The First Mitogenomes of the Subfamily Odontiinae (Lepidoptera, Crambidae) and Phylogenetic Analysis of Pyraloidea
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Genomic DNA Extraction
2.2. High throughout Sequencing
2.3. Data Assemble and Annotation
2.4. Statistics of the Odontiine Mitochondrial Genomes
2.5. Phylogenetic Analyses
3. Results and Discussion
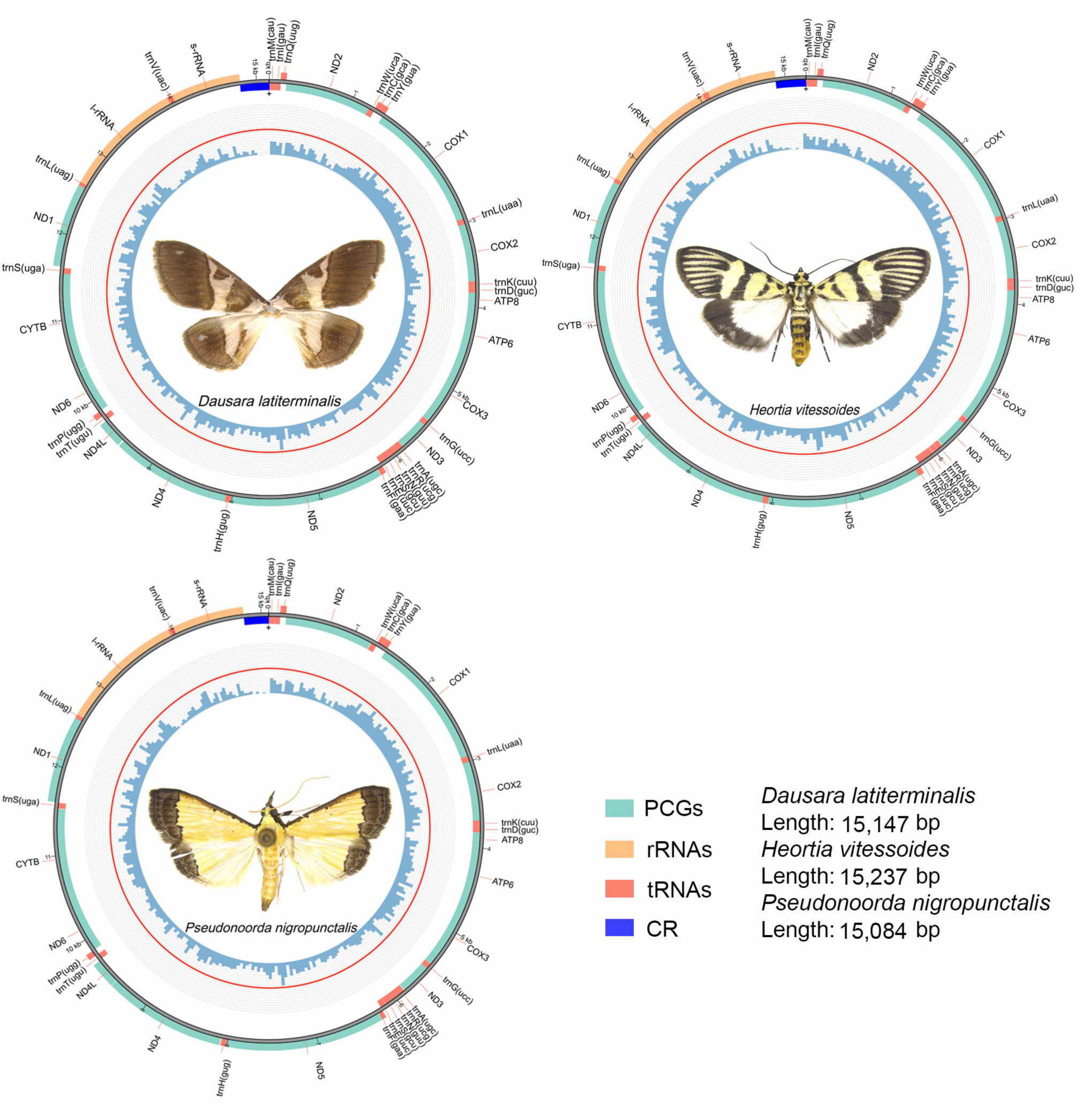
3.1. Mitogenome Organization and Nucleotide Composition of Odontiinae
