CRISPR-Mediated Endogenous Activation of Fibroin Heavy Chain Gene Triggers Cellular Stress Responses in Bombyx mori Embryonic Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
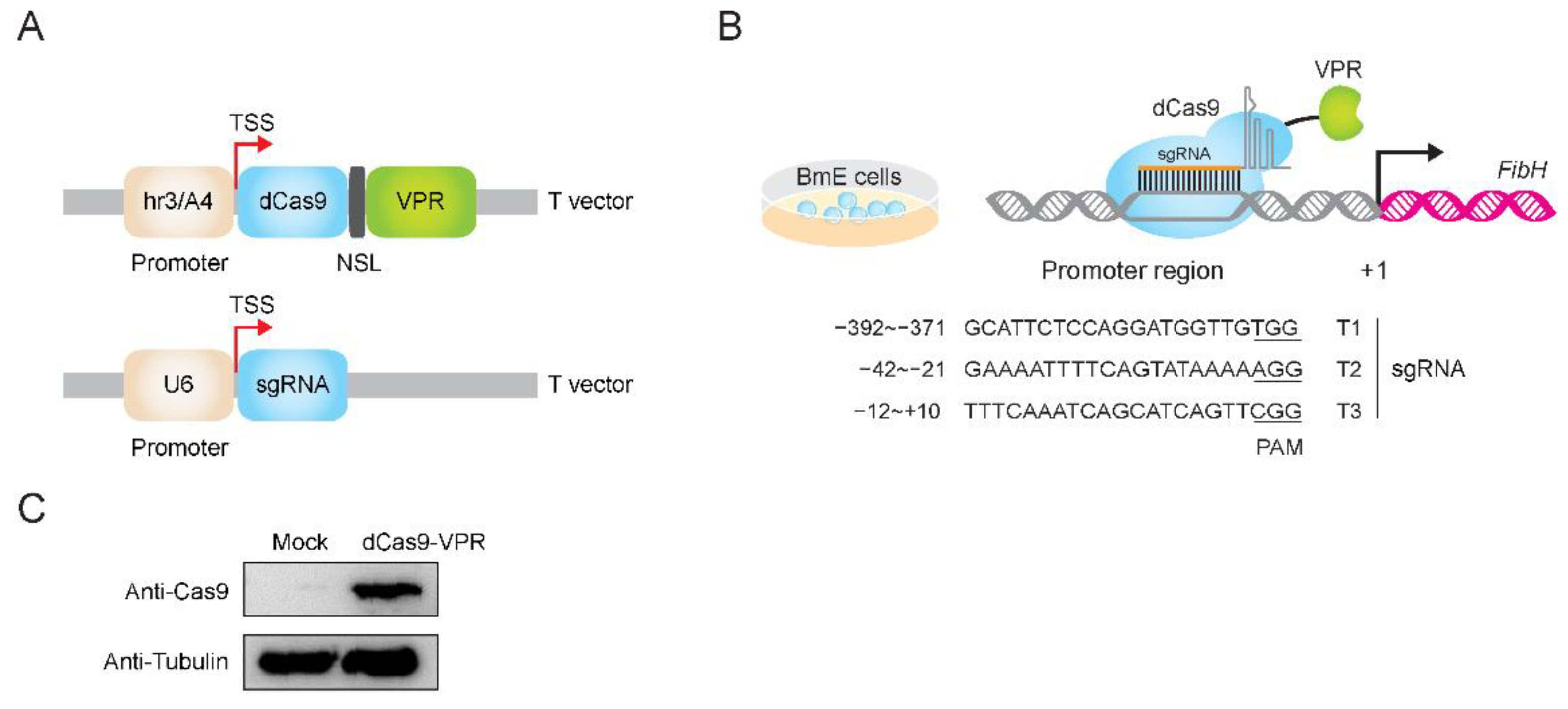
2.1. Vector Construction
2.2. Cell Transfection
2.3. Western Blot Assay
2.4. Total RNA Extraction
2.5. Quantitative RT-PCR
2.6. Autophagy Assay
2.7. Statistical Analysis
3. Results
3.1. Construction of dCas9 and sgRNA Expression System
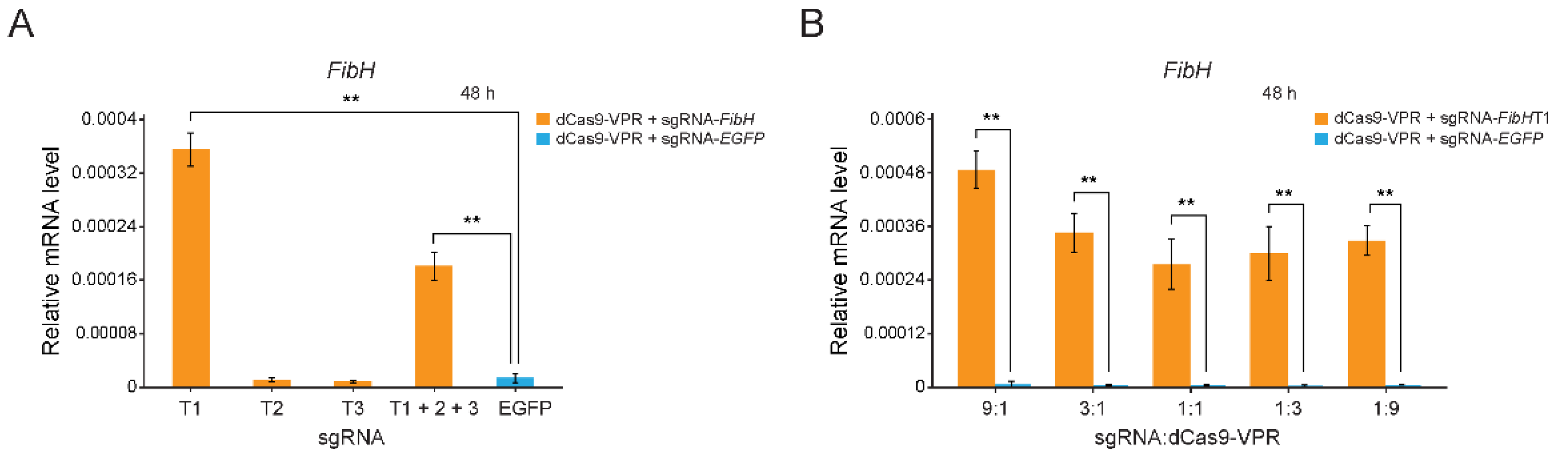
3.2. The Endogenous FibH Gene of BmE Cells Was Activated by CRISPRa System
3.3. Cellular Stress Responses-Related Genes Were Significantly Up-Regulated
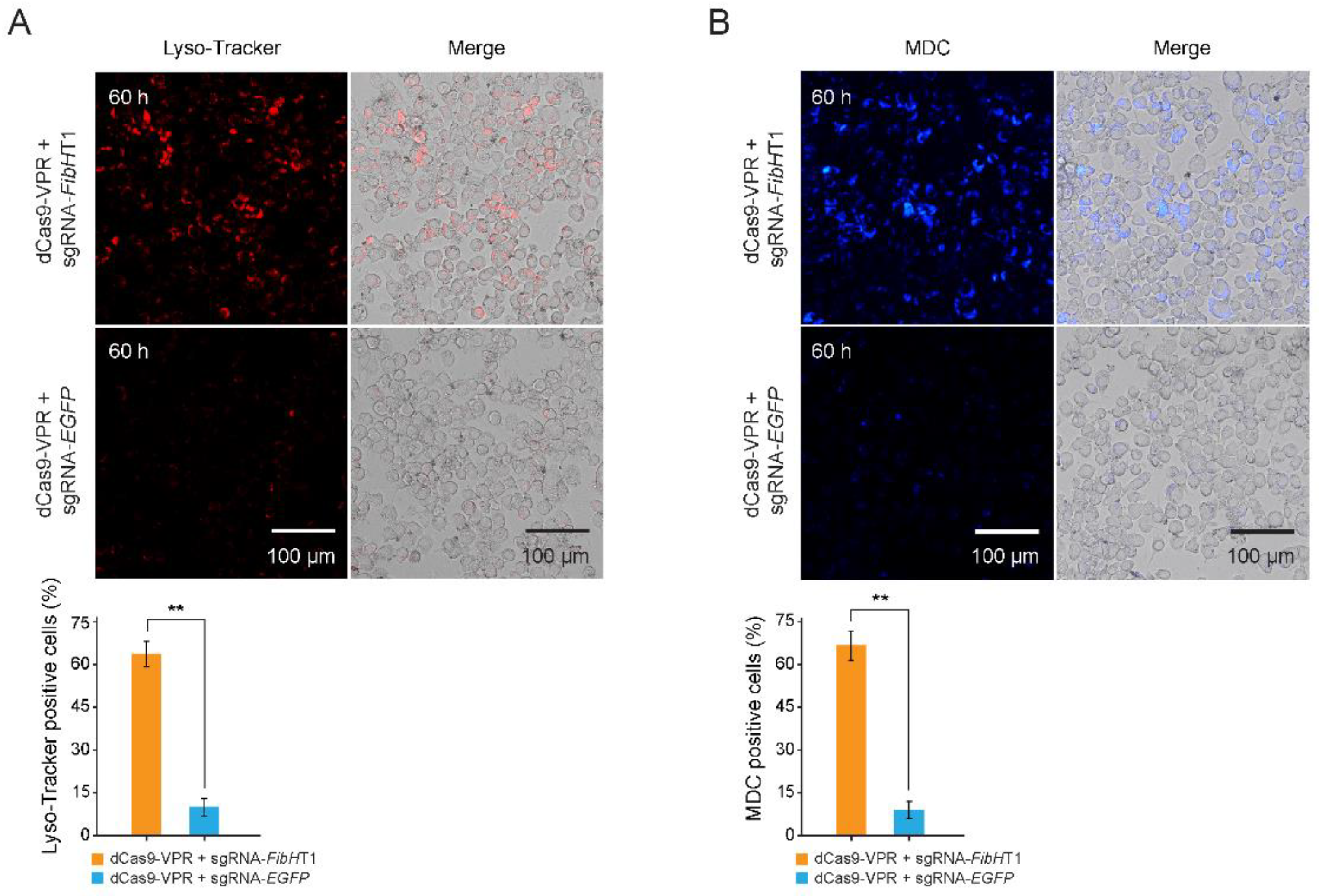
3.4. Autophagy Was Triggered in FibH-Activated BmE Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jiang, W.Y.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Olarte, R.D.; Bravo Rodriguez, R.; Morales-Rios, E. Genome Editing in Bacteria: CRISPR-Cas and Beyond. Microorganisms 2021, 9, 844. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.P.; Wang, G.H.; Ma, S.Y.; Xie, X.D.; Wu, X.W.; Zhang, X.T.; Wu, Y.Q.; Zhao, P.; Xia, Q.Y. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant. Mol. Biol. 2015, 87, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhang, T.; Xu, B.; Jia, L.; Xiao, B.; Liu, H.; Liu, L.; Yan, H.; Xia, Q. CRISPR/Cas9-Mediated Mutagenesis of Carotenoid Cleavage Dioxygenase 8 (CCD8) in Tobacco Affects Shoot and Root Architecture. Int. J. Mol. Sci. 2018, 19, 1062. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J.L. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Rep. 2013, 4, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.S.; Ren, M.D.; Wang, Z.X.; Zhang, B.; Rong, Y.K.S.; Jiao, R.J.; Gao, G.J. Highly Efficient Genome Modifications Mediated by CRISPR/Cas9 in Drosophila. Genetics 2013, 195, 289–291. [Google Scholar] [CrossRef] [Green Version]
- Gantz, V.M.; Akbari, O.S. Gene editing technologies and applications for insects. Curr. Opin. Insect Sci. 2018, 28, 66–72. [Google Scholar] [CrossRef]
