolbachia. Popcorn proliferates primarily in postmitotic tissues of the adult, causing death of the host fly. Despite its ovarian transmission, popcorn does not cause CI in the original Drosophila strain described by Min and Benzer [72], but the cell-line adapted variant does cause CI in Drosophila [43] and in transinfected mosquitoes [10]. Genome comparisons suggest that virulence of wMelPop is caused by copy number variation of a 21 kb “octomom” region encoding eight genes of unknown function [77,78].
The strain called wMelPop-CLA designates a “cell-line adapted” version of wMelPop, created by infecting Aa23(T) cells with wMelPop from w1118 eggs [43]. Although the rationale for adapting wMelPop to a cell line related to difficulties in using Wolbachia from infected tissues directly to establish a stable transovarial infection in a novel host, transfer of Wolbachia into an insect cell line can also be problematic. For example, establishment of wMelPop in Aa23(T) cells was successful in only two of 68 attempts [43]. After adaptation to mosquito cells, wMelPop-CLA was reintroduced into Drosophila embryos, in which it exhibited a diminution, but not complete loss, of properties related to life shortening, bacterial density, and CI, relative to the parental strain [43]. The life-shortening effect of wMelPop was of interest because reduced longevity of vector species would be expected to reduce pathogen transmission [79], but wMelPop-CLA, and to a lesser extent wMel, have deleterious effects on Ae. aegypti eggs and larval development [35,80], which have remained stable, at least over 10 years [81]. Other mosquito cell lines that maintain wMelPop-CLA include RML-12 (Ae. albopictus) and Mos-55 (An. gambiae).
A third variant from D. melanogaster, wMelCS, was isolated from Canton S flies [78]. Genomic studies indicate that wMelCS arose prior to wMel, and is ancestral to wMelPop. In nature, wMel appears to be replacing wMelCS [82]. The extent to which the Wolbachia genome undergoes changes during growth in cultured cells has been evaluated in a genetic comparison of wMel strains, in which five genetic differences were detected after introduction into cell lines: an IS5 insertion, a multi-gene deletion, two point mutations, and a 10 bp deletion [78]. Because comparable changes did not occur with cell line-derived Wolbachia transinfected into mosquitoes, it is possible that these genetic changes were triggered by cross-species transfer. Nevertheless, the changes are not known to affect essential genes or cause major genomic rearrangements, suggesting to a first approximation that use of genetically manipulated Wolbachia produced in cell lines will remain suitable for control applications. Although genome evolution of tissue-derived Wolbachia in cross-species transfers has not yet been investigated, recent in-depth comparisons suggest that relative to wRi (from Drosophila simulans) and wPip (from Cx. pipiens mosquitoes), wMelCS appears to be the best candidate for transinfection into Ae. aegypti for field release [35].
The Ae. albopictus cell line NIAS-AeAl-2 supports the Hemipteran Wolbachia strain, wStri, which was transferred from ovaries of the planthopper Laodelphax striatellus; this line also supports lepidopteran strains wKue, wCauA, and wCauB [47,48]. More recently, we found that wStri establishes a particularly robust infection in clonal C7-10 Aedes albopictus cells [53], and used the resulting C/wStri1 line to develop flow cytometric quantitation of Wolbachia infections [83]. The wStri strain also replicates in tick cell lines [42]. Strain wCauA provides an interesting model for investigating a single Wolbachia strain that causes two distinct phenotypes, CI and male killing, in different hosts [84,85], a phenomenon also described in Drosophila subquinaria [86]
7. Aedes aegypti Cells and New Viruses
Aside from a single transfer into Ae. aegypti Aa20 (Mos-20) cells [62,63], the Aag2 line is most commonly used in Wolbachia studies, including many that focus on Wolbachia’s antiviral effects. Aag2 cells are a subpopulation of cells isolated in the Peleg laboratory (Israel Institute for Biological Research, Ness-Ziona, Israel) that we adapted to Eagle’s medium [87] and designated Aag2 because the precise identity of Peleg’s culture (line 59 vs. line 364) was uncertain. Metaphase spreads from Aag2 cells contain a characteristic chromosome fragment, and the cells can be distinguished from Ae. albopictus cells by electrophoretic mobility of small heat shock proteins. Aag2 cells host an inapparent infection with the insect-specific flavivirus called Cell Fusing Agent (CFA), which was discovered when Ae. aegypti cells were co-cultured with Ae. albopictus cells [88]. For virological studies, this endogenous virus made Ae. aegypti cells less desirable, relative to Ae. albopictus cells, for classical studies with mosquito-borne viruses [89,90]. Although the generation of the clonal C7-10 and C6/36 lines included steps to exclude viruses [89], more recent proteomic studies do not exclude the presence of endogenous viruses in C7-10 cells [91].
In Aag2 cells, Wolbachia inhibits CFA replication [92], in addition to the replication of a newly described positive-sense RNA negev-like virus [57], but the antiviral effect may differ in field populations of mosquitoes [93]. More recently, a bunyavirus called Phasi Charoen-like virus was also found persistently to infect Aag2 cells, but is not inhibited by Wolbachia, possibly reflecting its negative-sense RNA genome [61,94]. Similarly, a negative-sense RNA anphevirus found in Aedes cell lines and mosquitoes is not inhibited by Wolbachia [95].