3.2. Protein-Coding Genes of Odontiinae
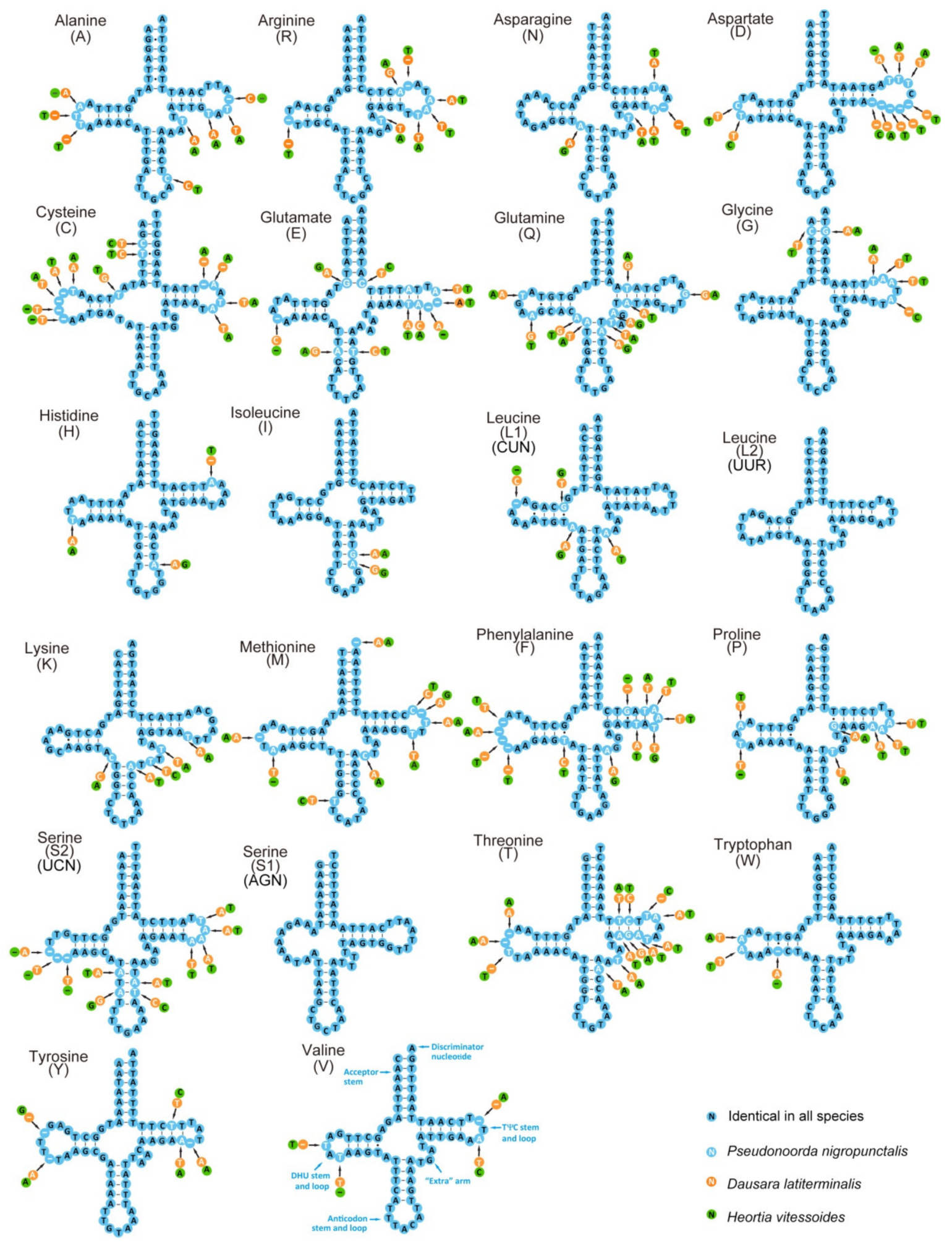
3.3. RNA Genes of Odontiinae
3.4. A + T-Rich Region of Odontiinae
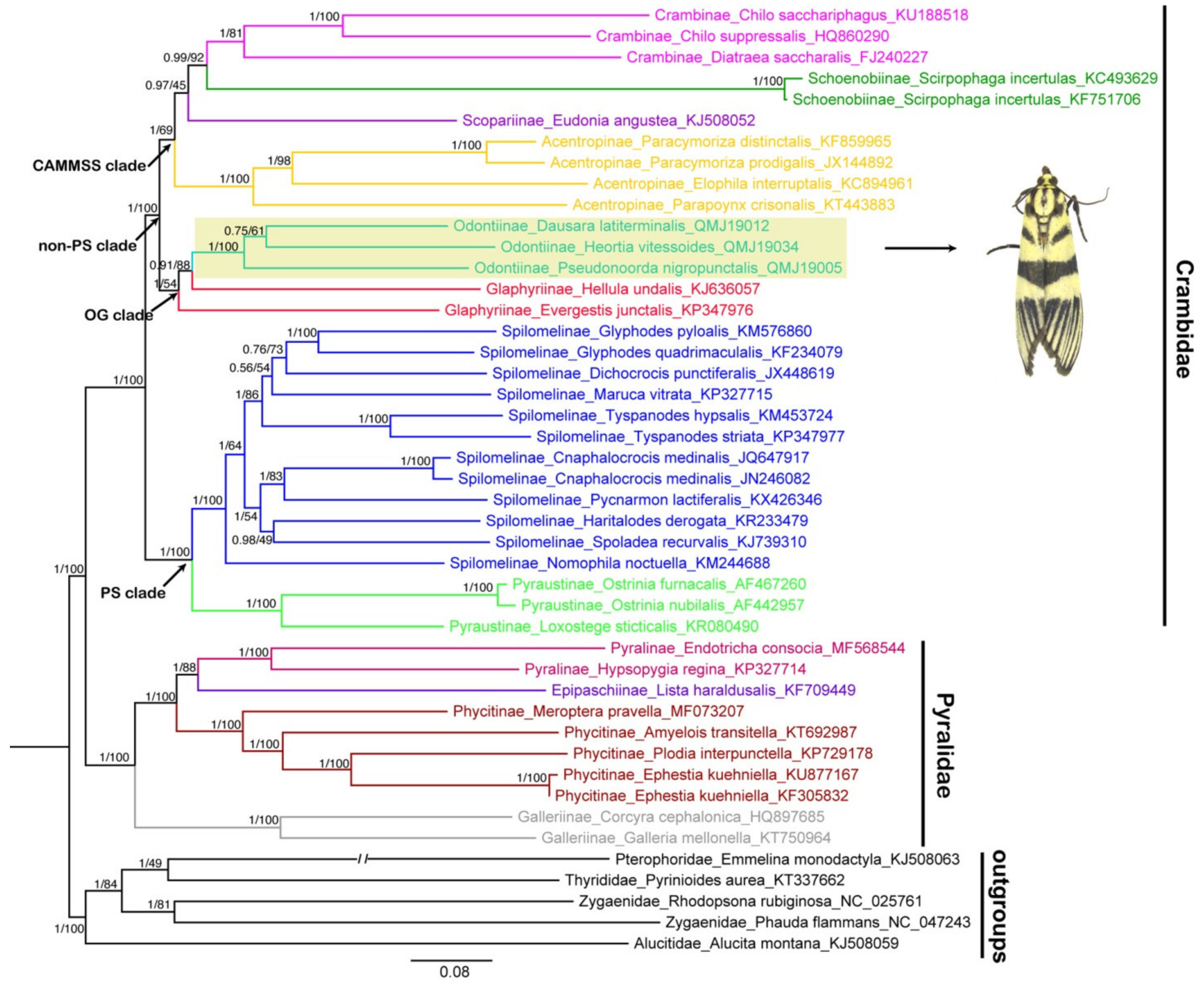
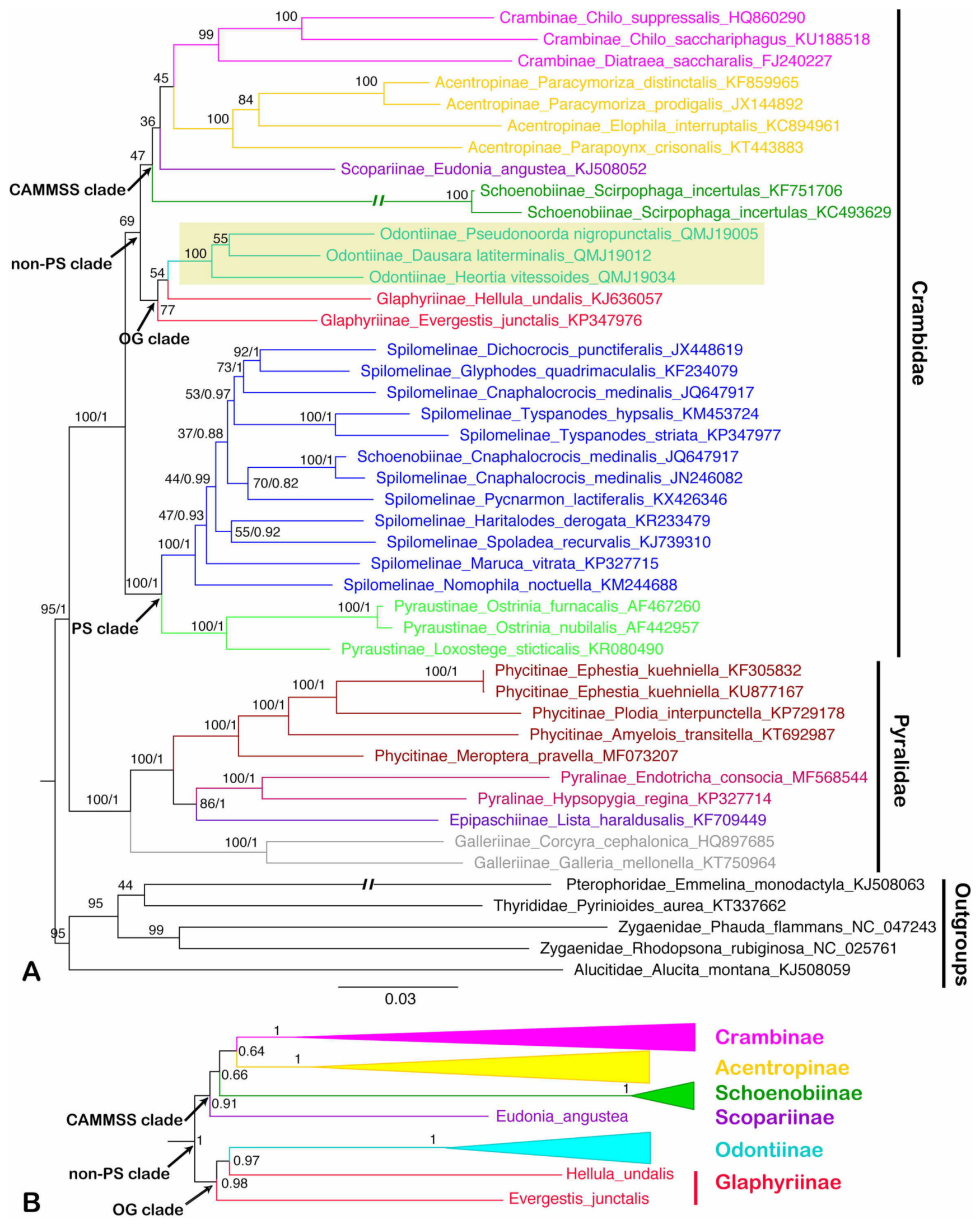
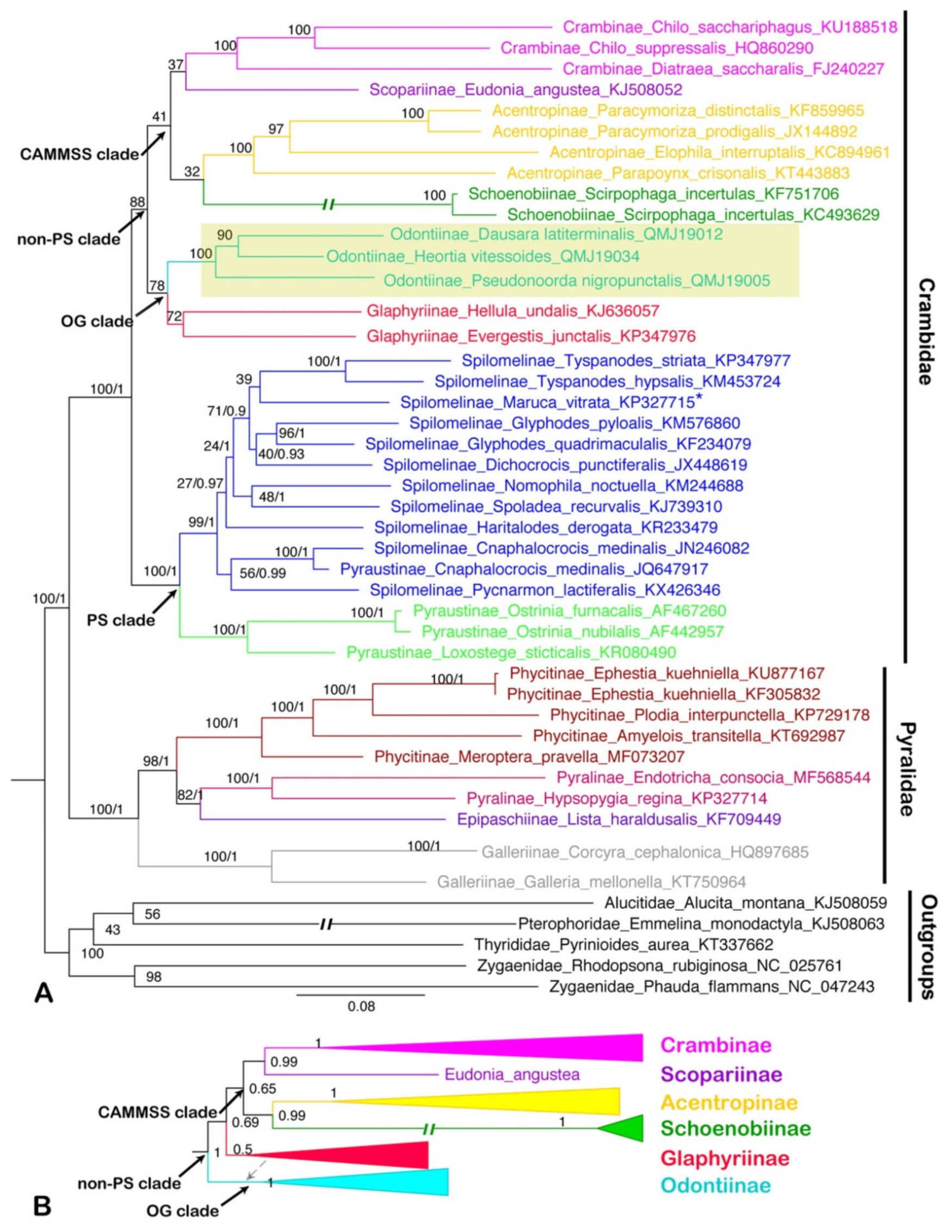
3.5. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munroe, E.; Solis, M.A. Pyraloidea. In Lepidoptera, Moths and Butterflies, Volume 1: Evolution, Systematics, and Biogeography. Handbook of Zoology; Kristensen, N., Ed.; Walter de Gruyter: Berlin, Germany; New York, NY, USA, 1999; Volume IV, pp. 233–256. ISBN 3-11-015704-7. [Google Scholar]
- Solis, M.A. Phylogenetic studies and modern classification of the Pyraloidea (Lepidoptera). Rev. Colomb. Entomol. 2007, 33, 1–8. [Google Scholar] [CrossRef]
- Hayden, J.E. Revision of Cliniodes Guenée (Lepidoptera: Crambidae: Odontiinae). Ann. Carnegie Mus. 2011, 79, 231–347. [Google Scholar] [CrossRef]
- Léger, T.; Mally, R.; Neinhuis, C.; Nuss, M. Refining the phylogeny of Crambidae with complete sampling of subfamilies (Lepidoptera, Pyraloidea). Zool. Scr. 2021, 50, 84–99. [Google Scholar] [CrossRef]
- Nuss, M.; Landry, B.; Mally, R.; Vegliante, F.; Tränkner, A.; Bauer, F.; Hayden, J.; Segerer, A.; Schouten, R.; Li, H.; et al. Global Information System on Pyraloidea. Available online: http://www.pyraloidea.org (accessed on 30 January 2021).