- Hruscha, A.; Krawitz, P.; Rechenberg, A.; Heinrich, V.; Hecht, J.; Haass, C.; Schmid, B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 2013, 140, 4982–4987. [Google Scholar] [CrossRef] [Green Version]
- Jao, L.E.; Wente, S.R.; Chen, W.B. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, P.; Yang, L.H.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 2020, 16, 465–480. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Ma, S.Y.; Wang, X.G.; Chang, J.S.; Gao, J.; Shi, R.; Zhang, J.D.; Lu, W.; Liu, Y.; Zhao, P.; et al. Highly efficient multiplex targeted mutagenesis and genomic structure variation in Bombyx mori cells using CRISPR/Cas9. Insect Biochem. Mol. Biol. 2014, 49, 35–42. [Google Scholar] [CrossRef]
- Ma, S.; Chang, J.; Wang, X.; Liu, Y.; Zhang, J.; Lu, W.; Gao, J.; Shi, R.; Zhao, P.; Xia, Q. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci. Rep. 2014, 4, 4489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.Y.; Liu, Y.; Liu, Y.Y.; Chang, J.S.; Zhang, T.; Wang, X.G.; Shi, R.; Lu, W.; Xia, X.J.; Zhao, P.; et al. An integrated CRISPR Bombyx mori genome editing system with improved efficiency and expanded target sites. Insect Biochem. Mol. Biol. 2017, 83, 13–20. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, U326–U365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.G.; Ma, S.Y.; Chang, J.S.; Shi, R.; Wang, R.L.; Zhao, P.; Xia, Q.Y. Programmable activation of Bombyx gene expression using CRISPR/dCas9 fusion systems. Insect Sci. 2019, 26, 983–990. [Google Scholar] [CrossRef]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef]
- Omenetto, F.G.; Kaplan, D.L. New opportunities for an ancient material. Science 2010, 329, 528–531. [Google Scholar] [CrossRef] [Green Version]
- Shao, Z.; Vollrath, F. Surprising strength of silkworm silk. Nature 2002, 418, 741. [Google Scholar] [CrossRef]
- Xia, Q.; Li, S.; Feng, Q. Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu. Rev. Entomol. 2014, 59, 513–536. [Google Scholar] [CrossRef]