8. Other Dipteran, Lepidopteran and Tick Cell Lines
Newly established dipteran cell lines that support Wolbachia include those derived from the sandfly, Lutzomyia longipalpis, the biting midge, Culicoides sonorensis, and the horn fly, Hematobia irritans. Two lepidopteran lines, from the moths Heliothis zea and Spodoptera frugiperda, support wStri, wRi, and wCauB, whereas cell lines from the ticks Ixodes scapularis, Ixodes ricinus, and Riphicephalus microplus support wAlbB and wStri. Uniquely, establishment of wCfe_F in Ixodes scapularis lines raises the expectation that members of supergroups other than A and B will eventually be cultured in vitro [42]. Filarial strains of Wolbachia have not yet been cultured; by default, infected mosquito cell lines have been used as surrogates to screen for potential anti-filarial drugs [39,40,41].
9. Studies with wAlbB and wStri in C7-10 Cell Derivatives
My own lab has worked extensively with the Aedes albopictus C7-10 cell line [89,96,97] and Wolbachia strains wAlbB [44,55] and wStri [47,53,56]. C7-10 is a clonal population developed as a standard “wild-type” cell line adapted to a modified Eagle’s medium that lacks undefined components, such as yeastolate or lactalbumin hydrolysate, that are included in many insect-specific media formulations. Because Eagle’s medium has a defined chemical composition, apart from the supplemental serum, it facilitates use of the techniques and approaches of somatic cell genetics to isolate viral and host cell mutants [90,96,97]. This medium is bicarbonate-buffered, and cells are maintained at 28 to 30 °C in a 5% CO2 atmosphere. The persistently-infected line called C/wStri1 was produced by inoculating wStri from NIAS-AeAl-2 cells [47] into a growing population of C7-10 cells [53].
Although C/wStri1 cells are primarily used in current studies, Aa23 cells infected with wAlbB provided the starting point for our investigations [98]. Aa23 cells represent a mixed cell population from embryonic tissue that maintains a somewhat variable level of wAlbB in individual cells and between populations of cells [44]. In our hands, Aa23 cells readily adapted to Eagle’s medium containing 20% heat-inactivated fetal bovine serum. In contrast to clonal C7-10 cells, Aa23 cells grew as patchy monolayers that tended to form solid aggregates as cultures aged; these cytological characteristics persisted after elimination of Wolbachia with tetracycline [98]. Because the solid clusters reach sizes visible to the naked eye, it seemed likely that cells within a cluster vary in metabolic activity as direct contact with the culture medium decreases. In addition, it seemed possible that internal cells may die and/or be cannibalized by cells growing closer to the surface of the cluster. To minimize cell clusters, we often used trypsin and/or passed resuspended cells through a 40 μm nylon mesh before plating. Although additional isolates of Aa23 lines were available in liquid nitrogen storage in the lab of Dr. Robert Tesh (University of Texas Medical Branch, Galveston, TX, USA), inspection of these lines (Fallon, 2005, unpublished) suggested that none was more tractable than the Aa23 line described by O’Neill and coworkers [44]. It remains to be learned whether the tendency of Aa23 cells to aggregate is relevant to recovery of a Wolbachia-infected line, and whether the desirability of cell lines that grow as attached, relatively uniform monolayers may have contributed to inadvertent loss of infection with continued passage of long-established Ae. albopictus cell lines.
Aa23 cells grow relatively slowly with a doubling time of 4–5 days, compared to 20 h with C7-10 cells. Counting Aa23 cells using a Coulter electronic cell counter can be facilitated by adding NP-40 to the culture medium to release cell nuclei, rather than attempting to disrupt clusters of tightly-associated cells [98]. Alternatively, the number of cells can be evaluated using an assay based on conversion of methylthiazole tetrazolium (MTT) to a colored formazan product. Use of this approach suggested that the Aa23 doubling time increases from 2 to 3 days as the cells approach stationary phase, and that addition of tetracycline to suppress Wolbachia replication does not improve the doubling time [99].
Difficulties in obtaining accurate cell counts and enumerating wAlbB in Aa23 cells led us to transfer wAlbB from Aa23 cells into a TK-6 thymidine kinase-deficient derivative of C7-10 cells [55]. Because they are resistant to 5-bromodeoxyuridine (BrdU) [100], we reasoned that TK-6 cells provided a means of selecting against Aa23 cells, if donor cells are present in the infected culture. Although selection was not needed, the wAlbB infection was lost after five months in TK-6 cells. Nevertheless, analysis of radiolabeled protein profiles in wAlbB-infected TK-6 cells led to the observation that the ubiquitin/proteasome pathway may play an important role in the interaction between Wolbachia and its host cell [55], and contributed to later work elucidating the roles of a two-gene operon encoding CidA/CidB proteins in cytoplasmic incompatibility [74,75,76].
