Transcriptome Profiling of Starvation in the Peripheral Chemosensory Organs of the Crop Pest Spodoptera littoralis Caterpillars
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
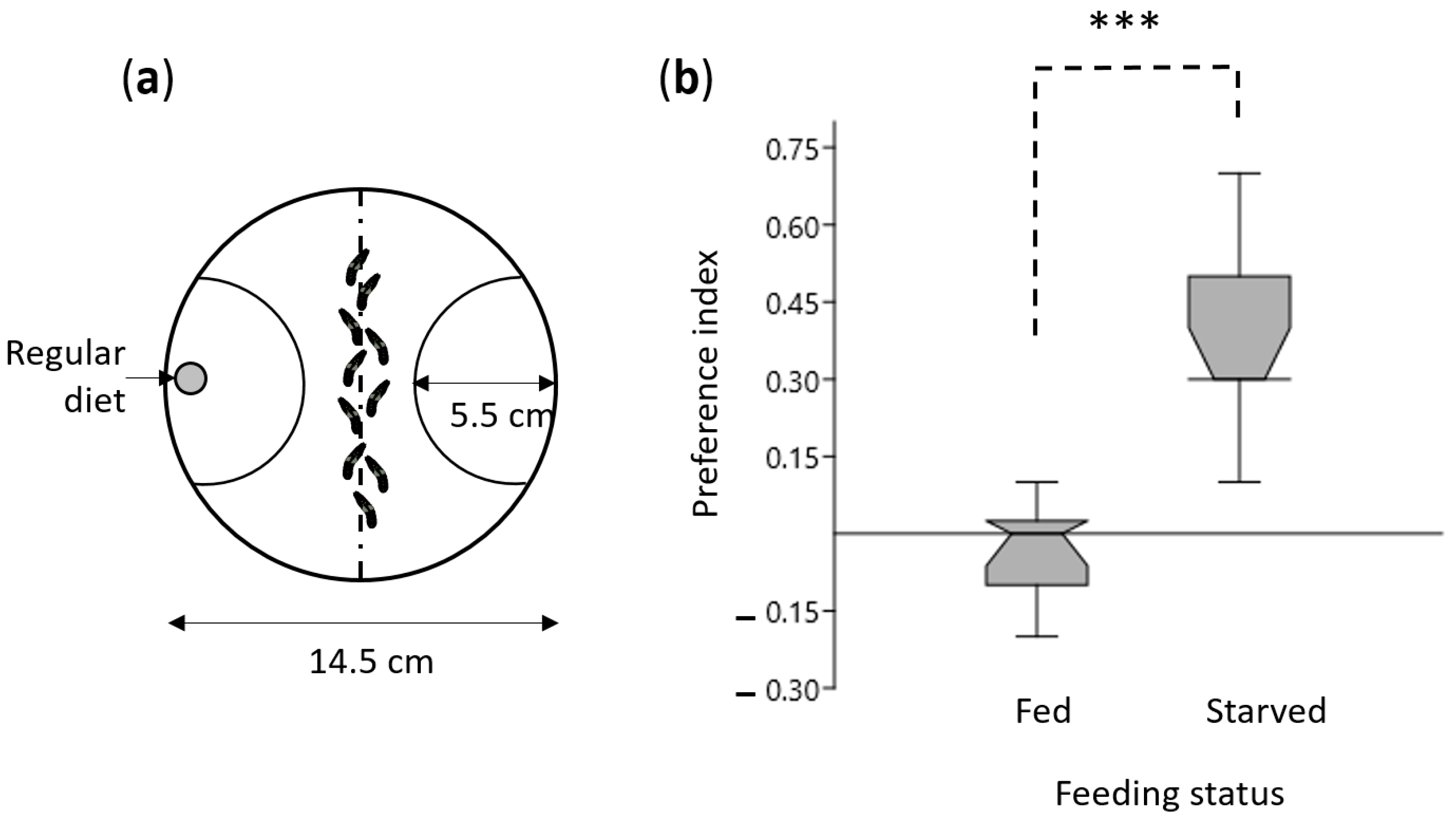
2.1. Insect Rearing and Behavioral Assay
2.2. Tissue Collection and Illumina Sequencing
2.3. Illumina Read Alignment and Statistical Analysis
2.4. Validation of RNA-Seq Data by Reverse Transcription Quantitative PCR
3. Results and Discussion
3.1. Starvation Enhanced Olfactory Behavior in S. littoralis Larvae
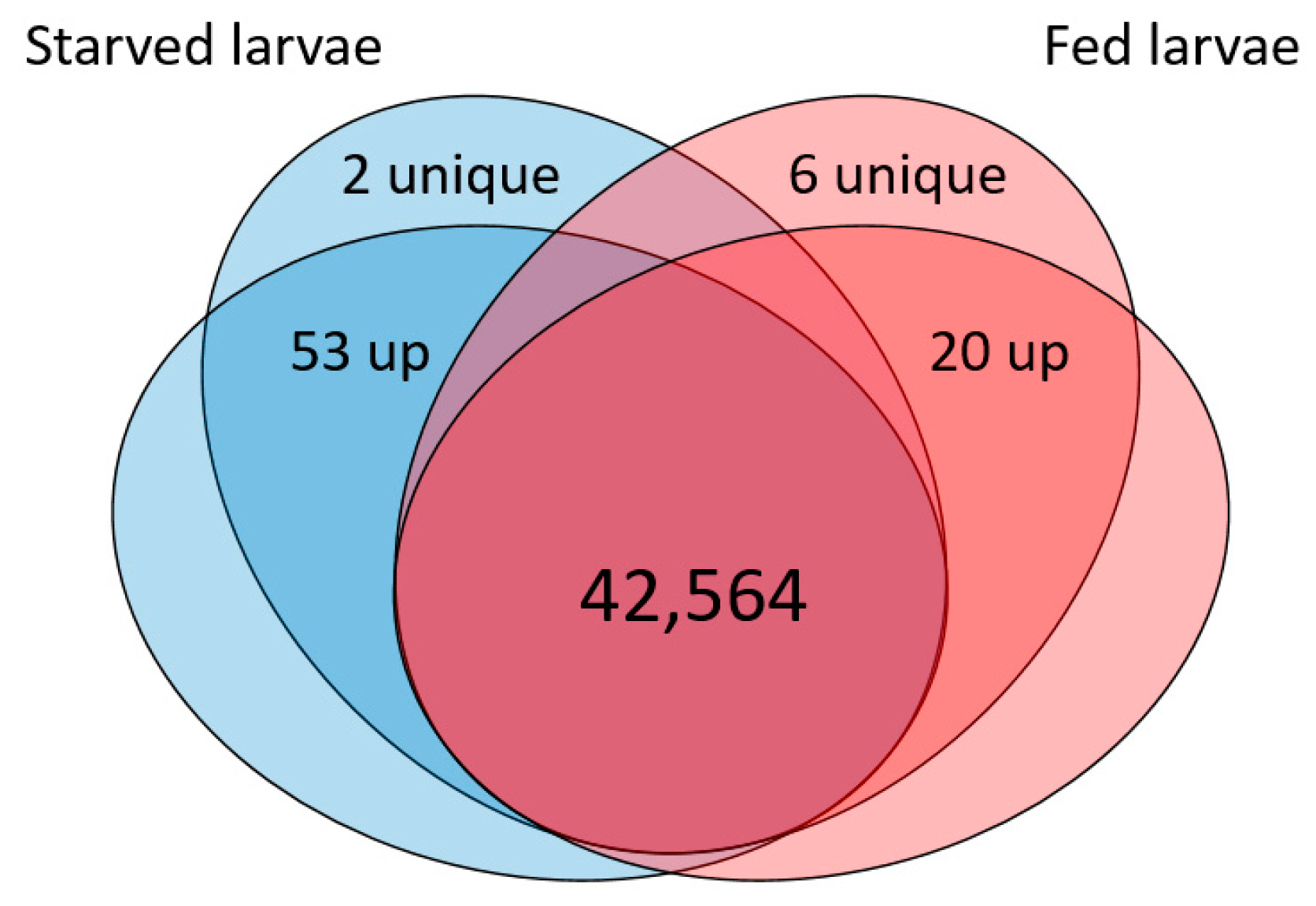
3.2. Starvation Leads to Up- and Down-Regulation of 81 Transcripts in Larval Chemosensory Organs
3.3. Starvation Affects Different Biological Functions
3.4. Starvation Modulates Peri-Receptor-Encoding but Not Chemosensory Receptor-Encoding Transcripts
3.5. qPCR Validates the RNA-Seq Expression Profiling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dobzhansky, T.; Cooper, D.M.; Phaff, H.J.; Knapp, E.P.; Carson, H.L. Studies on the ecology of Drosophila in the yosemite region of California: 4.Differential attraction of species of Drosophila to different species of yeasts. Ecology 1956, 37, 544–550. [Google Scholar] [CrossRef]
- Ruebenbauer, A.; Schlyter, F.; Hansson, B.S.; Lofstedt, C.; Larsson, M.C. Genetic variability and robustness of host odor preference in Drosophila melanogaster. Curr. Biol. 2008, 18, 1438–1443. [Google Scholar] [CrossRef]
- Massolt, E.T.; van Haard, P.M.; Rehfeld, J.F.; Posthuma, E.F.; van der Veer, E.; Schweitzer, D.H. Appetite suppression through smelling of dark chocolate correlates with changes in ghrelin in young women. Regul. Pept. 2010, 161, 81–86. [Google Scholar] [CrossRef]
- Aime, P.; Duchamp-Viret, P.; Chaput, M.A.; Savigner, A.; Mahfouz, M.; Julliard, A.K. Fasting increases and satiation decreases olfactory detection for a neutral odor in rats. Behav. Brain Res. 2007, 179, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, J.J.; de Wied, D.; Adan, R.A. Neuropeptides, food intake and body weight regulation: A hypothalamic focus. Peptides 2002, 23, 2283–2306. [Google Scholar] [CrossRef]
- Savigner, A.; Duchamp-Viret, P.; Grosmaitre, X.; Chaput, M.; Garcia, S.; Ma, M.; Palouzier-Paulignan, B. Modulation of spontaneous and odorant-evoked activity of rat olfactory sensory neurons by two anorectic peptides, insulin and leptin. J. Neurophysiol. 2009, 101, 2898–2906. [Google Scholar] [CrossRef]
