Differential Gene Expression in the Heads of Behaviorally Divergent Culex pipiens Mosquitoes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mosquito Populations
2.2. Host Landing Assay
2.3. Chicken vs. Human Choice Landing Assays: Data Analysis
2.4. RNA Sequencing of Mosquito Heads
2.5. Read-Filtering and Quality Control
2.6. Differential Gene Expression Analysis
2.7. Splice Site Variation
2.8. Candidate Sensory Gene Identification
3. Results
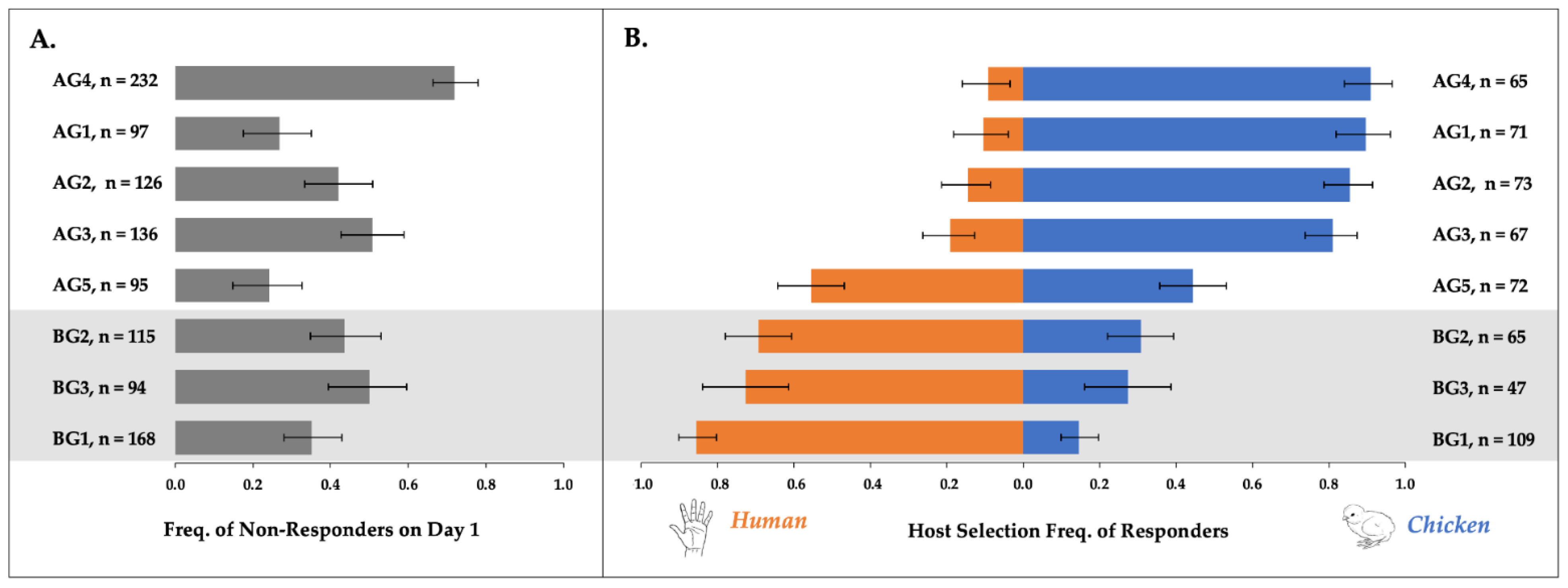
3.1. Host Response Rates and Preferences of Cx. pipiens
3.1.1. Population-Level Day One Overall Response Rates
3.1.2. Multi-Day Host Responses by Individual Females
3.1.3. Multi-Day Host Switching by Individual Females
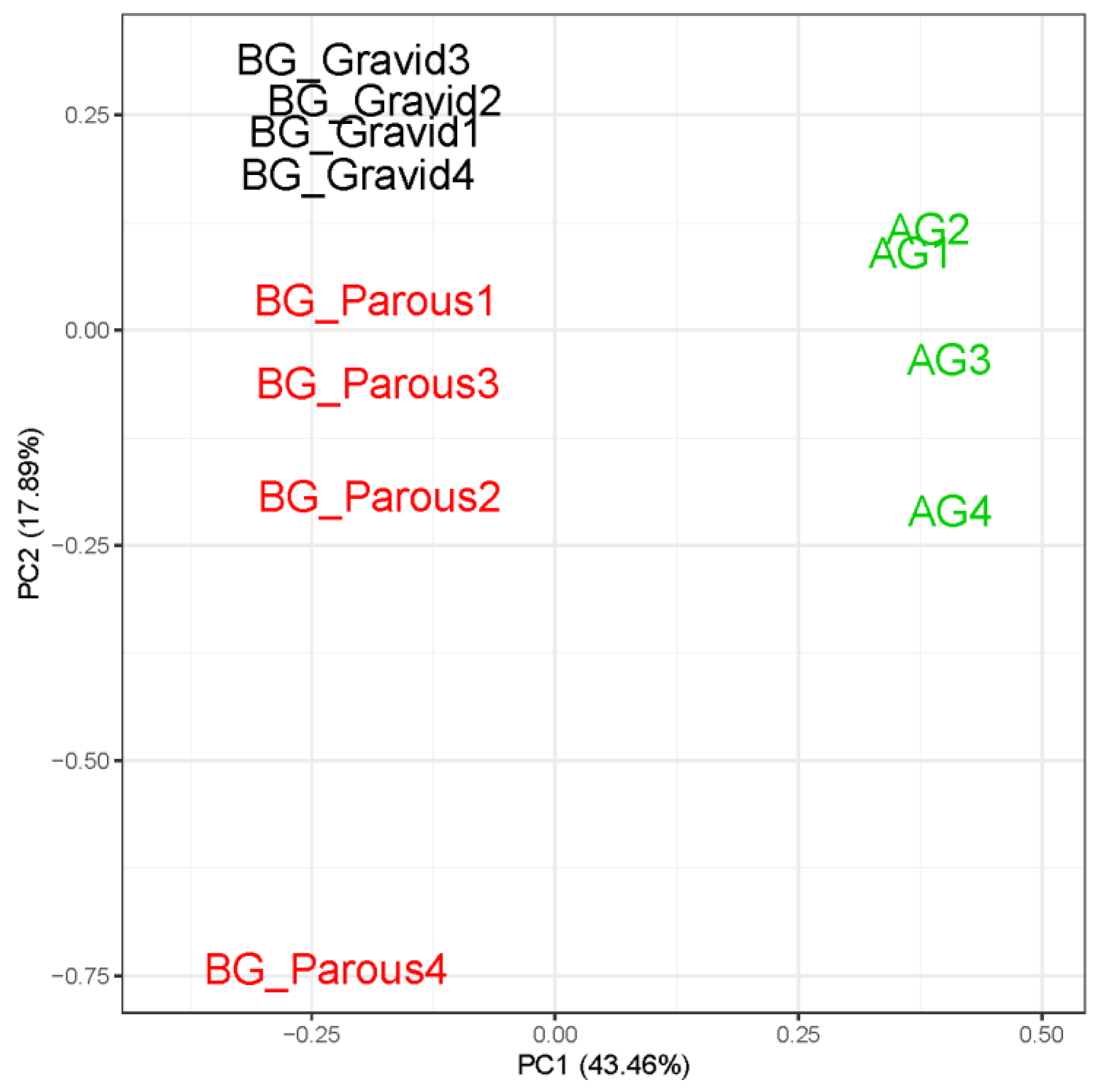
3.2. Read Quality and Sample Clustering
3.3. Differential Gene Expression in the Heads of Behaviorally Divergent Females
3.4. Splice Site Variation
3.5. Candidate Sensory Gene Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takken, W.; Verhulst, N. Host preferences of blood-feeding mosquitoes. Annu. Rev. Entomol. 2013, 58, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Garrett-Jones, C. Prognosis for interruption of malaria transmission through assessment of the mosquito’s vectorial capacity. Nature 1964, 204, 1173–1175. [Google Scholar] [CrossRef] [PubMed]
- Friend, W.G.; Smith, J.J.B. Factors affecting feeding by bloodsucking insects. Annu. Rev. Entomol. 1997, 22, 309–331. [Google Scholar] [CrossRef]
- Bowen, M.F. The sensory physiology of host-seeking behavior in mosquitoes. Annu. Rev. Entomol. 1991, 36, 139–158. [Google Scholar] [CrossRef]
- van Breugel, F.; Riffell, J.; Fairhall, A.; Dickinson, M.H. Mosquitoes use vision to associate odor plumes with thermal targets. Curr. Biol. 2015, 25, 2123–2129. [Google Scholar] [CrossRef]
- Cardé, R.T. Multi-cue integration: How female mosquitoes locate a human host. Curr. Biol. 2015, 25, R793–R795. [Google Scholar] [CrossRef]
- Kennedy, J.S. The visual responses of flying mosquitoes. Proc. Zool. Soc. 1940, 109, 221–242. [Google Scholar] [CrossRef]
- Reeves, W.C. Field Studies on Carbon Dioxide as a Possible Host Simulant to Mosquitoes. Proc. Soc. Exp. Biol. Med. 1951, 77, 64–66. [Google Scholar] [CrossRef]
- Gillies, M.T. The role of carbon dioxide in host-finding by mosquitoes (Diptera: Culicidae): A review. Bull. Entomol. Res. 1980, 70, 525–532. [Google Scholar] [CrossRef]
- Smallegange, R.C.; Schmied, W.H.; van Roey, K.J.; Verhulst, N.O.; Spitzen, J.; Mukabana, W.R.; Takken, W. Sugar-fermenting yeast as an organic source of carbon dioxide to attract the malaria mosquito Anopheles gambiae. Malar. J. 2010, 9, 292. [Google Scholar] [CrossRef]
- McMeniman, C.J.; Corfas, R.A.; Matthews, B.J.; Ritchie, S.A.; Vosshall, L.B. Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell 2014, 156, 1060–1071. [Google Scholar] [CrossRef]