In its native host, the planthopper Laodelphax striatellus, wStri causes CI, as does wPip in Cx. pipiens mosquitoes. Although we have had little success with wPip, transfer of wStri from NIAS-AeAl-2 cells to C7-10 cells was highly successful [53], and established a long-term, stable infection with which we have worked for several years. On two separate occasions, however, C/wStri1 cells lost the infection after approximately 130 subcultures, suggesting that with continued passage, the cells suppress Wolbachia. Factors that affect stability, such as changes in the ratio of Wolbachia to host cells over time, remain to be systematically examined. These observations support the possibility that there is an unknown fitness cost to maintaining Wolbachia in cell lines, consistent with the absence of Wolbachia in long-established Ae. albopictus lines. As new Wolbachia-infected lines are developed, investigators should bear in mind the importance of periodically storing cells in liquid nitrogen to recover early-passage cultures, against which changes over time can be evaluated.
A major advance in our ability to work with Wolbachia in cultured cells is flow cytometry, which allows simultaneous evaluation of cell and bacterial particles (Figure 1).
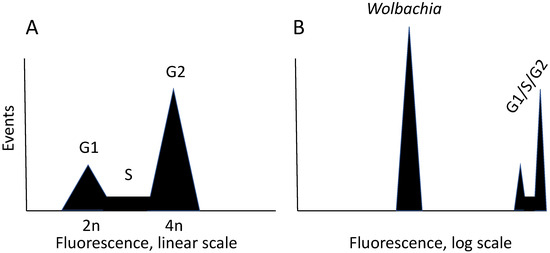
Figure 1.
Use of a flow cytometer for evaluating Wolbachia abundance. Flow cytometry involves staining a subcellular component, such as DNA, with a fluorescent dye. For DNA, propidium iodide is commonly used. In the instrument, stained cells flow past a detector that records the dye amount, which is displayed on a histogram. Panel (A) shows a schematic representation of a typical image obtained with a growing culture of cells. In a mixed population of cells, individual cells independently traverse the cell cycle. Immediately after mitosis, a cell has a diploid content of DNA, which is called G1. As DNA is synthesized, DNA content increases. This increase is represented by the S (synthesis) phase of the cycle. When synthesis is complete, cells have a G2 DNA content. The fluorescence in G2 is double that of G1, as is the total amount of DNA in a cell. In a cell cycle histogram, the X-axis is on a linear scale, and areas under the curves are proportional to the percentage of the population in that particular phase of the cycle. (B). To detect Wolbachia in the same sample, the position of host cell cycle events is electronically shifted to the far right of a histogram with the X-axis on a log scale, allowing particles with lower amounts of DNA to be detected. Details of conditions for visualizing Wolbachia by flow cytometry are described elsewhere [83].
Using this approach, we have determined that the most infectious Wolbachia are those newly-released from fragile host cells, which constitute about 1% of the total Wolbachia population in C/wStri1 cells. Establishment of a robust level of Wolbachia in a naive cell line requires about 6 days, with an infection ratio of 80 to 100 bacteria per cell. Reminiscent of Aa23 cells, mild aggregation of recipient cells is a typical response of C7-10 cells to infection [56]. Current experiments suggest that wStri replicates better in stationary, as opposed to growing, cells, and we have recently found that high yields of infectious Wolbachia can be recovered from mitotically inactivated feeder layers generated by treatment with mitomycin C [101]. In addition to determining conditions that are favorable to establishment of new infections, developing a cell line in which the Wolbachia infection itself confers a selective advantage will constitute an important resource for recovery of genetically modified Wolbachia.
In addition to changes in the Wolbachia phenotype and/or virulence that accompany long-term maintenance outside the wildtype host, much remains to be learned about optimal conditions for maintaining Wolbachia in vitro. The relationships between cell cycle parameters, doubling time, and ploidy of cultured cells, relative to conditions that support Wolbachia in natural host tissues, have not been systematically explored, nor have effects of nutrient requirements and medium composition. In future studies it will be important to undertake transcriptomic and proteomic analyses to identify genes and gene products that regulate infection and replication of Wolbachia in cell lines. Insect cell culture itself remains a developing field, relative to accomplishments that have been achieved with vertebrate cell lines and embryonic stem cells.
10. Why Cultured Cells?
If the streamlined Wolbachia genome can be genetically engineered in the future, propagation of the altered genome will require efficient reintroduction into a host cell to allow replication and expansion of transformant populations. Use of cell lines offers a practical means of producing the large quantities of Wolbachia that will be needed to develop transformation protocols that are sufficiently robust for use in basic research and pest control applications. Although isolated examples of successful transformation of intracellular microorganisms such as Coxiella burnetti, the pathogen that causes Q fever, have been achieved, these remain labor intensive and have low frequencies of success [102]. Nevertheless, over the past two decades, remarkable progress towards cell-free culture of Coxiella has been achieved, despite its streamlined 2 Mb genome [103,104]. These successes underscore the importance of detailed attention to culture conditions and metabolic activities of obligate intracellular microbes. Wolbachia lacks pathogenicity to humans, and its genome is more extensively streamlined, relative to that of Coxiella. Nevertheless, the long evolutionary history of Wolbachia’s interaction with invertebrate hosts and its adaptations for germline transmission contribute to the value of Wolbachia as a model system for understanding the biology of obligate intracellular bacteria in invertebrate cells and manipulating their biology for control of insect pests.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data described in this review article are available in referenced publications.