- Chen, Z.; Li, D.; Wang, L.; Li, Y.; Huang, X.; Qin, C. Studies on biological characteristics of Heortia vitessoides Moore on Aquilaris sinensis. China Plant Protect. 2011, 11, 10–14. [Google Scholar]
- Zhang, J.; Zhang, R.; Zeng, H.; Zhang, Z.; Wu, Q. Study on biological characteristics and occurrence regularity of Pseudonoorda minor. J. Southwest For. Univ. 2009, 29, 71–74. [Google Scholar]
- Regier, J.; Mitter, C.; Solis, M.; Hayden, J.; Landry, B.; Nuss, M.; Simonsen, T.; Yen, S.-H.; Zwick, A.; Cummings, M. A molecular phylogeny for the pyraloid moths (Lepidoptera: Pyraloidea) and its implications for higher-level classification. Syst. Entomol. 2012, 37, 635–656. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef]
- Wang, I.J. Recognizing the temporal distinctions between landscape genetics and phylogeography. Mol. Ecol. 2010, 19, 2605–2608. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Lees, D.C.; Simonsen, T.J. Towards a mitogenomic phylogeny of Lepidoptera. Mol. Phylogenet. Evol. 2014, 79, 169–178. [Google Scholar] [CrossRef]
- Li, H.; Leavengood, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.; Zhou, X.; Cai, W. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. R. Soc. B 2017, 284, 20171223. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, F.; Lan, X.; You, P. Complete mitochondrial genomes of three Spilomelinae species and a preliminary phylogenetic analysis of the Pyraloidea (Insecta: Lepidoptera). Chin. J. Appl. Entomol. 2017, 54, 22–34. [Google Scholar]
- Chen, S.; Zhang, H.; Wu, S.; You, P. Sequencing and analysis of the complete mitochondrial genome of Diaphania perspectalis and Pleuroptya inferior (Insecta: Lepidoptera). J. Shaanxi Norm. Univ. 2018, 45, 65–75. [Google Scholar]
- Zhu, W.; Yan, J.; Song, J.; You, P. The first mitochondrial genomes for Pyralinae (Pyralidae) and Glaphyriinae (Crambidae), with phylogenetic implications of Pyraloidea. PLoS ONE 2018, 13, e0194672. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Jiang, X.; Hou, X.; Yang, H.; Chen, W. The mitochondrial genome of Ephestia elutella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA Part B 2018, 3, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Song, L.; Mao, J.; Shi, Y.; Wu, C.; Zhang, Y.; Huang, L.; Peng, W.; Liu, X. Complete mitochondrial genome of the soybean leaffolder, Omiodes indicata (Lepidoptera: Pyraloidea: Crambidae), and phylogenetic analysis for Pyraloidea. Int. J. Biol. Macromol. 2018, 115, 53–60. [Google Scholar] [CrossRef]
- Yang, L.; Dai, J.; Gao, Q.; Yuan, G.; Liu, J.; Sun, Y.; Wang, L.; Qian, C.; Zhu, B.; Liu, C.; et al. Characterization of the complete mitochondrial genome of Orthaga olivacea Warre (Lepidoptera Pyralidae) and comparison with other Lepidopteran insects. PLoS ONE 2020, 15, e0227831. [Google Scholar] [CrossRef]
- Meng, G.; Li, Y.; Yang, C.; Liu, S. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, L.; Chen, J.; You, P. The complete mitochondrial genome of Amesia sanguiflua (Lepidoptera, Zygaenidae). Mitochondrial DNA Part B 2020, 5, 988–989. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, J.; Gao, Q.; Wilson, J.J.; Zhang, A. Mitochondrial phylogeny and comparative mitogenomics of closely related pine moth pests (Lepidoptera: Dendrolimus). PeerJ 2019, 7, e7317. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; You, P. Complete mitochondrial genome of Camptochilus aurea (Lepidoptera: Thyrididae). Mitochondrial DNA 2015, 27, 4528–4529. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Ali, M.; Almaden, J.; Balchan, N.; Bennici, C.E.; Bhasin, J.; Brown, C.; Carlson, H.; Chavda, A.; Deckert, J.; Eastman, T.; et al. The complete mitochondrial genome of the lesser aspen webworm moth Meroptera pravella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA Part B 2017, 2, 344–346. [Google Scholar] [CrossRef]
- Traut, W.; Vogel, H.; Glöckner, G.; Hartmann, E.; Heckel, D.G. High-throughput sequencing of a single chromosome: A moth W chromosome. Chromosome Res. 2013, 21, 491–505. [Google Scholar] [CrossRef]
- Chang, Z.; Shen, Q. The complete mitochondrial genome of the navel orangeworm Amyelois transitella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA Part A 2016, 27, 4561–4562. [Google Scholar] [CrossRef]
- Liu, Q.; Chai, X.; Bian, D.; Zhou, C.; Tang, B. The complete mitochondrial genome of Plodia interpunctella (Lepidoptera: Pyralidae) and comparison with other Pyraloidea insects. Genome 2015, 59, 37–49. [Google Scholar] [CrossRef]