- Hawkes, F.; Gibson, G. Seeing is believing: The nocturnal malarial mosquito Anopheles coluzzii responds to visual host-cues when odour indicates a host is nearby. Parasit Vectors 2016, 9, 320. [Google Scholar] [CrossRef]
- Dekker, T.; Cardé, R.T. Moment-to-moment flight manoeuvres of the female yellow fever mosquito (Aedes aegypti L.) in response to plumes of carbon dioxide and human skin odour. J. Exp. Biol. 2011, 214, 3480–3494. [Google Scholar] [CrossRef]
- Takken, W.; Knols, B.G.J.; Otten, H. Interactions between physical and olfactory cues in the host-seeking behaviour of mosquitoes: The role of relative humidity. Ann. Trop. Med. Parasit. 1997, 91 (Suppl. 1), S119–S120. [Google Scholar] [CrossRef]
- DeGennaro, M.; McBride, C.S.; Seeholzer, L.; Nakagawa, T.; Dennis, E.J.; Goldman, C.; Jasinskiene, N.; James, A.A.; Vosshall, M.B. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 2013, 498, 487–491. [Google Scholar] [CrossRef]
- Suh, E.; Bohbot, J.D.; Zweibel, L.J. Peripheral olfactory signaling in insects. Curr. Opin. Insect. Sci. 2014, 6, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Montell, C.; Zwiebel, L.J. Mosquito Sensory Systems. In Advances in Insect Physiology; Academic Press: Cambridge, MA, USA, 2016; Volume 51, pp. 293–328. [Google Scholar]
- Corfas, A.; Vosshall, L. The cation channel TRPA1 tunes mosquito thermotaxis to host temperatures. Elife 2015, 4, e11750. [Google Scholar] [CrossRef]
- Zwiebel, L.J.; Takken, W. Olfactory regulation of mosquito–host interactions. Insect Biochem. Mol. 2004, 34, 645–652. [Google Scholar] [CrossRef]
- Pelletier, J.; Leal, W.S. Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PLoS ONE 2009, 4, e6237. [Google Scholar] [CrossRef]
- Silbering, A.F.; Rytz, R.; Grosjean, Y.; Abuin, L.; Ramdya, P.; Jefferis, G.S.; Benton, R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 2011, 31, 13357–13375. [Google Scholar] [CrossRef]
- Harbach, R.E. Culex pipiens: Species versus species complex–taxonomic history and perspective. J. Am. Mosq. Control Assoc. 2012, 28, 10–23. [Google Scholar] [CrossRef]
- Fritz, M.L.; Walker, E.D.; Miller, J.R.; Severson, D.W.; Dworkin, I. Divergent host preferences of above-and below-ground Culex pipiens mosquitoes and their hybrid offspring. Med. Vet. Entomol. 2015, 29, 115–123. [Google Scholar] [CrossRef]