Acknowledgments
Work in the author’s lab is currently supported by the University of Minnesota Agricultural Experiment Station, St. Paul, MN, USA. I thank former students for participation in this work and the NIH for past financial support. I dedicate this review to the memory of Que Lan, 1959–2014.
Conflicts of Interest
The author declares no conflict of interest.
References
- Hertig, M.; Wolbach, S.B. Studies on Rickettsia-like micro-organisms in insects. J. Med. Res. 1924, 44, 329–374. [Google Scholar]
- Hertig, M. The rickettsia, Wolbachia pipientis (Gen. Et SP.N.) and associated inclusions of the mosquito Culex pipiens. Parasitology 1936, 28, 453–486. [Google Scholar] [CrossRef]
- Hilgenboecker, K.; Hammerstein, P.; Schlattmann, P.; Telschow, A.; Werren, J.H. How many species are infected with Wolbachia? —A statistical analysis of current data. FEMS Microbiol. Lett. 2008, 281, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Zug, R.; Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 2012, 7, e38544. [Google Scholar] [CrossRef]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Landmann, F. The Wolbachia Endosymbionts. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.J.; Bandi, C.; Hoerauf, A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 2005, 60, 245–284. [Google Scholar] [PubMed]
- Fenn, K.; Conlon, C.; Jones, M.; Quail, M.A.; Holroyd, N.E.; Parkhill, J.; Blaxter, M. Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog. 2006, 2, e94. [Google Scholar] [CrossRef]
- Johnston, K.L.; Ford, L.; Umareddy, I.; Townson, S.; Specht, S.; Pfarr, K.; Hoerauf, A.; Altmeyer, R.; Taylor, M.J. Repurposing of approved drugs from the human pharmacopoeia to target Wolbachia endosymbionts of onchocerciasis and lymphatic filariasis. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Joshi, D. Genetic control of dengue and malaria using Wolbachia. In Genetic Control of Malaria and Dengue; Adelman, Z.N., Ed.; Academic Press: London, UK, 2016; Chapter 14; pp. 305–333. [Google Scholar] [CrossRef]
- Gross, D.; Schäfer, G. 100th anniversary of the death of Ricketts: Howard Taylor Ricketts (1871–1910). The namesake of the Rickettsiaceae family. Microbes Infect. 2011, 13, 10–13. [Google Scholar] [CrossRef]
- Bernardes Filho, F.; Avelleira, J.C. Henrique da Rocha Lima. An. Bras. Dermatol. 2015, 90, 364–366. [Google Scholar] [CrossRef]
- Conlon, J.M. The Historical Impact of Epidemic Typhus. Available online: https://www.montana.edu/historybug/documents/TYPHUS-Conlon.pdf (accessed on 30 June 2021).
- Dumler, J.S.; Barbet, A.F.; Bekker, C.P.; Dasch, G.A.; Palmer, G.H.; Ray, S.C.; Rikihisa, Y.; Rurangirwa, F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001, 51, 2145–2165. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Weill, M. Wolbachia infection influences the development of Culex pipiens embryo in incompatible crosses. Heredity 2006, 96, 493–500. [Google Scholar] [CrossRef]
- Sinkins, S.P.; Walker, T.; Lynd, A.R.; Steven, A.R.; Makepeace, B.L.; Godfray, H.C.; Parkhill, J. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature 2005, 436, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, M.; Atyame, C.; Beji, M.; Justy, F.; Cohen-Gonsaud, M.; Sicard, M.; Weill, M. Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat. Commun. 2018, 9, 319. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, M.; Hochstrasser, M. The biochemistry of cytoplasmic incompatibility caused by endosymbiotic bacteria. Genes 2020, 11, 852. [Google Scholar] [CrossRef]
- Laven, H. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 1967, 216, 383–384. [Google Scholar] [CrossRef]
- Curtis, C.F.; Sinkins, S.P. Wolbachia as a possible means of driving genes into populations. Parasitology 1998, 116, S111–S115. [Google Scholar] [CrossRef]
- Yen, J.; Barr, A. New hypothesis of the cause of cytoplasmic incompatibility in Culex pipiens L. Nature 1971, 232, 657–658. [Google Scholar] [CrossRef]
- Yen, J.H.; Barr, A.R. The etiological agent of cytoplasmic incompatibility in Culex pipiens. J. Invertebr. Pathol. 1973, 2, 242–250. [Google Scholar] [CrossRef]
- Wu, M.; Sun, L.V.; Vamathevan, J.; Riegler, M.; Deboy, R.; Brownlie, J.C.; McGraw, E.A.; Martin, W.; Esser, C.; Ahmadinejad, N.; et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol. 2014, 2, e69. [Google Scholar] [CrossRef]