- Andersson, M.; Johansson, J.; Rising, A. Silk Spinning in Silkworms and Spiders. Int. J. Mol. Sci. 2016, 17, 1290. [Google Scholar] [CrossRef] [Green Version]
- Aramwit, P.; Siritientong, T.; Srichana, T. Potential applications of silk sericin, a natural protein from textile industry by-products. Waste Manag. Res. 2012, 30, 217–224. [Google Scholar] [CrossRef]
- Inoue, S.; Tanaka, K.; Arisaka, F.; Kimura, S.; Ohtomo, K.; Mizuno, S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6:6:1 molar ratio. J. Biol. Chem. 2000, 275, 40517–40528. [Google Scholar] [CrossRef] [Green Version]
- Takei, F.; Kikuchi, Y.; Kikuchi, A.; Mizuno, S.; Shimura, K. Further evidence for importance of the subunit combination of silk fibroin in its efficient secretion from the posterior silk gland cells. J. Cell Biol. 1987, 105, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Mori, K.; Tanaka, K.; Kikuchi, Y.; Waga, M.; Waga, S.; Mizuno, S. Production of a chimeric fibroin light-chain polypeptide in a fibroin secretion-deficient naked pupa mutant of the silkworm Bombyx mori. J. Mol. Biol. 1995, 251, 217–228. [Google Scholar] [CrossRef]
- Tanaka, K.; Kajiyama, N.; Ishikura, K.; Waga, S.; Kikuchi, A.; Ohtomo, K.; Takagi, T.; Mizuno, S. Determination of the site of disulfide linkage between heavy and light chains of silk fibroin produced by Bombyx mori. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1999, 1432, 92–103. [Google Scholar] [CrossRef]
- Inoue, S.; Tanaka, K.; Tanaka, H.; Ohtomo, K.; Kanda, T.; Imamura, M.; Quan, G.X.; Kojima, K.; Yamashita, T.; Nakajima, T.; et al. Assembly of the silk fibroin elementary unit in endoplasmic reticulum and a role of L-chain for protection of alpha1,2-mannose residues in N-linked oligosaccharide chains of fibrohexamerin/P25. Eur. J. Biochem. 2004, 271, 356–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, W.; Chen, Y.; Lin, Y.; Xia, Q. Developmental and transcriptomic features characterize defects of silk gland growth and silk production in silkworm naked pupa mutant. Insect Biochem. Mol. Biol. 2019, 111, 103175. [Google Scholar] [CrossRef]