- Kilpatrick, M.; Daszak, P.; Jones, M.J.; Marra, P.P.; Kramer, L.D. Host heterogeneity dominates West Nile virus transmission. Proc. Royal. Soc. B 2006, 273, 2327–2333. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Molaei, G.; Andreadis, T.G. Genetic insights into the population structure of Culex pipiens (Diptera: Culicidae) in the northeastern United States by using microsatellite analysis. Am. J. Trop. Med. Hyg. 2008, 79, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, M.; Czajka, C.; Börstler, J.; Melaun, C.; Jöst, H.; von Thien, H.; Badusche, M.; Becker, N.; Schmidt-Chanasit, J.; Kruger, A.; et al. First nationwide surveillance of Culex pipiens complex and Culex torrentium mosquitoes demonstrated the presence of Culex pipiens biotype pipiens/molestus hybrids in Germany. PLoS ONE 2013, 8, e71832. [Google Scholar] [CrossRef]
- Fonseca, D.M.; Keyghobadi, N.; Malcolm, C.A.; Mehmet, C.; Schaffner, F.; Mogi, M.; Fleischer, R.; Wilkerson, R.C. Emerging vectors in the Culex pipiens complex. Science 2004, 303, 1535–1538. [Google Scholar] [CrossRef]
- Farajollahi, A.; Fonseca, D.M.; Kramer, L.D.; Kilpatrick, A.M. “Bird biting” mosquitoes and human disease: A review of the role of Culex pipiens complex mosquitoes in epidemiology. Infect. Genet. Evol. 2011, 11, 1577–1585. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Sousa, C.A.; Vicente, J.L.; Pinho, L.; Calderón, I.; Arez, E.; Almeida, A.; Donnelly, M.J.; Pinto, J. Feeding patterns of molestus and pipiens forms of Culex pipiens (Diptera: Culicidae) in a region of high hybridization. Parasit Vectors 2013, 6, 93. [Google Scholar] [CrossRef]
- Yurchenko, A.A.; Masri, R.A.; Khrabrova, N.V.; Sibataev, A.K.; Fritz, M.L.; Sharakhova, M.V. Genomic differentiation and intercontinental population structure of mosquito vectors Culex pipiens pipiens and Culex pipiens molestus. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Spielman, A.; Wong, J. Environmental control of ovarian diapause in Culex pipiens. Ann. Entomol. Soc. 1973, 66, 905–907. [Google Scholar] [CrossRef]
- Vinogradova, E.B. Culex Pipiens Pipiens Mosquitoes: Taxonomy, Distribution, Ecology, Physiology, Genetics, Applied Importance and Control (No. 2); Pensoft Publishers: Sofia, Bulgaria, 2000. [Google Scholar]
- Mori, A.; Romero-Severson, J.; Severson, D.W. Genetic basis for reproductive diapause is correlated with life history traits within the Culex pipiens complex. Insect Mol. Biol. 2007, 16, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Sim, C.; Denlinger, D.L. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex Pipiens. Proc. Natl. Acad. Sci. USA 2008, 105, 6777–6781. [Google Scholar] [CrossRef]
- Denlinger, D.L.; Armbruster, P.A. Mosquito diapause. Annu. Rev. Entomol. 2014, 59, 73–93. [Google Scholar] [CrossRef] [PubMed]
- Byrne, K.; Nichols, R.A. Culex pipiens in London Underground tunnels: Differentiation between surface and subterranean populations. Heredity 1999, 82, 7–15. [Google Scholar] [CrossRef]
- Mutebi, J.P.; Savage, H.M. Discovery of Culex pipiens pipiens form molestus in Chicago. J. Am. Mosq. Control Assoc. 2009, 25, 500–503. [Google Scholar] [CrossRef]