- Paraskevopoulos, C.; Bordenstein, S.R.; Wernegreen, J.J.; Werren, J.H.; Bourtzis, K. Toward a Wolbachia multilocus sequence typing system: Discrimination of Wolbachia strains present in Drosophila species. Curr. Microbiol. 2006, 53, 388–395. [Google Scholar] [CrossRef]
- Lefoulon, E.; Clark, T.; Borveto, F.; Perriat-Sanguinet, M.; Moulia, C.; Slatko, B.E.; Gavote, L. Pseudoscorpion Wolbachia symbionts: Diversity and evidence for a new supergroup S. BMC Micro. 2020, 20, 188. [Google Scholar] [CrossRef] [PubMed]
- Lefoulon, E.; Clark, T.; Guerrero, R.; Canizales, I.; Cardenas-Callirgos, J.M.; Junker, K.; Vallarino-Lhermitte, N.; Makepeace, B.L.; Darby, A.C.; Foster, J.M.; et al. Diminutive, degraded but dissimilar: Wolbachia genomes from filarial nematodes do not conform to a single paradigm. Microb. Genom. 2020, 6, mgen000487. [Google Scholar] [CrossRef]
- Hellemans, S.; Kaczmarek, N.; Marynowska, M.; Calusinska, M.; Roisin, Y.; Fournier, D. Bacteriome-associated Wolbachia of the parthenogenetic termite Cavitermes Tuberosus. FEMS Microbiol. Ecol. 2019, 95, fiy235. [Google Scholar] [CrossRef] [PubMed]
- Saridaki, A.; Bourtzis, K. Wolbachia: More than just a bug in insects genitals. Curr. Opin. Microbiol. 2010, 13, 67–72. [Google Scholar] [CrossRef]
- Li, Y.Y.; Floate, K.D.; Fields, P.G.; Pang, B.-P. Review of treatment methods to remove Wolbachia bacteria from arthropods. Symbiosis 2014, 62, 1–15. [Google Scholar] [CrossRef]
- Kajtoch, L.; Kolasa, M.; Kubisz, D.; Gutowski, J.M.; Scibior, R.; Mazur, A.; Holecova, M. Using host species traits to understand the Wolbachia infection distribution across terrestrial beetles. Sci. Rep. 2019, 9, 847. [Google Scholar] [CrossRef]
- Almeida, F.; Moura, A.S.; Cardoso, A.F.; Winter, C.E.; Bijovsky, A.T.; Suesdek, L. Effects of Wolbachia on fitness of Culex quinquefasciatus (Diptera: Culicidae). Infect. Genet. Evol. 2011, 11, 2138–2143. [Google Scholar] [CrossRef]
- Dean, M.D. A Wolbachia-associated fitness benefit depends on genetic background in Drosophila simulans. Proc. Biol. Sci. 2006, 273, 1415–1420. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Ross, P.A.; Rasic, G. Wolbachia strains for disease control: Ecological and evolutionary considerations. Evol. Appl. 2015, 8, 751–768. [Google Scholar] [CrossRef]
- Sinkins, S.; Gould, F. Gene drive systems for insect disease vectors. Nat. Rev. Genet. 2006, 7, 427–435. [Google Scholar] [CrossRef]
- Fraser, J.E.; De Bruyne, J.T.; Iturbe-Ormaetxe, I.; Stepnell, J.; Burns, R.L.; Flores, H.A.; O’Neill, S.L. Novel Wolbachia-transinfected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotypes. PLoS Pathog. 2017, 13, e1006751. [Google Scholar] [CrossRef]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef]
- Walker, T.; Johnson, P.H.; Moreira, L.A.; Iturbe-Ormaetxe, I.; Frentiu, F.D.; McMeniman, C.J.; Leong, Y.S.; Dong, Y.; Axford, J.; Kriesner, P.; et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 2011, 476, 450–453. [Google Scholar] [CrossRef]
- Zhang, D.; Lees, R.S.; Xi, Z.; Gilles, J.R.L.; Bourtzis, K. Combining the sterile insect technique with Wolbachia-based approaches: II- A safer approach to Aedes albopictus population suppression programmes, designed to minimize the consequences of inadvertent female release. PLoS ONE 2015, 10, e0135194. [Google Scholar] [CrossRef]
- Hermans, P.G.; Hart, C.A.; Trees, A.J. In vitro activity of antimicrobial agents against the endosymbiont Wolbachia pipientis. J. Antimicrob. Chemother. 2001, 47, 659–664. [Google Scholar] [CrossRef]
- Clare, R.H.; Cook, D.A.; Johnston, K.L.; Ford, L.; Ward, S.A.; Taylor, M.J. Development and validation of a high-throughput anti-Wolbachia whole-cell screen: A route to macrofilaricidal drugs against onchocerciasis and lymphatic filariasis. J. Biomol. Screen 2015, 20, 64–69. [Google Scholar] [CrossRef]
- Serbus, L.R.; Landmann, F.; Bray, W.M.; White, P.M.; Ruybal, J.; Lokey, R.S.; Debec, A.; Sullivan, W.A. cell-based screen reveals that the albendazole metabolite, albendazole sulfone, targets Wolbachia. PLoS Pathog. 2012, 8, e1002922. [Google Scholar] [CrossRef]
- Khoo, J.J.; Kurtti, T.J.; Husin, N.A.; Beliavskaia, A.; Lim, F.S.; Zulkifli, M.M.S.; Al-Khafaji, A.M.; Hartley, C.; Darby, A.C.; Hughes, G.L.; et al. Isolation and propagation of laboratory strains and a novel flea-derived field strain of Wolbachia in tick cell lines. Microorganisms 2020, 8, 988. [Google Scholar] [CrossRef]