- Prud’homme, M.J.; Lacroix, M.C.; Badonnel, K.; Gougis, S.; Baly, C.; Salesse, R.; Caillol, M. Nutritional status modulates behavioural and olfactory bulb Fos responses to isoamyl acetate or food odour in rats: Roles of orexins and leptin. Neuroscience 2009, 162, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Baly, C.; Aioun, J.; Badonnel, K.; Lacroix, M.C.; Durieux, D.; Schlegel, C.; Salesse, R.; Caillol, M. Leptin and its receptors are present in the rat olfactory mucosa and modulated by the nutritional status. Brain Res. 2007, 1129, 130–141. [Google Scholar] [CrossRef]
- Takken, W.; van Loon, J.J.; Adam, W. Inhibition of host-seeking response and olfactory responsiveness in Anopheles gambiae following blood feeding. J. Insect Physiol. 2001, 47, 303–310. [Google Scholar] [CrossRef]
- Fox, A.N.; Pitts, R.J.; Robertson, H.M.; Carlson, J.R.; Zwiebel, L.J. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc. Natl. Acad. Sci. USA 2001, 98, 14693–14697. [Google Scholar] [CrossRef] [PubMed]
- Siju, K.P.; Hill, S.R.; Hansson, B.S.; Ignell, R. Influence of blood meal on the responsiveness of olfactory receptor neurons in antennal sensilla trichodea of the yellow fever mosquito, Aedes aegypti. J. Insect Physiol. 2010, 56, 659–665. [Google Scholar] [CrossRef]
- Biessmann, H.; Nguyen, Q.K.; Le, D.; Walter, M.F. Microarray-based survey of a subset of putative olfactory genes in the mosquito Anopheles gambiae. Insect Mol. Biol. 2005, 14, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Rinker, D.C.; Pitts, R.J.; Zhou, X.; Suh, E.; Rokas, A.; Zwiebel, L.J. Blood meal-induced changes to antennal transcriptome profiles reveal shifts in odor sensitivities in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2013, 110, 8260–8265. [Google Scholar] [CrossRef]
- Piesik, D.; Rochat, D.; van der Pers, J.; Marion-Poll, F. Pulsed odors from maize or spinach elicit orientation in European corn borer neonate larvae. J. Chem. Ecol. 2009, 35, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Krashes, M.J.; DasGupta, S.; Vreede, A.; White, B.; Armstrong, J.D.; Waddell, S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell 2009, 139, 416–427. [Google Scholar] [CrossRef]
- Farhadian, S.F.; Suarez-Farinas, M.; Cho, C.E.; Pellegrino, M.; Vosshall, L.B. Post-fasting olfactory, transcriptional, and feeding responses in Drosophila. Physiol. Behav. 2012, 105, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Farhan, A.; Gulati, J.; Grobetae-Wilde, E.; Vogel, H.; Hansson, B.S.; Knaden, M. The CCHamide 1 receptor modulates sensory perception and olfactory behavior in starved Drosophila. Sci. Rep. 2013, 3, 2765. [Google Scholar] [CrossRef]