- Hu, W.; Lu, W.; Wei, L.; Zhang, Y.; Xia, Q. Molecular nature of dominant naked pupa mutation reveals novel insights into silk production in Bombyx mori. Insect Biochem. Mol. Biol. 2019, 109, 52–62. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.H.; Xiao, S.Q.; Chen, M.; Hong, X.J.; Lu, C. Establishment and characterization of two embryonic cell lines of Bombyx mori. Vitr. Cell. Dev. Biol. Anim. 2007, 43, 101–104. [Google Scholar] [CrossRef]
- Wang, X.; Ma, S.; Liu, Y.; Lu, W.; Sun, L.; Zhao, P.; Xia, Q. Transcriptional repression of endogenous genes in BmE cells using CRISPRi system. Insect Biochem. Mol. Biol. 2019, 111, 103172. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A.; Bukau, B.; Sommer, T. Protein quality control in the cytosol and the endoplasmic reticulum: Brothers in arms. Mol. Cell 2010, 40, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, Z.Y.; Li, W.F.; Li, Q.R.; Deng, X.J.; Yang, W.Y.; Cao, Y.; Zhou, C.Z. Systematic cloning and analysis of autophagy-related genes from the silkworm Bombyx mori. BMC Mol. Biol. 2009, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Eisoldt, L.; Thamm, C.; Scheibel, T. Review the role of terminal domains during storage and assembly of spider silk proteins. Biopolymers 2012, 97, 355–361. [Google Scholar] [CrossRef]
- He, Y.X.; Zhang, N.N.; Li, W.F.; Jia, N.; Chen, B.Y.; Zhou, K.; Zhang, J.; Chen, Y.; Zhou, C.Z. N-Terminal domain of Bombyx mori fibroin mediates the assembly of silk in response to pH decrease. J. Mol. Biol. 2012, 418, 197–207. [Google Scholar] [CrossRef]