- McMeniman, C.J.; Lane, A.M.; Fong, A.W.; Voronin, D.A.; Iturbe-Ormaetxe, I.; Yamada, R.; McGraw, E.A.; O’Neill, S.L. Host adaptation of a Wolbachia strain after long-term serial passage in mosquito cell lines. Appl. Environ. Microbiol. 2008, 74, 6964–6969. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.L.; Pettigrew, M.M.; Sinkins, S.P.; Braig, H.R.; Andreadis, T.G.; Tesh, R.B. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 1997, 6, 33–39. [Google Scholar] [CrossRef]
- Voronin, D.; Tran-Van, V.; Potier, P.; Mavingui, P. Transinfection and growth discrepancy of Drosophila Wolbachia strain wMel in cell lines of the mosquito Aedes albopictus. J. Appl. Microbiol. 2010, 108, 2133–2141. [Google Scholar] [CrossRef]
- Dobson, S.L.; Marsland, E.J.; Veneti, Z.; Bourtzis, K.; O’Neill, S.L. Characterization of Wolbachia host range via the in vitro establishment of infections. Appl. Environ. Microbiol. 2002, 68, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Noda, H.; Miyoshi, T.; Koizumi, Y. In vitro cultivation of Wolbachia in insect and mammalian cell lines. Vitr. Cell. Dev. Biol. Anim. 2002, 38, 423–427. [Google Scholar] [CrossRef]
- Furukawa, S.; Tanaka, K.; Fukatsu, T.; Sasaki, T. In vitro infection of Wolbachia in insect cell lines. Appl. Entomol. Zool. 2008, 43, 519–525. [Google Scholar] [CrossRef][Green Version]
- Raquin, V.; Valiente Moro, C.; Saucereau, Y.; Tran, F.-H.; Potier, P.; Mavingui, P. Native Wolbachia from Aedes albopictus blocks Chikungunya virus infection in cellulo. PLoS ONE 2015, 10, e0125066. [Google Scholar] [CrossRef]
- Fenollar, F.; La Scola, B.; Inokuma, H.; Dumler, J.S.; Taylor, M.J.; Raoult, D. Culture and phenotypic characterization of a Wolbachia pipientis isolate. J. Clin. Microbiol. 2003, 41, 5434–5441. [Google Scholar] [CrossRef] [PubMed]
- Frentiu, F.D.; Robinson, J.; Young, P.R.; McGraw, E.A.; O’Neill, S.L. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS ONE 2010, 5, e13398. [Google Scholar] [CrossRef]
- Ekwudu, O.; Devine, G.J.; Aaskov, J.G.; Frentiu, F.D. Wolbachia strain wAlbB blocks replication of flaviviruses and alphaviruses in mosquito cell culture. Parasites Vectors 2020, 13, 54. [Google Scholar] [CrossRef] [PubMed]
- Fallon, A.M.; Baldridge, G.D.; Higgins, L.-A.; Witthuhn, B.A. Wolbachia from the planthopper Laodelphax striatellus establishes a robust, persistent, streptomycin-resistant infection in clonal mosquito cells. Vitr. Cell. Dev. Biol. Anim. 2013, 49, 66–73. [Google Scholar] [CrossRef]
- Venard, C.M.; Crain, P.R.; Dobson, S.L. SYTO11 staining vs. FISH staining: A comparison of two methods to stain Wolbachia pipientis in cell cultures. Lett. Appl. Microbiol 2011, 52, 168–176. [Google Scholar] [CrossRef]
- Fallon, A.M.; Witthuhn, B.A. Proteasome activity in a naïve mosquito cell line infected with Wolbachia pipientis wAlbB. Vitr. Cell. Dev. Biol. Anim. 2009, 45, 460–466. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fallon, A.M. Conditions facilitating infection of mosquito cell lines with Wolbachia, an obligate intracellular bacterium. Vitr. Cell. Dev. Biol. Anim. 2019, 55, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.; Parry, R.; Asgari, S. Effect of Wolbachia wAlbB on a positive-sense RNA negev-like virus: A novel virus persistently infecting Aedes albopictus mosquitoes and cells. J. Gen. Virol. 2020, 101, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Bian, G.; Pan, X.; Xi, Z. Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Negl. Trop. Dis. 2012, 6, e1754. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.; Audsley, M.D.; Di Giallonardo, F.; Kerton, E.J.; Young, P.R.; Holmes, E.C.; McGraw, E.A. Sustained Wolbachia-mediated blocking of dengue virus isolates following serial passage in Aedes aegypti cell culture. Virus Evol. 2019, 8, vez012. [Google Scholar] [CrossRef] [PubMed]
- Terradas, G.; Joubert, D.A.; McGraw, E.A. The RNAi pathway plays a small part in Wolbachia-mediated blocking of dengue virus in mosquito cells. Sci. Rep. 2017, 7, 43847. [Google Scholar] [CrossRef]
- McLean, B.J.; Dainty, K.R.; Flores, H.A.; O’Neill, S.L. Differential suppression of persistent insect specific viruses in trans-infected wMel and wMelPop-CLA Aedes-derived mosquito lines. Virology 2019, 527, 141–145. [Google Scholar] [CrossRef]
- Etebari, K.; Asad, S.; Zhang, G.; Asgari, S. Identification of Aedes aegypti long intergenic non-coding RNAs and their association with Wolbachia and dengue virus infection. PLoS Negl. Trop. Dis. 2016, 10, e0005069. [Google Scholar] [CrossRef]