- Kothera, L.; Godsey, M.; Mutebi, J.P.; Savage, H.M. A Comparison of Aboveground and Belowground Populations of Culex pipiens (Diptera: Culicidae) Mosquitoes in Chicago, Illinois, and New York City, New York, Using Microsatellites. J. Med. Entomol. 2010, 47, 805–813. [Google Scholar] [CrossRef]
- Becker, N.; Jöst, A.; Weitzel, T. The Culex pipiens complex in Europe. J. Am. Mosq. Control Assoc. 2012, 28, 53–67. [Google Scholar] [CrossRef]
- Reisen, W.K. The contrasting bionomics of Culex mosquitoes in western North America. J. Am. Mosq. Control Assoc. 2012, 28, 82–91. [Google Scholar] [CrossRef]
- Reusken, C.B.E.M.; De Vries, A.; Buijs, J.; Braks, M.A.H.; Den Hartog, W.; Scholte, E.J. First evidence for presence of Culex pipiens biotype molestus in the Netherlands, and of hybrid biotype pipiens and molestus in northern Europe. J. Vector Ecol. 2010, 35, 210–212. [Google Scholar] [CrossRef]
- Kim, S.; Trocke, S.; Sim, C. Comparative studies of stenogamous behaviour in the mosquito Culex pipiens complex. Med. Vet. Entomol. 2018, 32, 427–435. [Google Scholar] [CrossRef]
- Spielman, A. The inheritance of autogeny in the Culex pipiens complex of mosquitoes. Am. J. Hyg. 1957, 65, 404–425. [Google Scholar]
- Spielman, A. Population structure in the Culex pipiens complex of mosquitos. Bull. World Health Organ. 1967, 37, 271. [Google Scholar]
- Strickman, D.; Fonseca, D.M. Autogeny in Culex pipiens complex mosquitoes from the San Francisco Bay Area. Am. J. Trop. Med. Hyg. 2012, 87, 719–726. [Google Scholar] [CrossRef][Green Version]
- Gao, Q.; Su, F.; Zhou, Y.B.; Chu, W.; Cao, H.; Song, L.L.; Leng, P.E. Autogeny, Fecundity, and Other Life History Traits of Culex pipiens molestus (Diptera: Culicidae) in Shanghai, China. J. Med. Entomol. 2019, 56, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Apperson, C.S.; Harrison, B.A.; Unnasch, T.R.; Hassan, H.K.; Irby, W.S.; Savage, H.M.; Aspen, S.E.; Watson, D.; Rueda, L.M.; Engber, B.R.; et al. Host-feeding habits of Culex and other mosquitoes (Diptera: Culicidae) in the Borough of Queens in New York City, with characters and techniques for identification of Culex mosquitoes. J. Med. Entomol. 2002, 39, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Apperson, C.S.; Hassan, H.K.; Harrison, B.A.; Savage, H.M.; Aspen, S.E.; Farajollahi, A.R.Y.; Unnasch, T.R. Host feeding patterns of established and potential mosquito vectors of West Nile virus in the eastern United States. Vector Borne Zoonotic Dis. 2004, 4, 71–82. [Google Scholar] [CrossRef]
- Molaei, G.; Andreadis, T.G.; Armstrong, P.M.; Anderson, J.F.; Vossbrinck, C.R. Host feeding patterns of Culex mosquitoes and West Nile virus transmission, northeastern United States. Emerg. Infect. Dis. 2006, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, A.M.; Kramer, L.D.; Jones, M.J.; Marra, P.P.; Daszak, P.; Fonseca, D.M. Genetic influences on mosquito feeding behavior and the emergence of zoonotic pathogens. Am. J. Trop Med. Hyg. 2007, 77, 667–671. [Google Scholar] [CrossRef]
