Variants in the Mitochondrial Genome Sequence of Rhyzopertha dominica (Fabricius) (Coleoptera: Bostrycidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Strain
2.2. Extraction of Nucleic Acids and Sequencing of the Mitochondrial Genome
2.3. Assembly and Annotation of the Mitochondrial Genome
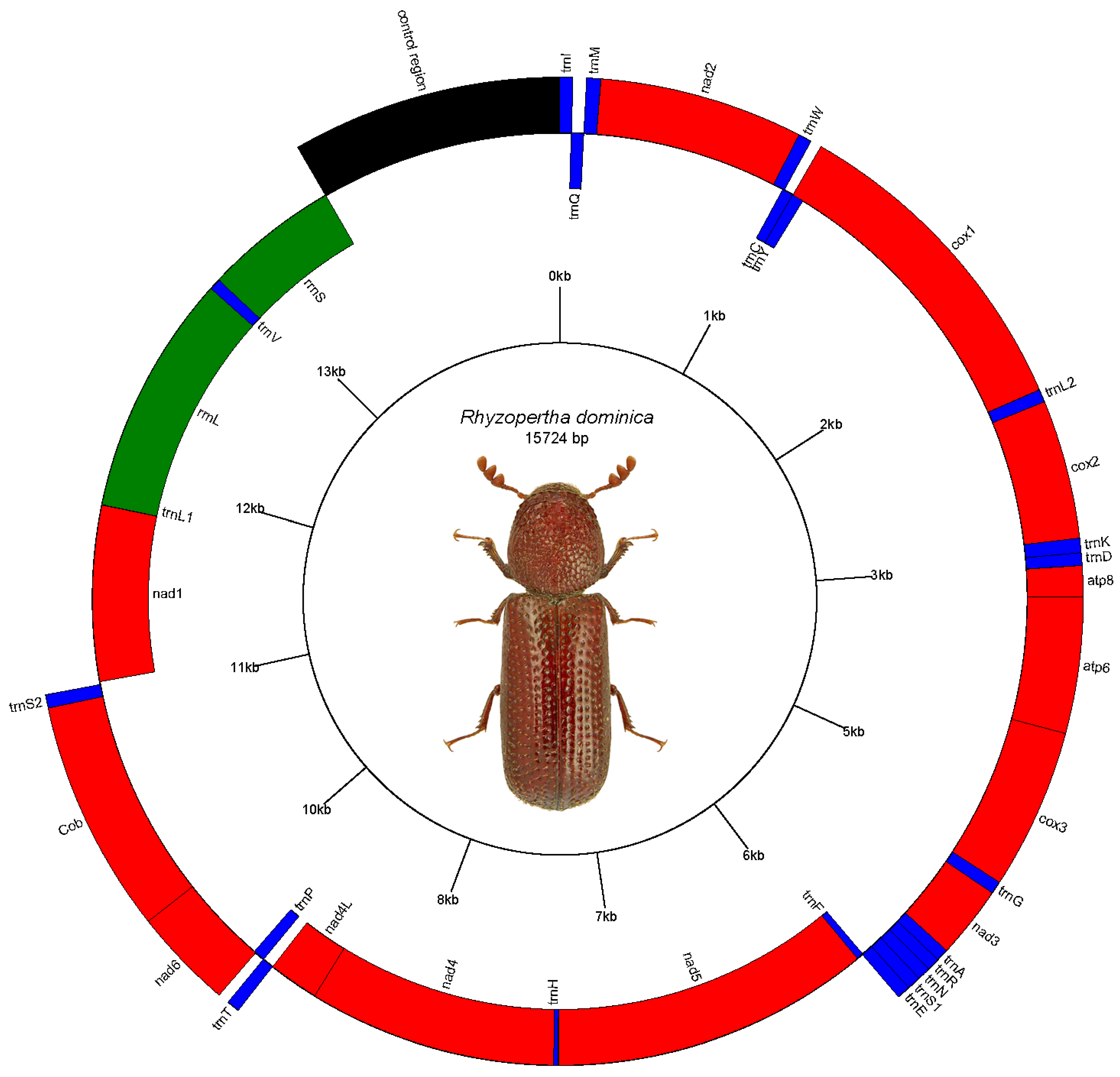
3. Results
3.1. RNA Sequences
3.2. Protein Coding Sequences
3.3. Control Region
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edde, P.A. A Review of the Biology and Control of Rhyzopertha dominica (F.) the Lesser Grain Borer. J. Stored Prod. Res. 2012, 48, 1–18. [Google Scholar] [CrossRef]
- Afful, E.; Elliott, B.; Nayak, M.K.; Phillips, T.W. Phosphine Resistance in North American Field Populations of the Lesser Grain Borer, Rhyzopertha dominica (Coleoptera: Bostrichidae). J. Econ. Entomol. 2018, 111, 463–469. [Google Scholar] [CrossRef]
- Schlipalius, D.I.; Chen, W.; Collins, P.J.; Nguyen, T.; Reilly, P.E.B.; Ebert, P.R. Gene Interactions Constrain the Course of Evolution of Phosphine Resistance in the Lesser Grain Borer, Rhyzopertha dominica. Heredity 2008, 100, 506–516. [Google Scholar] [CrossRef]
- Mau, Y.S.; Collins, P.J.; Daglish, G.J.; Nayak, M.K.; Pavic, H.; Ebert, P.R. The Rph1 Gene Is a Common Contributor to the Evolution of Phosphine Resistance in Independent Field Isolates of Rhyzopertha dominica. PLoS ONE 2012, 7, e31541. [Google Scholar] [CrossRef] [PubMed]
- Schlipalius, D.I.; Valmas, N.; Tuck, A.G.; Jagadeesan, R.; Ma, L.; Kaur, R.; Goldinger, A.; Anderson, C.; Kuang, J.; Zuryn, S.; et al. A Core Metabolic Enzyme Mediates Resistance to Phosphine Gas. Science 2012, 338, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Schlipalius, D.I.; Tuck, A.G.; Jagadeesan, R.; Nguyen, T.; Kaur, R.; Subramanian, S.; Barrero, R.; Nayak, M.; Ebert, P.R. Variant Linkage Analysis Using de Novo Transcriptome Sequencing Identifies a Conserved Phosphine Resistance Gene in Insects. Genetics 2018, 209, 281–290. [Google Scholar] [CrossRef]
- Nakakita, H. The Mode of Action of Phosphine. J. Pestic. Sci. 1987, 12, 299–309. [Google Scholar] [CrossRef][Green Version]