- Kurtti, T.J.; Munderloh, U.G. Mosquito cell culture. In Advanced Cell Culture; Maramorosch, K., Ed.; Academic Press: London, UK, 1984; pp. 259–302. [Google Scholar]
- Rasgon, J.L.; Ren, X.; Petridis, M. Can Anopheles gambiae be infected with Wolbachia pipientis? Insights from an in vitro system. Appl. Environ. Microbiol. 2006, 72, 7718–7722. [Google Scholar] [CrossRef]
- Andrianova, B.V.; Goriacheva, I.I.; Aleksandrov, I.D.; Gorelova, T.V. Establishment of a new continuous cell line of Drosophila melanogaster strain infected by the intracellular endosymbiotic bacterium Wolbachia pipientis under natural conditions. Genetika 2010, 46, 14–17. (In Russian) [Google Scholar] [CrossRef]
- White, P.M.; Pietri, J.E.; Debec, A.; Russell, S.; Patel, B.; Sullivan, W. Mechanisms of horizontal cell-to-cell transfer of Wolbachia spp. in Drosophila melanogaster. Appl. Environ. Microbiol. 2017, 83, e03425-16. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Gonçalves, D.; Iturbe-Ormaetxe, I.; Martins-da-Silva, A.; Tellieria, E.L.; Rocha, M.N.; Traub-Cseko, Y.M.; O’Neill, S.L.; Sant’Anna, M.R.V.; Moreira, L.A. Wolbachia introduction into Lutzomyia longipalpis (Diptera: Psychodidae) cell lines and its effects on immune-related gene expression and interaction with Leishmania infantum. Parasites Vectors 2019, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Jasperson, D.; Cohnstaedt, L.W.; Brelsford, C.L. Transfection of Culicoides sonorensis biting midge cell lines with Wolbachia pipientis. Parasites Vectors 2019, 12, 483. [Google Scholar] [CrossRef] [PubMed]
- Madhav, M.; Brown, G.; Morgan, J.A.; Asgari, S.; McGraw, E.A.; Munderloh, U.G.; Kurtti, T.J.; James, P. Wolbachia successfully replicate in a newly established horn fly, Haematobia irritans irritans (L.) (Diptera: Muscidae) cell line. Pest Manag. Sci. 2020, 76, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Kittayapong, P.; Baimai, V.; O’Neill, S. Field prevalence of Wolbachia in the mosquito Aedes Albopictus. Am. J. Trop. Med. Hyg. 2002, 66, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Ruang-areerate, T.; Kittayapong, P.; McGraw, E.A.; Baimai, V.; O’Neill, S.L. Wolbachia Replication and Host Cell Division in Aedes albopictus. Curr. Microbiol. 2004, 49, 10–12. [Google Scholar] [CrossRef]
- Min, K.T.; Benzer, S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. USA 1997, 94, 10792–10796. [Google Scholar] [CrossRef]
- Walker, T.; Jeffries, C.L.; Mansfield, K.L.; Johnson, N. Mosquito cell lines: History, isolation, availability and application to assess the threat of arboviral transmission in the United Kingdom. Parasites Vectors 2014, 7, 382. [Google Scholar] [CrossRef]
- Beckmann, J.F.; Fallon, A.M. Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: Implications for cytoplasmic incompatibility. Insect Biochem. Mol. Biol. 2013, 43, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, J.F.; Ronau, J.A.; Hochstrasser, M.A. Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2017, 2, 17007. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, J.F.; Sharma, G.D.; Mendez, L.; Chen, H.; Hochstrasser, M. The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. Elife 2019, 8, e50026. [Google Scholar] [CrossRef] [PubMed]
- Chrostek, E.; Teixeira, L. Mutualism breakdown by amplification of Wolbachia genes. PLoS Biol. 2015, 13, e1002065. [Google Scholar] [CrossRef]
- Woolfit, M.; Iturbe-Ormaetxe, I.; Brownlie, J.C.; Walker, T.; Riegler, M.; Seleznev, A.; Popovici, J.; Rancès, E.; Wee, B.A.; Pavlides, J.; et al. Genomic evolution of the pathogenic Wolbachia strain, wMelPop. Genome Biol. Evol. 2013, 5, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- McMeniman, C.J.; Lane, R.V.; Cass, B.N.; Fong, A.W.; Sidhu, M.; Wang, Y.F.; O’Neill, S.L. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 2009, 323, 141–144. [Google Scholar] [CrossRef]
- Farnesi, L.C.; Belinato, T.A.; Gesto, J.S.M.; Martins, A.J.; Bruno, R.V.; Moreira, L.A. Embryonic development and egg viability of wMel-infected Aedes aegypti. Parasites Vectors 2019, 12, 211. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.A.; Axford, J.K.; Callahan, A.G.; Richardson, K.M.; Hoffmann, A.A. Persistent deleterious effects of a deleterious Wolbachia infection. PLoS Negl. Trop. Dis. 2020, 14, e0008204. [Google Scholar] [CrossRef]