- Lebreton, S.; Trona, F.; Borrero-Echeverry, F.; Bilz, F.; Grabe, V.; Becher, P.G.; Carlsson, M.A.; Nassel, D.R.; Hansson, B.S.; Sachse, S.; et al. Feeding regulates sex pheromone attraction and courtship in Drosophila females. Sci. Rep. 2015, 5, 13132. [Google Scholar] [CrossRef]
- Bracker, L.B.; Siju, K.P.; Varela, N.; Aso, Y.; Zhang, M.; Hein, I.; Vasconcelos, M.L.; Grunwald Kadow, I.C. Essential role of the mushroom body in context-dependent CO(2) avoidance in Drosophila. Curr. Biol. 2013, 23, 1228–1234. [Google Scholar] [CrossRef]
- Reisenman, C.E. Hunger is the best spice: Effects of starvation in the antennal responses of the blood-sucking bug Rhodnius prolixus. J. Insect Physiol. 2014, 71, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Nassel, D.R.; Enell, L.E.; Santos, J.G.; Wegener, C.; Johard, H.A. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 2008, 9, 90. [Google Scholar] [CrossRef]
- Root, C.M.; Ko, K.I.; Jafari, A.; Wang, J.W. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 2011, 145, 133–144. [Google Scholar] [CrossRef]
- Slankster, E.; Kollala, S.; Baria, D.; Dailey-Krempel, B.; Jain, R.; Odell, S.R.; Mathew, D. Mechanism underlying starvation-dependent modulation of olfactory behavior in Drosophila larva. Sci. Rep. 2020, 10, 3119. [Google Scholar] [CrossRef] [PubMed]
- Zinke, I.; Schutz, C.S.; Katzenberger, J.D.; Bauer, M.; Pankratz, M.J. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J. 2002, 21, 6162–6173. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, K.; Takahashi, A.; Nishimura, A.; Itoh, M.; Takano-Shimizu, T.; Ozaki, M. Characteristics of genes up-regulated and down-regulated after 24 h starvation in the head of Drosophila. Gene 2009, 446, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Moskalev, A.; Zhikrivetskaya, S.; Krasnov, G.; Shaposhnikov, M.; Proshkina, E.; Borisoglebsky, D.; Danilov, A.; Peregudova, D.; Sharapova, I.; Dobrovolskaya, E.; et al. A comparison of the transcriptome of Drosophila melanogaster in response to entomopathogenic fungus, ionizing radiation, starvation and cold shock. BMC Genom. 2015, 16, S8. [Google Scholar] [CrossRef]
- Ko, K.I.; Root, C.M.; Lindsay, S.A.; Zaninovich, O.A.; Shepherd, A.K.; Wasserman, S.A.; Kim, S.M.; Wang, J.W. Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. Elife 2015, 4, e08298. [Google Scholar] [CrossRef] [PubMed]