- Pimentel, M.A.G.; Faroni, L.R.D.; Tótola, M.R.; Guedes, R.N.C. Phosphine Resistance, Respiration Rate and Fitness Consequences in Stored-Product Insects. Pest Manag. Sci. 2007, 63, 876–881. [Google Scholar] [CrossRef]
- Oppert, B.; Guedes, R.N.C.; Aikins, M.J.; Perkin, L.; Chen, Z.; Phillips, T.W.; Zhu, K.Y.; Opit, G.P.; Hoon, K.; Sun, Y.; et al. Genes Related to Mitochondrial Functions Are Differentially Expressed in Phosphine-Resistant and -Susceptible Tribolium castaneum. BMC Genom. 2015, 16, 968. [Google Scholar] [CrossRef]
- Taanman, J.-W. The Mitochondrial Genome: Structure, Transcription, Translation and Replication. Biochim. Biophys. Acta BBA-Bioenerg. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, M.H.L. Drosophila melanogaster Mitochondrial DNA, a Novel Organization and Genetic Code. Nature 1983, 304, 234–240. [Google Scholar] [CrossRef]
- Boore, J.L. Animal Mitochondrial Genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of Animal Mitochondrial DNA: Relevance for Population Biology and Systematics. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Ouyang, B.; Bai, Y.; Chen, J.; Wang, J.; Liang, S. Characterization of the Complete Mitochondrial Genome of Lesser Grain Borer Rhyzopertha dominica Fabricius (Insecta: Coleoptera: Bostrichidae) from Jingziguan. Mitochondrial DNA Part B 2019, 4, 3952–3953. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De Novo Assembly of Organelle Genomes from Whole Genome Data. Nucleic Acids Res. 2016, gkw955. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Conant, G.C.; Wolfe, K.H. GenomeVx: Simple Web-Based Creation of Editable Circular Chromosome Maps. Bioinformatics 2008, 24, 861–862. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB Bioinformatics Resource Portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.m.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.B.C.; Vlk, J.L.; Kapatral, V. Comparative Insect Mitochondrial Genomes: Differences despite Conserved Genome Synteny. Afr. J. Biotechnol. 2006, 5, 1308–1318. [Google Scholar]