- Riegler, M.; Sidhu, M.; Miller, W.J.; O’Neill, S.L. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 2005, 15, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Fallon, A.M. Flow cytometric evaluation of the intracellular bacterium, Wolbachia pipientis, in mosquito cells. J. Microbiol. Methods. 2014, 107, 119–125. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sasaki, T.; Kubo, T.; Ishikawa, H. Interspecific transfer of Wolbachia between two lepidopteran insects expressing cytoplasmic incompatibility: A Wolbachia variant naturally infecting Cadra cautella causes male killing in Ephestia Kuehniella. Genetics 2002, 162, 1313–1319. [Google Scholar] [CrossRef]
- Sasaki, T.; Massaki, N.; Kubo, T. Wolbachia variant that induces two distinct reproductive phenotypes in different hosts. Heredity 2005, 95, 389–393. [Google Scholar] [CrossRef]
- Jaenike, J. Spontaneous emergence of a new Wolbachia phenotype. Evolution 2007, 61, 2244–2252. [Google Scholar] [CrossRef]
- Lan, Q.; Fallon, A.M. Small heat shock proteins distinguish between two mosquito species and confirm identity of their cell lines. Am. J. Trop. Med. Hyg. 1990, 43, 669–676. [Google Scholar] [CrossRef]
- Stollar, V.; Thomas, V.L. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology 1975, 64, 367–377. [Google Scholar] [CrossRef]
- Fallon, A.M. Transfection of cultured mosquito cells. In Molecular Biology of Insect Disease Vectors; Crampton, J.M., Beard, C.B., Louis, C., Eds.; Chapman and Hall: New York, NY, USA, 1997; pp. 430–443. [Google Scholar]
- Stollar, V. Insect-transmitted vertebrate viruses: Alphatogaviruses. Vitr. Cell. Dev. Biol. 1993, 29, 289–295. [Google Scholar] [CrossRef]
- Baldridge, G.; Higgins, L.; Witthuhn, B.; Markowski, T.; Baldridge, A.; Armien, A.; Fallon, A. Proteomic analysis of a mosquito host cell response to persistent Wolbachia infection. Res. Microbiol. 2017, 168, 609–625. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Etebari, K.; Asgari, S. Wolbachia suppresses cell fusing agent virus in mosquito cells. J. Gen. Virol. 2016, 97, 3427–3432. [Google Scholar] [CrossRef] [PubMed]
- Amuzu, H.E.; Tsyganov, K.; Koh, C.; Herbert, R.I.; Powell, D.R.; McGraw, E.A. Wolbachia enhances insect-specific flavivirus infection in Aedes aegypti mosquitoes. Ecol. Evol. 2018, 8, 5441–5454. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Sreenu, V.B.; Mottram, T.; McFarlane, M. Wolbachia restricts insect-specific flavivirus infection in Aedes aegypti cells. J. Gen. Virol. 2016, 97, 3024–3029. [Google Scholar] [CrossRef]
- Parry, R.; Asgari, S. Aedes Anphevirus: An insect-specific virus distributed worldwide in Aedes aegypti mosquitoes that has complex interplays with Wolbachia and dengue virus infection in cells. J. Virol. 2018, 92, e00224-18. [Google Scholar] [CrossRef]
- Sarver, N.; Stollar, V. Sindbis virus-induced cytopathic effect in clones of Aedes albopictus (Singh) cells. Virology 1977, 80, 390–400. [Google Scholar] [CrossRef]
- Fallon, A.M.; Kurtti, T.J. Cultured cells as a tool for analysis of gene expression. In Biology of Disease Vectors; Marquardt, W.C., Ed.; Elsevier: New York, NY, USA, 2005; Volume 2, pp. 539–549. [Google Scholar]
- Fallon, A.M. Cytological properties of an Aedes albopictus mosquito cell line infected with Wolbachia strain wAlbB. Vitr. Cell. Dev. Biol. Anim. 2008, 44, 154–161. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fallon, A.M.; Hellestad, V.J. Standardization of a colorimetric method to quantify growth and metabolic activity of Wolbachia-infected mosquito cells. Vitr. Cell. Dev. Biol. Anim. 2008, 44, 351–356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazzacano, C.A.; Fallon, A.M. Thymidine kinase deficient mutants of Aedes albopictus mosquito cells. Vitr. Cell. Dev. Biol. Anim. 1992, 28, 455–458. [Google Scholar] [CrossRef]
- Fallon, A.M. Assessment of mitotically inactivated mosquito cell feeder layers produced with mitomycin C. Vitr. Cell. Dev. Biol. Anim. 2021, 57, 583–586. [Google Scholar] [CrossRef]
- Beare, P.A.; Howe, D.; Cockrell, D.C.; Omsland, A.; Hansen, B.; Heinzen, R.A. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J. Bacteriol. 2009, 191, 1369–1381. [Google Scholar] [CrossRef]
- Omsland, A.; Beare, P.A.; Hill, J.; Cockrell, D.C.; Howe, D.; Hansen, B.; Samuel, J.E.; Heinzen, R.A. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl. Environ. Microbiol. 2011, 77, 3720–3725. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Vallejo-Esquerra, E.; Omsland, A. Use of axenic culture tools to study Coxiella burnetii. Curr. Protoc. Microbiol. 2008, 50, e52. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).