- Park, Y.; Park, C.; Hong, S.; Jung, B.; Ibal, J.C.; Park, G.; Shin, J. The complete mitochondrial genome sequence of the greater wax moth Galleria mellonella (Insecta, Lepidoptera, Pyralidae): Sequence and phylogenetic analysis comparison based on whole mitogenome. Mitochondrial DNA Part B 2017, 2, 714–715. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Zhao, J.; Su, T.; Luo, A.; Fan, R.; Chen, M.; Wu, C.; Zhu, C. The complete mitochondrial genome of the rice moth, Corcyra cephalonica. J. Insect Sci. 2012, 12, 72. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Yu, H.; Li, P.; You, P. The complete mitochondrial genome of Lista haraldusalis (Lepidoptera: Pyralidae). Mitochondrial DNA 2015, 26, 853–854. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wang, A.; Hong, G.; Cao, Y.; Wei, Z. Complete mitochondrial genome of Chilo suppressalis (Walker) (Lepidoptera: Crambidae). Mitochondrial DNA 2011, 22, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Qu, F.; Yang, Z.; Zhang, X.; Yue, B. Structural characteristics and phylogenetic analysis of the mitochondrial genome of the rice leafroller, Cnaphalocrocis medinalis (Lepidoptera: Crambidae). Mol. Biol. Rep. 2014, 41, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Coates, B.S.; Sumerford, D.V.; Hellmich, R.L.; Lewis, L.C. Partial mitochondrial genome sequences of Ostrinia nubilalis and Ostrinia furnicalis. Int. J. Biol. Sci. 2005, 1, 13–18. [Google Scholar] [CrossRef]
- Cao, S.; Yu, W.; Sun, M.; Du, Y. Characterization of the complete mitochondrial genome of Tryporyza incertulas, in comparison with seven other Pyraloidea moths. Gene 2014, 533, 356–365. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Kim, S.; Kim, I. Complete mitochondrial genome of an aquatic moth, Elophila interruptalis (Lepidoptera: Crambidae). Mitochondrial DNA 2014, 25, 275–277. [Google Scholar] [CrossRef]
- Ye, F.; Shi, Y.; Xing, L.; Yu, H.; You, P. The complete mitochondrial genome of Paracymoriza prodigalis (Leech, 1889) (Lepidoptera), with a preliminary phylogenetic analysis of Pyraloidea. Aquat. Insects 2013, 35, 71–88. [Google Scholar] [CrossRef]
- Ye, F.; You, P. The complete mitochondrial genome of Paracymoriza distinctalis (Lepidoptera: Crambidae). Mitochondrial DNA Part A 2016, 27, 28–29. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Ahn, S.; Kim, I. Complete mitochondrial genome of the grass moth Glyphodes quadrimaculalis (Lepidoptera: Crambidae). Mitochondrial DNA 2015, 26, 247–249. [Google Scholar] [CrossRef]
- Chen, S.; Li, F.; Lan, X.; You, P. The complete mitochondrial genome of Pycnarmon lactiferalis (Lepidoptera: Crambidae). Mitochondrial DNA Part B 2016, 1, 638–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, P.; You, P. The complete mitochondrial genome of Tyspanodes hypsalis (Lepidoptera: Crambidae). Mitochondrial DNA Part A 2016, 27, 1821–1822. [Google Scholar] [CrossRef]
- Chai, H.; Du, Y.; Zhai, B. Characterization of the complete mitochondrial genomes of Cnaphalocrocis medinalis and Chilo suppressalis (Lepidoptera: Pyralidae). Int. J. Biol. Sci. 2012, 8, 561–579. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Kim, M.; Kim, I. Description of new mitochondrial genomes (Spodoptera litura, Noctuoidea and Cnaphalocrocis medinalis, Pyraloidea) and phylogenetic reconstruction of Lepidoptera with the comment on optimization schemes. Mol. Biol. Rep. 2013, 40, 6333–6349. [Google Scholar] [CrossRef]
- Wu, Q.; Gong, Y.; Shi, B.; Gu, Y.; Wei, S. The complete mitochondrial genome of the yellow peach moth Dichocrocis punctiferalis (Lepidoptera: Pyralidae). Mitochondrial DNA 2013, 24, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, Y.; Xiao, L.; Tan, Y.; Bai, L. Complete mitochondrial genome of Cotton Leaf Roller Haritalodes derogata (Lepidoptera: Crambidae). Mitochondrial DNA Part A 2016, 27, 2833–2834. [Google Scholar] [CrossRef]
- Tang, M.; Tan, M.; Meng, G.; Yang, S.; Su, X.; Liu, S.; Song, W.; Li, Y.; Wu, Q.; Zhang, A.; et al. Multiplex sequencing of pooled mitochondrial genomes—A crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014, 42, e166. [Google Scholar] [CrossRef]
- He, S.; Zou, Y.; Zhang, L.; Ma, W.; Zhang, X.; Yue, B. The complete mitochondrial genome of the beet webworm, Spoladea recurvalis (Lepidoptera: Crambidae) and its phylogenetic implications. PLoS ONE 2015, 10, e0129355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, J.; Geng, S.; Yang, J.; Zhang, X.; An, Y.; Li, C.; Cui, H.; Li, X.; Wang, Y. The first mitochondrial genome for Phaudidae (Lepidoptera) with phylogenetic analyses of Zygaenoidea. Int. J. Biol. Macromol. 2020, 149, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 53942. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004, 21, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Rota-Stabelli, O.; Yang, Z.; Telford, M.J. MtZoa: A general mitochondrial amino acid substitutions model for animal evolutionary studies. Mol. Phylogenet. Evol. 2009, 52, 268–272. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A. Tracer v1.6. Available online: http://tree.bio.ed.ac.uk/software/tracer/ (accessed on 6 September 2020).
- Rambaut, A. Figtree 1.4.3. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 18 August 2020).