- Huang, S.; Hamer, G.L.; Molaei, G.; Walker, E.D.; Goldberg, T.L.; Kitron, U.D.; Andreadis, T.G. Genetic variation associated with mammalian feeding in Culex pipiens from a West Nile virus epidemic region in Chicago, Illinois. Vector Borne Zoonotic Dis. 2009, 9, 637–642. [Google Scholar] [CrossRef]
- Simpson, J.E.; Folsom-O’Keefe, C.M.; Childs, J.E.; Simons, L.E.; Andreadis, T.G.; Diuk-Wasser, M.A. Avian host-selection by Culex pipiens in experimental trials. PLoS ONE 2009, 4, e7861. [Google Scholar] [CrossRef]
- Simpson, J.E.; Hurtado, P.J.; Medlock, J.; Molaei, G.; Andreadis, T.G.; Galvani, A.P.; Diuk-Wasser, M.A. Vector host-feeding preferences drive transmission of multi-host pathogens: West Nile virus as a model system. Proc. Royal. Soc. B 2012, 279, 925–933. [Google Scholar] [CrossRef]
- Savage, H.M.; Kothera, L. The Culex pipiens complex in the Mississippi River basin: Identification, distribution, and bloodmeal hosts. J. Am. Mosq. Control Assoc. 2012, 28, 93–99. [Google Scholar] [CrossRef]
- Faraji, A.; Gaugler, R. Experimental host preference of diapause and non-diapause induced Culex pipiens pipiens (Diptera: Culicidae). Parasit Vectors 2015, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- Kothera, L.; Mutebi, J.P.; Kenney, J.L.; Saxton-Shaw, K.; Ward, M.P.; Savage, H.M. Bloodmeal, host selection, and genetic admixture analyses of Culex pipiens complex (Diptera: Culicidae) mosquitoes in Chicago, IL. J. Med. Entomol. 2020, 57, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Klowden, J. Initiation and termination of host-seeking inhibition in Aedes aegypti during oöcyte maturation. J. Insect Phys. 1981, 27, 799–803. [Google Scholar] [CrossRef]
- Klowden, J.; Arden, O. Humoral inhibition of host-seeking in Aedes aegypti during oöcyte maturation. J. Insect Phys. 1979, 25, 231–235. [Google Scholar] [CrossRef]
- Kassim, N.F.A.; Webb, C.E.; Russell, R.C. Is the expression of autogeny by Culex molestus Forskal (Diptera: Culicidae) influenced by larval nutrition or by adult mating, sugar feeding, or blood feeding? J. Vector Ecol. 2012, 37, 162–171. [Google Scholar] [CrossRef]
- Reeves, W.C. Quantitative Field Studies on a Carbon Dioxide Chemotropism of Mosquitoes. Am. J. Trop. Med. Hyg. 1953, 2, 325–331. [Google Scholar] [CrossRef]
- Bates, D.; Maechler, M.; Bolker, B. lme4: Linear Mixed-Effects Models Using S4 Classes; R Package: Los Angeles, CA, USA, 2011. [Google Scholar]
- Zeileis, A.; Hothorn, T. Diagnostic Checking in Regression Relationships. R News 2, 7–10. 2002. Available online: https://CRAN.R-project.org/doc/Rnews/ (accessed on 9 January 2018).