- Liu, Q.-N.; Bian, D.-D.; Jiang, S.-H.; Li, Z.-X.; Ge, B.-M.; Xuan, F.-J.; Yang, L.; Li, C.-F.; Zhang, D.-Z.; Zhou, C.-L.; et al. The Complete Mitochondrial Genome of the Red Flour Beetle, Tribolium dastaneum (Coleoptera: Tenebrionidae). Mitochondrial DNA 2016, 27, 1525–1527. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Tamura, K.; Aotsuka, T. Replication Origin of Mitochondrial DNA in Insects. Genetics 2005, 171, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Yang, J.O.; Sung, J.-Y.; Lee, J.-Y.; Park, J.S.; Lee, H.-S.; Lee, B.-H.; Ren, Y.; Lee, D.-W.; Lee, S.-E. Minimization of Energy Transduction Confers Resistance to Phosphine in the Rice Weevil, Sitophilus oryzae. Sci. Rep. 2019, 9, 14605. [Google Scholar] [CrossRef] [PubMed]
| Gene | Strand | Feature | Start, Stop | NC# (This Paper) | NC_042820 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Start | Stop | Length (bp) | Start | Stop | Length (bp) | ||||
| trnI | + | tRNA | 1 | 63 | 63 | 0 | 63 | 63 | |
| trnQ | − | tRNA | 61 | 129 | 69 | 60 | 129 | 69 | |
| trnM | + | tRNA | 129 | 197 | 69 | 128 | 197 | 69 | |
| nad2 | + | CDS | ATA, TAA | 198 | 1190 | 993 | 197 | 1190 | 993 |
| trnW | + | tRNA | 1189 | 1253 | 65 | 1188 | 1253 | 65 | |
| trnC | − | tRNA | 1246 | 1306 | 61 | 1245 | 1306 | 61 | |
| trnY | − | tRNA | 1306 | 1368 | 63 | 1305 | 1368 | 63 | |
| cox1 | + | CDS | ATA, TAA | 1370 | 2898 | 1529 | 1369 | 2900 | 1531 |
| trnL2 | + | tRNA | 2901 | 2962 | 62 | 2900 | 2962 | 62 | |
| cox2 | + | CDS | ATA, TCA | 2963 | 3640 | 678 | 2962 | 3638 | 676 |
| trnK | + | tRNA | 3639 | 3709 | 71 | 3638 | 3709 | 71 | |
| trnD | + | tRNA | 3709 | 3770 | 62 | 3708 | 3770 | 62 | |
| atp8 | + | CDS | ATT, TAA | 3771 | 3926 | 156 | 3770 | 3926 | 156 |
| atp6 | + | CDS | ATG, TAA | 3920 | 4585 | 666 | 3919 | 4585 | 666 |
| cox3 | + | CDS | ATG, TTA | 4585 | 5373 | 789 | 4584 | 5368 | 784 |
| trnG | + | tRNA | 5369 | 5430 | 62 | 5368 | 5430 | 62 | |
| nad3 | + | CDS | ATA, TAG | 5431 | 5784 | 354 | 5430 | 5784 | 354 |
| trnA | + | tRNA | 5783 | 5844 | 62 | 5782 | 5844 | 62 | |
| trnR | + | tRNA | 5844 | 5908 | 65 | 5843 | 5908 | 65 | |
| trnN | + | tRNA | 5908 | 5971 | 64 | 5907 | 5971 | 64 | |
| trnS | + | tRNA | 5971 | 6037 | 67 | 5970 | 6037 | 67 | |