- Meunier, N.; Belgacem, Y.H.; Martin, J.R. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J. Exp. Biol. 2007, 210, 1424–1434. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Marguerat, S.; Bahler, J. RNA-seq: From technology to biology. Cell. Mol. Life Sci. 2010, 67, 569–579. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; den Dunnen, J.T. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef] [PubMed]
- Poivet, E.; Gallot, A.; Montagné, N.; Glaser, N.; Legeai, F.; Jacquin-Joly, E. A comparison of the olfactory gene repertoires of adults and larvae in the noctuid moth Spodoptera littoralis. PLoS ONE 2013, 8, e60263. [Google Scholar] [CrossRef]
- Cain, M.L.; Eccleston, J.; Kareiva, P.M. The influence of food plant dispersion on caterpillar searching success. Ecol. Entomol. 1985, 10, 1–7. [Google Scholar] [CrossRef]
- Legeai, F.; Malpel, S.; Montagne, N.; Monsempes, C.; Cousserans, F.; Merlin, C.; Francois, M.C.; Maibeche-Coisne, M.; Gavory, F.; Poulain, J.; et al. An Expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: A resource for olfactory and pheromone detection research. BMC Genom. 2011, 12, 86. [Google Scholar] [CrossRef]
- Jacquin-Joly, E.; Legeai, F.; Montagné, N.; Monsempes, C.; François, M.C.; Poulain, J.; Gavory, F.; Walker III, W.B.; Hansson, B.S.; Larsson, M.C. Candidate chemosensory Genes In Female Antennae Of The Noctuid Moth Spodoptera littoralis. Int. J. Biol. Sci. 2012, 8, 1036. [Google Scholar] [CrossRef]
- Poitout, S.; Buès, R. Elevage de chenilles de vingt-huit espèces de Lépidoptères Noctuidae et de deux espèces d’Arctiidae sur milieu artificiel simple. Particularités de l’élevage selon les espèces. Ann. Zool. Ecol. Anim. 1974, 6, 431–441. [Google Scholar]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B-Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Poivet, E.; Rharrabe, K.; Monsempes, C.; Glaser, N.; Rochat, D.; Renou, M.; Marion-Poll, F.; Jacquin-Joly, E. The use of the sex pheromone as an evolutionary solution to food source selection in caterpillars. Nat. Commun. 2012, 3, 1047. [Google Scholar] [CrossRef]
- Rharrabe, K.; Jacquin-Joly, E.; Marion-Poll, F. Electrophysiological and behavioral responses of Spodoptera littoralis caterpillars to attractive and repellent plant volatiles. Front. Ecol. Evol. 2014, 2, 5. [Google Scholar] [CrossRef]
- Panagabko, C.; Morley, S.; Hernandez, M.; Cassolato, P.; Gordon, H.; Parsons, R.; Manor, D.; Atkinson, J. Ligand specificity in the CRAL-TRIO protein family. Biochemistry 2003, 42, 6467–6474. [Google Scholar] [CrossRef]