- Lindsay, S.W.; Adiamah, J.H.; Miller, J.E.; Pleass, R.J.; Armstrong, J.R.M. Variation in attractiveness of human subjects to malaria mosquitoes (Diptera: Culicidae) in The Gambia. J. Med. Entomol. 1993, 30, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.T.; Smallegange, R.C.; Van Loon, J.J.A.; Ter Braak, C.J.F.; Takken, W. Interindividual variation in the attractiveness of human odours to the malaria mosquito Anopheles gambiae ss. Med. Vet. Entomol. 2006, 20, 280–287. [Google Scholar] [CrossRef]
- Fritz, M.L.; Walker, E.D.; Yunker, A.J.; Dworkin, I. Daily blood feeding rhythms of laboratory-reared North American Culex pipiens. J. Circadian Rhythms 2014, 12, 1. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 January 2018).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Gelman, A.; Su, Y. Arm: Data Analysis Using Regression and Multilevel/Hierarchical Models. R Package Version 1.11-2. 2020. Available online: https://CRAN.R-project.org/package=arm (accessed on 10 January 2020).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Stephens, M. Ashr: Methods for Adaptive Shrinkage, Using Empirical Bayes. R Package Version 2.2-47. 2016. Available online: https://CRAN.R-project.org/package=ashr (accessed on 10 January 2020).
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucl. Acids Res. 2020. [Google Scholar] [CrossRef]
- Fischer, S.; Brunk, B.P.; Chen, F.; Gao, X.; Harb, O.S.; Iodice, J.B.; Stoeckert, C.J., Jr. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. 2011, 35, 6–12. [Google Scholar]
- Hamer, G.L.; Kitron, U.D.; Brawn, J.D.; Loss, S.R.; Ruiz, M.O.; Goldberg, T.L.; Walker, E.D. Culex pipiens (Diptera: Culicidae): A bridge vector of West Nile virus to humans. J. Med. Entomol. 2008, 45, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Hamer, G.L.; Kitron, U.D.; Goldberg, T.L.; Brawn, J.D.; Loss, S.R.; Ruiz, M.O.; Hayes, D.; Walker, E.D. Host selection by Culex pipiens mosquitoes and West Nile virus amplification. Am. J. Trop. Med. Hyg. 2009, 80, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Gillies, M.T. Experiments on host selection in the Anopheles gambiae complex. Ann. Trop. Med. Parasit. 1967, 61, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Mukwaya, L.G. Genetic control of feeding preferences in the mosquitoes Aedes (Stegomyia) simpsoni and aegypti. Physiol. Entomol. 1977, 2, 133–145. [Google Scholar] [CrossRef]
- McBride, C.S. Genes and odors underlying the recent evolution of mosquito preference for humans. Curr. Biol. 2016, 26, R41–R46. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.; Ohmori, D.; Yamakura, F.; Suzuki, K. Changes in free amino acid concentration in the hemolymph of the female Culex pipiens pallens (Diptera: Culicidae), after a blood meal. J. Med. Entomol. 1990, 27, 302–308. [Google Scholar] [CrossRef]
- Sterkel, M.; Perdomo, H.D.; Guizzo, M.G.; Barletta, A.B.F.; Nunes, R.D.; Dias, F.A.; Sorgine, M.; Oliveira, P.L. Tyrosine detoxification is an essential trait in the life history of blood-feeding arthropods. Curr. Biol. 2016, 26, 2188–2193. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, Z.; Dai, Z.; Yang, Y.; Wang, J.; Wu, G. Glycine metabolism in animals and humans: Implications for nutrition and health. Amino Acids 2013, 45, 463–477. [Google Scholar] [CrossRef]
- Isoe, J.; Scaraffia, P.Y. Urea synthesis and excretion in Aedes aegypti mosquitoes are regulated by a unique cross-talk mechanism. PLoS ONE 2013, 8, e65393. [Google Scholar] [CrossRef]
- Leal, W.S.; Choo, Y.M.; Xu, P.; da Silva, C.S.; Ueira-Vieira, C. Differential expression of olfactory genes in the southern house mosquito and insights into unique odorant receptor gene isoforms. Proc. Natl. Acad. Sci. USA 2013, 110, 18704–18709. [Google Scholar] [CrossRef]