| trnE | + | tRNA | 6038 | 6101 | 64 | 6037 | 6101 | 64 | |
| trnF | − | tRNA | 6100 | 6163 | 64 | 6099 | 6163 | 64 | |
| nad5 | − | CDS | ATT, TAA | 6146 | 7871 | 1726 | 6163 | 7871 | 1708 |
| trnH | − | tRNA | 7872 | 7934 | 63 | 7871 | 7934 | 63 | |
| nad4 | − | CDS | ATG, TTA | 7936 | 9252 | 1317 | 7934 | 9252 | 1318 |
| nad4L | − | CDS | ATG, TAA | 9246 | 9518 | 273 | 9245 | 9518 | 273 |
| trnT | + | tRNA | 9521 | 9582 | 62 | 9520 | 9582 | 62 | |
| trnP | − | tRNA | 9583 | 9645 | 63 | 9582 | 9645 | 63 | |
| nad6 | + | CDS | ATT, TAA | 9647 | 10,129 | 483 | 9646 | 10,129 | 483 |
| cytb | + | CDS | ATG, TAG | 10,129 | 11,268 | 1140 | 10,128 | 11,268 | 1140 |
| trnS2 | + | tRNA | 11,267 | 11,332 | 66 | 11,266 | 11,332 | 66 | |
| nad1 | − | CDS | ATA, TAG | 11,350 | 12,303 | 954 | 11,349 | 12,300 | 951 |
| trnL1 | − | tRNA | 12,301 | 12,362 | 62 | 12,300 | 12,362 | 62 | |
| rrnL | − | rRNA | 12,379 | 13,580 | 1202 | 12,362 | 13,616 | 1254 | |
| trnV | − | tRNA | 13,618 | 13,679 | 62 | 13,616 | 13,678 | 62 | |
| rrnS | − | rRNA | 13,680 | 14,411 | 732 | 13,678 | 14,410 | 732 | |
| control region | + | 14,412 | 15,724 | 1312 | 14,410 | 15,859 | 1449 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perkin, L.C.; Smith, T.P.L.; Oppert, B. Variants in the Mitochondrial Genome Sequence of Rhyzopertha dominica (Fabricius) (Coleoptera: Bostrycidae). Insects 2021, 12, 387. https://doi.org/10.3390/insects12050387
Perkin LC, Smith TPL, Oppert B. Variants in the Mitochondrial Genome Sequence of Rhyzopertha dominica (Fabricius) (Coleoptera: Bostrycidae). Insects. 2021; 12(5):387. https://doi.org/10.3390/insects12050387
Chicago/Turabian StylePerkin, Lindsey C., Timothy P. L. Smith, and Brenda Oppert. 2021. "Variants in the Mitochondrial Genome Sequence of Rhyzopertha dominica (Fabricius) (Coleoptera: Bostrycidae)" Insects 12, no. 5: 387. https://doi.org/10.3390/insects12050387
APA StylePerkin, L. C., Smith, T. P. L., & Oppert, B. (2021). Variants in the Mitochondrial Genome Sequence of Rhyzopertha dominica (Fabricius) (Coleoptera: Bostrycidae). Insects, 12(5), 387. https://doi.org/10.3390/insects12050387