- Ma, H.; Zheng, X.; Peng, M.; Bian, H.; Chen, M.; Liu, Y.; Jiang, X.; Qin, L. Complete mitochondrial genome of the meadow moth, Loxostege sticticalis (Lepidoptera: Pyraloidea: Crambidae), compared to other Pyraloidea moths. J. Asia-Pac. Entomol. 2016, 19, 697–706. [Google Scholar] [CrossRef]
- Zhou, N.; Dong, Y.; Qiao, P.; Yang, Z. Complete mitogenomic structure and phylogenetic implications of the genus Ostrinia (Lepidoptera: Crambidae). Insects 2020, 11, 232. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.; Song, S.; Lim, P.; Chan, K.; Chow, W.; Eamsobhana, P. Complete mitochondrial genome of Bactrocera arecae (Insecta: Tephritidae) by next-generation sequencing and molecular phylogeny of Dacini tribe. Sci. Rep. 2015, 5, 15155. [Google Scholar] [CrossRef]
- Chen, P.; Zheng, B.; Liu, J.; Wei, S. Next-generation sequencing of two mitochondrial genomes from family pompilidae (Hymenoptera: Vespoidea) reveal novel patterns of gene arrangement. Int. J. Mol. Sci. 2016, 17, 1641. [Google Scholar] [CrossRef]
- Shi, Y.; Chu, Q.; Wei, D.; Qiu, Y.; Shang, F.; Dou, W.; Wang, J. The mitochondrial genome of booklouse, Liposcelis sculptilis (Psocoptera: Liposcelididae) and the evolutionary timescale of Liposcelis. Sci. Rep. 2016, 6, 30660. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ye, F. Comparative mitogenomic analyses of praying mantises (Dictyoptera, Mantodea): Origin and evolution of unusual intergenic gaps. Int. J. Biol. Sci. 2017, 13, 367–382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lavrov, D.V.; Brown, W.M.; Boore, J.L. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl. Acad. Sci. USA 2000, 97, 13738–13742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Szymura, J.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Minet, J. Les Pyraloidea et leurs principals divisions systématiques. Bull. Soc. Entomol. Fr. 1981, 86, 262–280. [Google Scholar]
- Léger, T.; Landry, B.; Nuss, M. Phylogeny, character evolution and tribal classification in Crambinae and Scopariinae (Lepidoptera, Crambidae). Syst. Entomol. 2019, 44, 757–776. [Google Scholar] [CrossRef]


| Subfamily | Taxa | GenBank Accession No. | References |
|---|---|---|---|
| Pyralidae | |||
| Pyralinae | Hypsopygia regina | KP327714 | Unpublished |
| Endotricha consocia | MF568544 | [18] | |
| Phycitinae | Meroptera pravella | MF073207 | [28] |
| Ephestia kuehniella | KF305832 | [29] | |
| Amyelois transitella | KT692987 | [30] | |
| Ephestia kuehniella | KU877167 | Unpublished | |
| Plodia interpunctella | KP729178 | [31] | |
| Galleriinae | Galleria mellonella | KT750964 | [32] |
| Corcyra cephalonica | HQ897685 | [33] | |
| Epipaschiinae | Lista haraldusalis | KF709449 | [34] |
| Crambidae | |||
| Odontiinae | Dausara latiterminalis | NW732137 | This study |
| Heortia vitessoides | NW732138 | This study | |
| Pseudonoorda nigropunctalis | NW732139 | This study | |
| Crambinae | Chilo suppressalis | HQ860290 | [35] |
| Chilo sacchariphagus | KU188518 | Unpublished | |
| Diatraea saccharalis | FJ240227 | [36] | |
| Pyraustinae | Loxostege sticticalis | KR080490 | Unpublished |
| Ostrinia furnacalis | AF467260 | [37] | |
| Ostrinia nubilalis | AF442957 | [37] | |
| Schoenobiinae | Scirpophaga incertulas | KC493629 | [38] |
| Scirpophaga incertulas | KF751706 | Unpublished | |
| Acentropinae | Elophila interruptalis | KC894961 | [39] |
| Parapoynx crisonalis | KT443883 | Unpublished | |
| Paracymoriza prodigalis | JX144892 | [40] | |
| Paracymoriza distinctalis | KF859965 | [41] | |
| Spilomelinae | Glyphodes quadrimaculalis | KF234079 | [42] |
| Pycnarmon lactiferalis | KX426346 | [43] | |
| Tyspanodes hypsalis | KM453724 | [44] | |
| Glyphodes pyloalis | KM576860 | Unpublished | |
| Tyspanodes striata | KP347977 | Unpublished | |
| Cnaphalocrocis medinalis | JN246082 | [45] | |
| Cnaphalocrocis medinalis | JQ647917 | [46] | |
| Dichocrocis punctiferalis | JX448619 | [47] | |
| Haritalodes derogata | KR233479 | [48] | |
| Maruca vitrata | KP327715 | Unpublished | |
| Nomophila noctuella | KM244688 | [49] | |
| Spoladea recurvalis | KJ739310 | [50] | |
| Tyspanodes striata | KP347977 | Unpublished | |
| Glaphyriinae | Evergestis junctalis | KP347976 | Unpublished |
| Scopariinae | Eudonia angustea | KJ508052 | [14] |
| Alucitidae | Alucita montana | KJ508059 | [14] |
| Pterophoridae | Emmelina monodactyla | KJ508063 | [14] |
| Thyrididae | Pyrinioides aurea | KT337662 | [25] |
| Zygaenidae | Phauda flammans | NC_047243 | [51] |