- Taparia, T.; Ignell, R.; Hill, S.R. Blood meal induced regulation of the chemosensory gene repertoire in the southern house mosquito. BMC Genom. 2017, 18, 393. [Google Scholar] [CrossRef]
- Isoe, J.; Petchampai, N.; Isoe, Y.E.; Co, K.; Mazzalupo, S.; Scaraffia, P.Y. Xanthine dehydrogenase-1 silencing in Aedes aegypti mosquitoes promotes a blood feeding–induced adulticidal activity. FASEB J. 2017, 31, 2276–2286. [Google Scholar] [CrossRef]
- Mattila, J.; Hietakangas, V. Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 2017, 207, 1231–1253. [Google Scholar]
- Beckstead, R.B.; Lam, G.; Thummel, C.S. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005, 6, R99. [Google Scholar] [CrossRef] [PubMed]
- Hagedorn, H.H.; O’Connor, J.D.; Fuchs, M.S.; Sage, B.; Schlaeger, D.A.; Bohm, M.K. The ovary as a source of alpha-ecdysone in an adult mosquito. Proc. Natl. Acad. Sci. USA 1975, 72, 3255–3259. [Google Scholar] [CrossRef]
- Dhara, A.; Eum, J.H.; Robertson, A.; Gulia-Nuss, M.; Vogel, K.J.; Clark, K.D.; Strand, M.R. Ovary ecdysteroidogenic hormone functions independently of the insulin receptor in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. 2013, 43, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Gulia-Nuss, M.; Eum, J.H.; Strand, M.R.; Brown, M.R. Ovary ecdysteroidogenic hormone activates egg maturation in the mosquito Georgecraigius atropalpus after adult eclosion or a blood meal. J. Exp. Biol. 2012, 215, 3758–3767. [Google Scholar] [CrossRef]
- Liu, C.; Pitts, R.; Bohbot, J.; Jones, P.; Wang, G. Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS Biol. 2010, 8, e72595. [Google Scholar] [CrossRef] [PubMed]
- Syed, Z.; Leal, W.S. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proc. Natl. Acad. Sci. USA 2009, 106, 18803–18808. [Google Scholar] [CrossRef] [PubMed]
- Price, D.C.; Fonseca, D.M. Genetic divergence between populations of feral and domestic forms of a mosquito disease vector assessed by transcriptomics. PeerJ 2015, 3, e807. [Google Scholar] [CrossRef]
- Hill, S.R.; Ghaninia, M.; Ignell, R. Blood meal induced regulation of gene expression in the maxillary palps, a chemosensory organ of the mosquito Aedes aegypti. Front. Ecol. Evol. 2019, 336. [Google Scholar] [CrossRef]
- Anholt, R.R.H. Chemosensation and evolution of Drosophila host plant selection. IScience 2020, 23, 100799. [Google Scholar] [CrossRef]
- Clyne, P.J.; Warr, C.G.; Freeman, M.R.; Lessing, D.; Kim, J.; Carlson, J.R. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron 1999, 22, 327–338. [Google Scholar] [CrossRef]
- Freeman, E.G.; Dahanukar, A. Molecular neurobiology of Drosophila taste. Curr. Opin. Neurobiol. 2015, 34, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Martin-Martin, I.; Paige, A.; Leon, P.C.V.; Gittis, A.G.; Kern, O.; Bonilla, B.; Chagas, A.C.; Ganesan, S.; Smith, L.B.; Garboczi, D.N.; et al. ADP binding by the Culex quinquefasciatus mosquito D7 salivary protein enhances blood feeding on mammals. Nat. Commun. 2020, 11, 2911. [Google Scholar] [CrossRef] [PubMed]

| Trait | Form Pipiens | Form Molestus | Citations |
|---|---|---|---|
| Overwintering behavior | Diapausing. | Does not diapause. | [31,32,33,34,35] |
| Breeding site | Breeds in above-ground aquatic habitats. | Breeds in below-ground aquatic habitats. | [36,37,38,39,40] |
| Mating behavior | Mates in swarms above ground. | Mates below ground in confined spaces. | [22,26,39,41,42] |