- Raikhel, N.V.; Lee, H.I.; Broekaert, W.F. Structure and Function of Chitin-Binding Proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 591–615. [Google Scholar] [CrossRef]
- Navarro-Cerrillo, G.; Hernandez-Martinez, P.; Vogel, H.; Ferre, J.; Herrero, S. A new gene superfamily of pathogen-response (repat) genes in Lepidoptera: Classification and expression analysis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2013, 164, 10–17. [Google Scholar] [CrossRef]
- Sarov-Blat, L.; So, W.V.; Liu, L.; Rosbash, M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell 2000, 101, 647–656. [Google Scholar] [CrossRef]
- Touhara, K.; Vosshall, L.B. Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 2009, 71, 307–332. [Google Scholar] [CrossRef]
- Silbering, A.F.; Rytz, R.; Grosjean, Y.; Abuin, L.; Ramdya, P.; Jefferis, G.S.; Benton, R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 2011, 31, 13357–13375. [Google Scholar] [CrossRef]
- Benton, R.; Vannice, K.S.; Gomez-Diaz, C.; Vosshall, L.B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 2009, 136, 149–162. [Google Scholar] [CrossRef]
- Chen, Q.; Pei, D.; Li, J.; Jing, C.; Wu, W.; Man, Y. The antenna transcriptome changes in mosquito Anopheles sinensis, pre- and post- blood meal. PLoS ONE 2017, 12, e0181399. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 2018, 93, 184–200. [Google Scholar] [CrossRef]
- Badonnel, K.; Denis, J.B.; Caillol, M.; Monnerie, R.; Piumi, F.; Potier, M.C.; Salesse, R.; Baly, C. Transcription profile analysis reveals that OBP-1F mRNA is downregulated in the olfactory mucosa following food deprivation. Chem. Senses 2007, 32, 697–710. [Google Scholar] [CrossRef][Green Version]
- Durand, N.; Carot-Sans, G.; Bozzolan, F.; Rosell, G.; Siaussat, D.; Debernard, S.; Chertemps, T.; Maibeche-Coisne, M. Degradation of pheromone and plant volatile components by a same odorant-degrading enzyme in the cotton leafworm, Spodoptera littoralis. PLoS ONE 2011, 6, e29147. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.; Carot-Sans, G.; Chertemps, T.; Bozzolan, F.; Party, V.; Renou, M.; Debernard, S.; Rosell, G.; Maïbèche-Coisne, M. Characterization of an antennal carboxylesterase from the pest moth Spodoptera littoralis degrading a host plant odorant. PLoS ONE 2010, 5, e15026. [Google Scholar] [CrossRef]
- Maïbeche-Coisné, M.; Nikonov, A.A.; Ishida, Y.; Jacquin-Joly, E.; Leal, W.S. Pheromone anosmia in a scarab beetle induced by in vivo inhibition of a pheromone-degrading enzyme. Proc. Natl. Acad. Sci. USA 2004, 101, 11459–11464. [Google Scholar] [CrossRef]