| Zygaenidae | Rhodopsona rubiginosa | NC_025761 | [49] |
| Feature | Strand | Position(from) | Position(to) | Length | Intergenic Nucleotides | Anticodon | Initial Codon | Stop Codon |
|---|---|---|---|---|---|---|---|---|
| trnM | J | 1/1/1 | 69/68/66 | 69/68/66 | 0/0/0 | CAT | ||
| trnI | J | 70/69/67 | 133/132/130 | 64/64/64 | −3/−3/−3 | GAT | ||
| trnQ | N | 131/130/128 | 199/198/196 | 69/69/69 | 0/0/0 | TTG | ||
| ND2 | J | 200/199/197 | 1213/1212/1210 | 1014/1014/1014 | 0/0/4 | ATT/ATC/ATT | TAA/TAA/TAA | |
| trnW | J | 1214/1213/1215 | 1280/1279/1281 | 67/67/67 | −8/−8/−8 | TCA | ||
| trnC | N | 1273/1272/1274 | 1339/1338/1337 | 67/67/64 | 0/0/0 | GCA | ||
| trnY | N | 1340/1339/1338 | 1406/1405/1402 | 67/67/65 | −8/8/0 | GTA | ||
| COX1 | J | 1399/1414/1403 | 2949/2952/2941 | 1551/1539/1539 | −5/−5/−5 | ATT/TTG/TTG | TAA/TAA/TAA | |
| trnL | J | 2945/2948/2937 | 3011/3014/3003 | 67/67/67 | 0/0/0 | TAA | ||
| COX2 | J | 3012/3015/3004 | 3693/3696/3720 | 682/682/717 | 0/0/−35 | ATG/ATG/ATG | T/T/TAA | |
| trnK | J | 3694/3697/3686 | 3763/3767/3755 | 70/71/70 | 0/1/0 | CTT | ||
| trnD | J | 3764/3769/3756 | 3829/3838/3821 | 66/70/66 | 0/0/0 | GTC | ||
| ATP8 | J | 3830/3839/3822 | 3991/4000/3983 | 162/162/162 | −7/−7/−7 | ATT/ATT/ATT | TAA/TAA/TAA | |
| ATP6 | J | 3985/3994/3977 | 4665/4674/4657 | 681/681/681 | −1/−1/−1 | ATG/ATG/ATG | TAA/TAA/TAA | |
| COX3 | J | 4665/4674/4657 | 5453/5462/5445 | 789/789/789 | 2/3/2 | ATG/ATG/ATG | TAA/TAA/TAA | |
| trnG | J | 5456/5466/5448 | 5521/5532/5514 | 66/67/67 | 0/0/0 | TCC | ||
| ND3 | J | 5522/5533/5515 | 5875/5886/5868 | 354/354/354 | 3/7/7 | ATT/ATT/ATT | TAA/TAA/TAA | |
| trnA | J | 5879/5894/5876 | 5944/5958/5940 | 66/65/65 | 0/2/0 | TGC | ||
| trnR | J | 5945/5961/5941 | 6007/6025/6003 | 63/65/63 | 0/4/0 | TCG | ||
| trnN | J | 6008/6030/6004 | 6073/6096/6069 | 66/67/66 | 0/0/0 | GTT | ||
| trnS | J | 6074/6097/6070 | 6139/6162/6135 | 66/66/66 | 1/0/1 | GCT | ||
| trnE | J | 6141/6163/6137 | 6206/6228/6201 | 66/66/65 | −2/12/4 | TTC | ||
| trnF | N | 6205/6241/6206 | 6270/6308/6270 | 66/68/65 | 4/−1/−17 | GAA | ||
| ND5 | N | 6275/6308/6254 | 8008/8043/8005 | 1734/1736/1752 | 0/0/0 | ATT/ATT/ATT | TAA/TTA/TAA | |
| trnH | N | 8009/8044/8006 | 8074/8109/8072 | 66/66/67 | −1/0/9 | GTG | ||
| ND4 | N | 8074/8110/8082 | 9415/9453/9422 | 1342/1344/1341 | 17/2/3 | ATG/ATG/ATG | TAA/TAA/TAA | |
| ND4L | N | 9433/9456/9426 | 9723/9749/9716 | 291/294/291 | 2/2/5 | ATG/ATG/ATA | TAA/TAA/TAA | |
| trnT | J | 9726/9752/9722 | 9790/9818/9786 | 65/67/65 | 0/0/0 | TGT | ||
| trnP | N | 9791/9819/9787 | 9856/9883/9852 | 66/65/66 | 0/0/0 | TGG | ||
| ND6 | J | 9857/9884/9853 | 10,390/10,465/10,386 | 534/582/534 | 4/0/−1 | ATA/ATC/ATA | TAA/TAA/TAA | |
| CYTB | J | 10,395/10,466/10,386 | 11,543/11,620/11,531 | 1149/1155/1146 | −2/3/4 | ATG/ATG/ATG | TAA/TAA/TAA | |
| trnS | J | 11,542/11,624/11,536 | 11,609/11,688/11,600 | 68/65/65 | 16/16/3 | TGA | ||
| ND1 | N | 11,626/11,705/11,604 | 12,564/12,643/12,554 | 939/939/951 | 1/1/0 | ATG/ATG/ATG | TAA/TAA/TAG | |
| trnL | N | 12,566/12,645/12,555 | 12,633/12,712/12,621 | 68/68/67 | −25/−19/−23 | TAG | ||
| l-rRNA | N | 12,609/12,694/12,599 | 13,977/14,057/13,967 | 1369/1364/1369 | −12/−12/−12 | |||
| trnV | N | 13,966/14,046/13,956 | 14,030/14,111/14,021 | 65/66/66 | −1/−1/−1 | TAC | ||
| s-rRNA | N | 14,030/14,111/14,021 | 14,812/14,890/14,804 | 783/780/784 | 0/0/0 | |||
| A + T-rich region | 14,901/15,057/14,891 | 15,235/15,404/15,170 | 334/348/280 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, M.; Zhao, H.; Yu, F.; Zhang, A.; Li, H. The First Mitogenomes of the Subfamily Odontiinae (Lepidoptera, Crambidae) and Phylogenetic Analysis of Pyraloidea. Insects 2021, 12, 486. https://doi.org/10.3390/insects12060486
Qi M, Zhao H, Yu F, Zhang A, Li H. The First Mitogenomes of the Subfamily Odontiinae (Lepidoptera, Crambidae) and Phylogenetic Analysis of Pyraloidea. Insects. 2021; 12(6):486. https://doi.org/10.3390/insects12060486
Chicago/Turabian StyleQi, Mujie, Huifeng Zhao, Fang Yu, Aibing Zhang, and Houhun Li. 2021. "The First Mitogenomes of the Subfamily Odontiinae (Lepidoptera, Crambidae) and Phylogenetic Analysis of Pyraloidea" Insects 12, no. 6: 486. https://doi.org/10.3390/insects12060486
APA StyleQi, M., Zhao, H., Yu, F., Zhang, A., & Li, H. (2021). The First Mitogenomes of the Subfamily Odontiinae (Lepidoptera, Crambidae) and Phylogenetic Analysis of Pyraloidea. Insects, 12(6), 486. https://doi.org/10.3390/insects12060486