- Zhou, C.Z.; Confalonieri, F.; Medina, N.; Zivanovic, Y.; Esnault, C.; Yang, T.; Jacquet, M.; Janin, J.; Duguet, M.; Perasso, R.; et al. Fine organization of Bombyx mori fibroin heavy chain gene. Nucleic Acids Res. 2000, 28, 2413–2419. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Liu, C.; Cheng, T.; Li, W.; Wang, N.; Xia, Q. Histomorphometric and transcriptomic features characterize silk glands’ development during the molt to intermolt transition process in silkworm. Insect Biochem. Mol. Biol. 2016, 76, 95–108. [Google Scholar] [CrossRef]
- Ellgaard, L.; Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003, 4, 181–191. [Google Scholar] [CrossRef]
- Gawron, K. Endoplasmic reticulum stress in chondrodysplasias caused by mutations in collagen types II and X. Cell Stress Chaperones 2016, 21, 943–958. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 426, 895–899. [Google Scholar] [CrossRef]
- Cui, Y.; Zhu, Y.; Lin, Y.; Chen, L.; Feng, Q.; Wang, W.; Xiang, H. New insight into the mechanism underlying the silk gland biological process by knocking out fibroin heavy chain in the silkworm. BMC Genom. 2018, 19, 215. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; Wang, X.; Ma, S.; Peng, Z.; Cao, Y.; Xia, Q. CRISPR-Mediated Endogenous Activation of Fibroin Heavy Chain Gene Triggers Cellular Stress Responses in Bombyx mori Embryonic Cells. Insects 2021, 12, 552. https://doi.org/10.3390/insects12060552
Hu W, Wang X, Ma S, Peng Z, Cao Y, Xia Q. CRISPR-Mediated Endogenous Activation of Fibroin Heavy Chain Gene Triggers Cellular Stress Responses in Bombyx mori Embryonic Cells. Insects. 2021; 12(6):552. https://doi.org/10.3390/insects12060552
Chicago/Turabian StyleHu, Wenbo, Xiaogang Wang, Sanyuan Ma, Zhangchuan Peng, Yang Cao, and Qingyou Xia. 2021. "CRISPR-Mediated Endogenous Activation of Fibroin Heavy Chain Gene Triggers Cellular Stress Responses in Bombyx mori Embryonic Cells" Insects 12, no. 6: 552. https://doi.org/10.3390/insects12060552
APA StyleHu, W., Wang, X., Ma, S., Peng, Z., Cao, Y., & Xia, Q. (2021). CRISPR-Mediated Endogenous Activation of Fibroin Heavy Chain Gene Triggers Cellular Stress Responses in Bombyx mori Embryonic Cells. Insects, 12(6), 552. https://doi.org/10.3390/insects12060552