- Durand, N.; Carot-Sans, G.; Chertemps, T.; Montagne, N.; Jacquin-Joly, E.; Debernard, S.; Maibeche-Coisne, M. A diversity of putative carboxylesterases are expressed in the antennae of the noctuid moth Spodoptera littoralis. Insect Mol. Biol. 2010, 19, 87–97. [Google Scholar] [CrossRef]
- Aanes, H.; Winata, C.; Moen, L.F.; Ostrup, O.; Mathavan, S.; Collas, P.; Rognes, T.; Alestrom, P. Normalization of RNA-sequencing data from samples with varying mRNA levels. PLoS ONE 2014, 9, e89158. [Google Scholar] [CrossRef]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
| FLAP | SLAP | |
|---|---|---|
| Number of raw reads | 1,947,899 | 2,389,809 |
| Number of processed reads | 1,807,931 | 2,171,664 |
| Number of mapped reads | 1,513,384 | 1,813,092 |
| Gene Name and ID | qPCR Relative Expression Level in Fed Larvae | qPCR Relative Expression Level in Starved Larvae | qPCR Relative Fold Change (Starved/Fed) | RNA-Seq Fold Change (Starved/Fed) |
|---|---|---|---|---|
| ODE (c6022) | 1.12 × 10−3 ± 3.85 × 10−4 | 5.09 × 10−3 ± 2.77 × 10−3 | 3.23 ± 1.00 | 6.00 |
| CSP (c997) | 0.06 ± 0.03 | 0.29 ± 0.20 | 6.98 ± 2.33 | 9.14 |
| CSP (c65324) | 0.13 ± 0.07 | 0.59 ± 0.33 | 5.42 ± 2.31 | 11.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poivet, E.; Gallot, A.; Montagné, N.; Senin, P.; Monsempès, C.; Legeai, F.; Jacquin-Joly, E. Transcriptome Profiling of Starvation in the Peripheral Chemosensory Organs of the Crop Pest Spodoptera littoralis Caterpillars. Insects 2021, 12, 573. https://doi.org/10.3390/insects12070573
Poivet E, Gallot A, Montagné N, Senin P, Monsempès C, Legeai F, Jacquin-Joly E. Transcriptome Profiling of Starvation in the Peripheral Chemosensory Organs of the Crop Pest Spodoptera littoralis Caterpillars. Insects. 2021; 12(7):573. https://doi.org/10.3390/insects12070573
Chicago/Turabian StylePoivet, Erwan, Aurore Gallot, Nicolas Montagné, Pavel Senin, Christelle Monsempès, Fabrice Legeai, and Emmanuelle Jacquin-Joly. 2021. "Transcriptome Profiling of Starvation in the Peripheral Chemosensory Organs of the Crop Pest Spodoptera littoralis Caterpillars" Insects 12, no. 7: 573. https://doi.org/10.3390/insects12070573
APA StylePoivet, E., Gallot, A., Montagné, N., Senin, P., Monsempès, C., Legeai, F., & Jacquin-Joly, E. (2021). Transcriptome Profiling of Starvation in the Peripheral Chemosensory Organs of the Crop Pest Spodoptera littoralis Caterpillars. Insects, 12(7), 573. https://doi.org/10.3390/insects12070573