| Reproduction | Requires a blood meal for egg production in the first gonotrophic cycle (anautogeny). | Does not require a blood meal for egg production in the first gonotrophic cycle (facultative autogeny). | [43,44,45,46] |
| Host preference | Primarily avian-seeking. | Primarily mammal-seeking. | [23,47,48,49,50,51,52,53,54,55,56] |
| BG Gravid vs. BG Parous | BG Parous vs. AG Nulliparous | BG Gravid vs. AG Nulliparous | |
|---|---|---|---|
| FC > 1.5 (up) | 10 | 705 | 761 |
| FC < 1.5 (down) | 6 | 689 | 641 |
| Total | 16 | 1394 | 1402 |
| Gene Family | Vector Base ID | Gene Name | Log2 FC (Shrunken) | padj | Orthology Group |
|---|---|---|---|---|---|
| Odorant receptor | CPIJ016433 | OR137 | −2.54 | 9.6 × 10−5 | OG6_163622 |
| Ionotropic receptor | CPIJ019300 | IR76b | 0.55 | 3.6 × 10−3 | OG6_126385 |
| Gustatory receptor | CPIJ011564 | NA | −1.22 | 2.4 × 10−2 | OG6_187005 |
| Odorant Binding Proteins | CPIJ001730 | OBP4 | −1.07 | 4.0 × 10−4 | OG6_201790 |
| CPIJ002108 | OBP108 | −0.83 | 4.0 × 10−2 | OG6_136613 | |
| CPIJ002109 | OBP107 | −1.11 | 1.1 × 10−2 | OG6_151055 | |
| CPIJ002111 | OBP110 | −0.60 | 2.6 × 10−2 | OG6_163322 | |
| CPIJ004145 | OBP64 | −4.73 | 7.6 × 10−16 | OG6_140589 | |
| CPIJ004634 | OBP102 | 0.49 | 6.1 × 10−3 | OG6_142050 | |
| CPIJ007604 | OBP1 | −0.49 | 2.1 × 10−2 | OG6_117872 | |
| CPIJ007617 | OBP2 | −0.65 | 6.4 × 10−3 | OG6_163124 | |
| CPIJ008793 | OBP6 | −0.55 | 2.0 × 10−4 | OG6_110106 | |
| CPIJ009568 | OBP8 | 0.64 | 1.4 × 10−4 | OG6_110106 | |
| CPIJ010367 | OBP55 | −1.09 | 1.0 × 10−12 | OG6_124323 | |
| CPIJ010787 | OBP51 | 2.30 | 9.1 × 10−11 | OG6_150797 | |
| CPIJ012716 | OBP17 | 0.89 | 2.1 × 10−2 | OG6_107904 | |
| CPIJ012717 | OBP18 | 1.07 | 2.4 × 10−2 | OG6_150726 | |
| CPIJ012719 | OBP20 | 3.18 | 5.4 × 10−19 | OG6_107904 | |
| Odorant Binding Proteins | CPIJ013976 | OBP10 | 8.24 | 6.2 × 10−15 | OG6_153567 |
| CPIJ014525 | OBP24 | 0.64 | 8.9 × 10−4 | OG6_107904 | |
| CPIJ016479 | OBP32 | −0.77 | 3.8 × 10−2 | OG6_117689 | |
| CPIJ016965 | OBP28 | 0.46 | 5.5 × 10−3 | OG6_128903 | |
| CPIJ016966 | OBP29 | −1.92 | 2.0 × 10−29 | OG6_128903 | |
| CPIJ019610 | OBP36 | −0.81 | 1.2 × 10−2 | OG6_167102 | |
| Sensory Neuron Membrane Protein | CPIJ014330 | SNMP1a | −0.62 | 5.0 × 10−3 | OG6_117830 |
| Chemosensory Proteins | CPIJ002605 | CSP2 | 1.42 | 1.9 × 10−12 | OG6_120812 |
| CPIJ002608 | CSP4 | 0.32 | 4.9 × 10−2 | OG6_107706 | |
| CPIJ002618 | CSP13 | 4.66 | 6.3 × 10−5 | OG6_120813 | |
| CPIJ002628 | CSP23 | 0.78 | 7.3 × 10−6 | OG6_162766 | |
| Opsins | CPIJ004067 | GPROP1 | −0.60 | 1.3 × 10−3 | OG6_163080 |
| CPIJ005000 | GPROP2 | 0.24 | 3.1 × 10−2 | OG6_120825 | |
| CPIJ009246 | GPROP3 | 0.19 | 4.5 × 10−2 | OG6_124296 | |
| CPIJ011571 | GPROP6 | 0.50 | 9.2 × 10−8 | OG6_104608 | |
| CPIJ011573 | GPROP7 | 0.54 | 4.7 × 10−3 | OG6_104608 | |
| CPIJ014334 | GPROP12 | 0.48 | 3.0 × 10−2 | OG6_105417 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noreuil, A.; Fritz, M.L. Differential Gene Expression in the Heads of Behaviorally Divergent Culex pipiens Mosquitoes. Insects 2021, 12, 271. https://doi.org/10.3390/insects12030271
Noreuil A, Fritz ML. Differential Gene Expression in the Heads of Behaviorally Divergent Culex pipiens Mosquitoes. Insects. 2021; 12(3):271. https://doi.org/10.3390/insects12030271
Chicago/Turabian StyleNoreuil, Anna, and Megan L. Fritz. 2021. "Differential Gene Expression in the Heads of Behaviorally Divergent Culex pipiens Mosquitoes" Insects 12, no. 3: 271. https://doi.org/10.3390/insects12030271
APA StyleNoreuil, A., & Fritz, M. L. (2021). Differential Gene Expression in the Heads of Behaviorally Divergent Culex pipiens Mosquitoes. Insects, 12(3), 271. https://doi.org/10.3390/insects12030271







