Five Silkworm 30K Proteins Are Involved in the Cellular Immunity against Fungi
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Bioinformatics Analysis
2.3. Immune Injection in Silkworm
2.4. Total RNA Isolation and cDNA Synthesis
2.5. Real-Time Quantitative Polymerase Chain Reaction
2.6. Expression and Purification of the Recombinant Protein
2.7. Binding Assay of Recombinant 30K Proteins
2.8. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis and Western Blotting
2.9. Immunofluorescence
2.10. Non-Reduce PAGE
2.11. In Vitro Encapsulation
3. Results
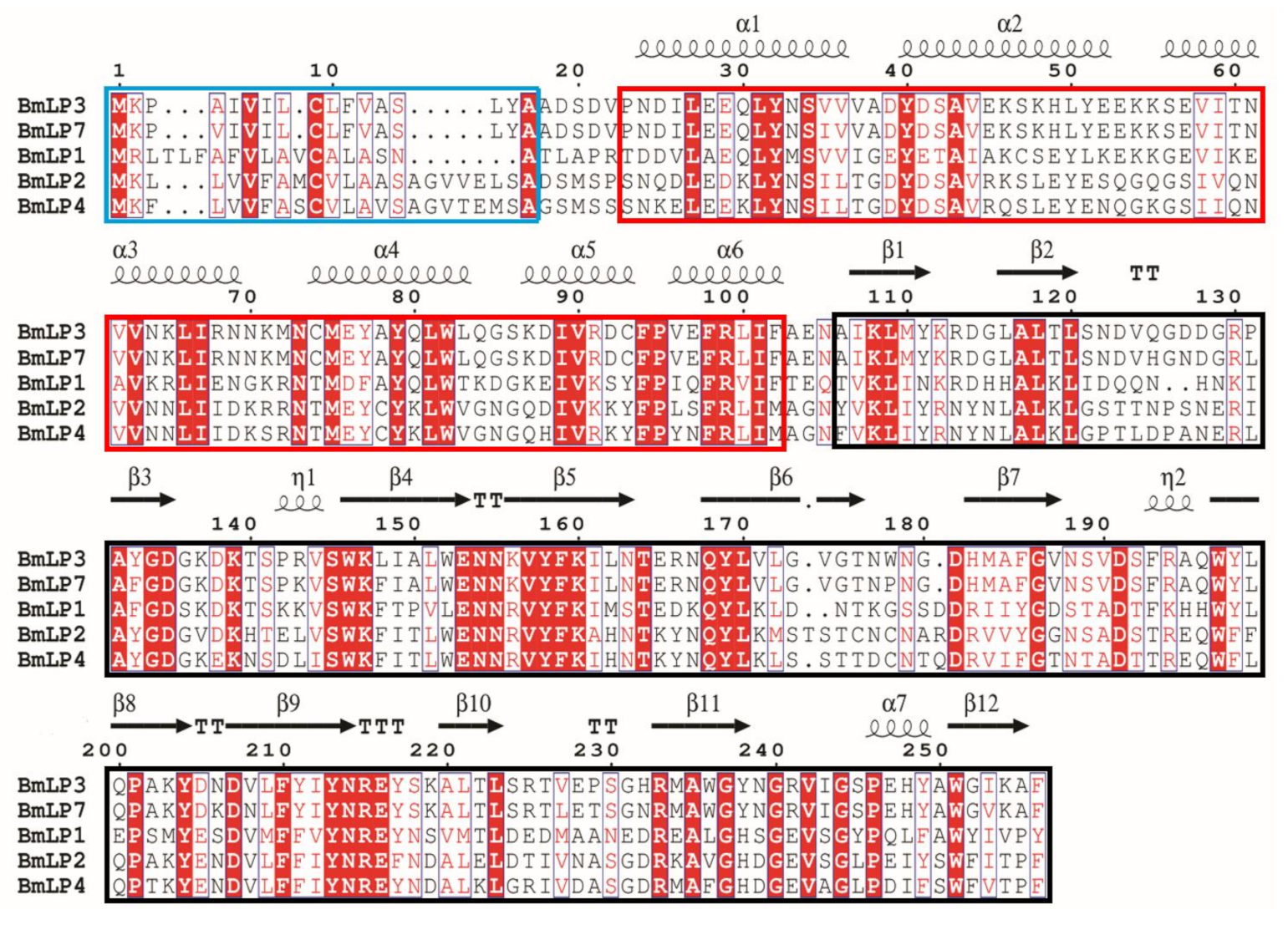
3.1. Sequence Analysis of Five 30K Proteins
3.2. PAMPs Upregulate the Expression of 30K Genes
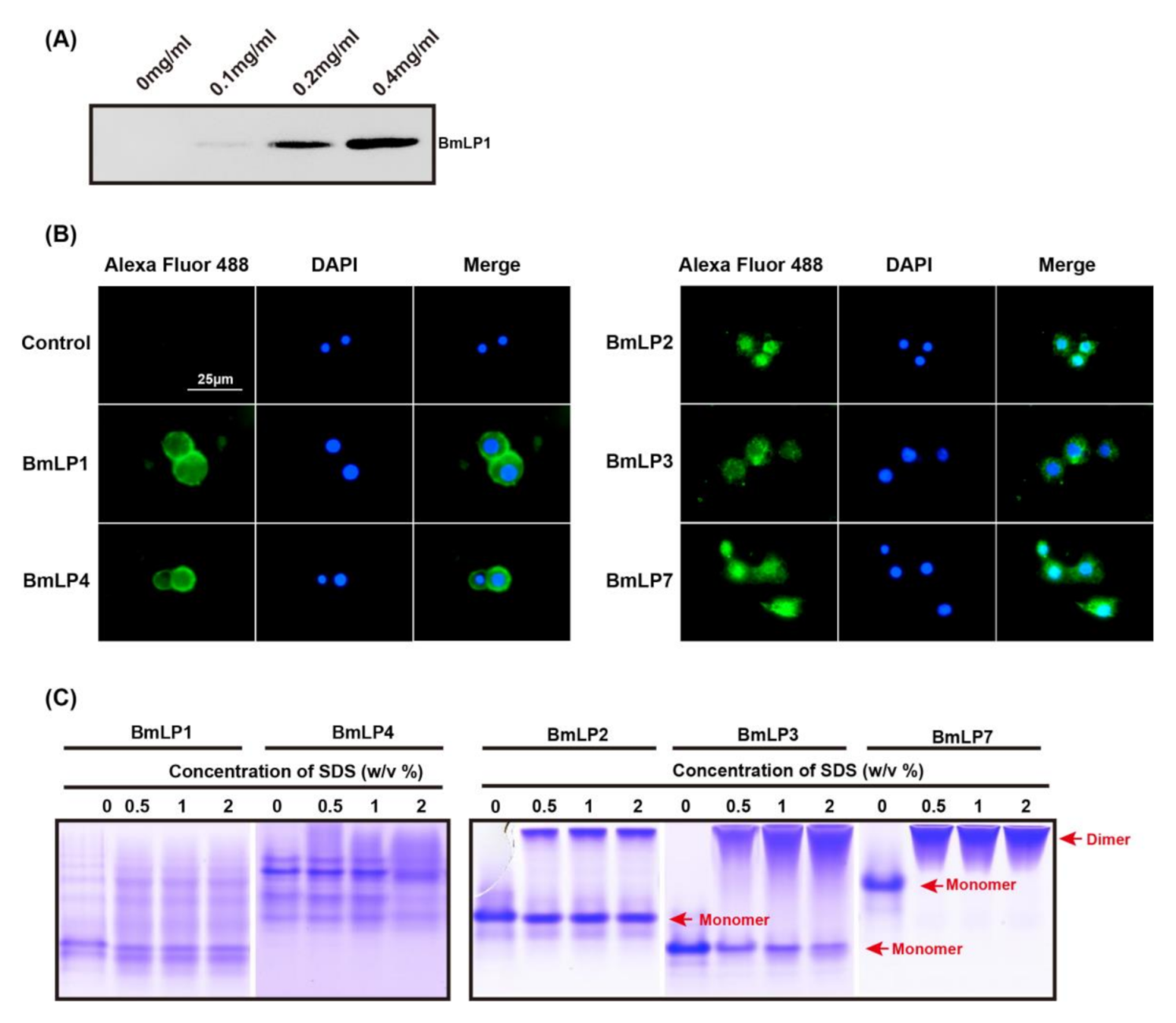
3.3. Expression and Purification of Recombinant 30K Proteins
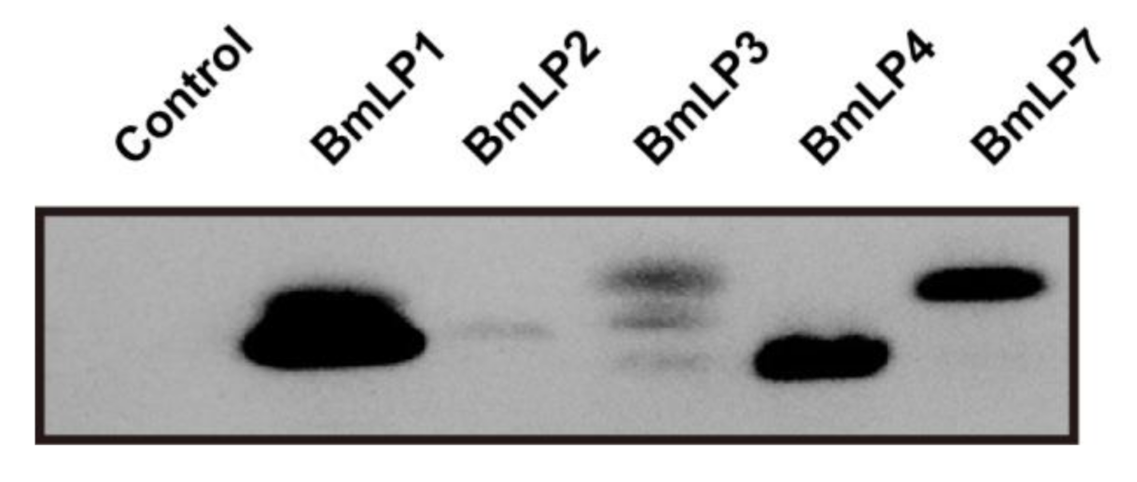
3.4. 30K Proteins Can Bind to Fungus
3.5. 30K Proteins Can Bind to Hemocytes
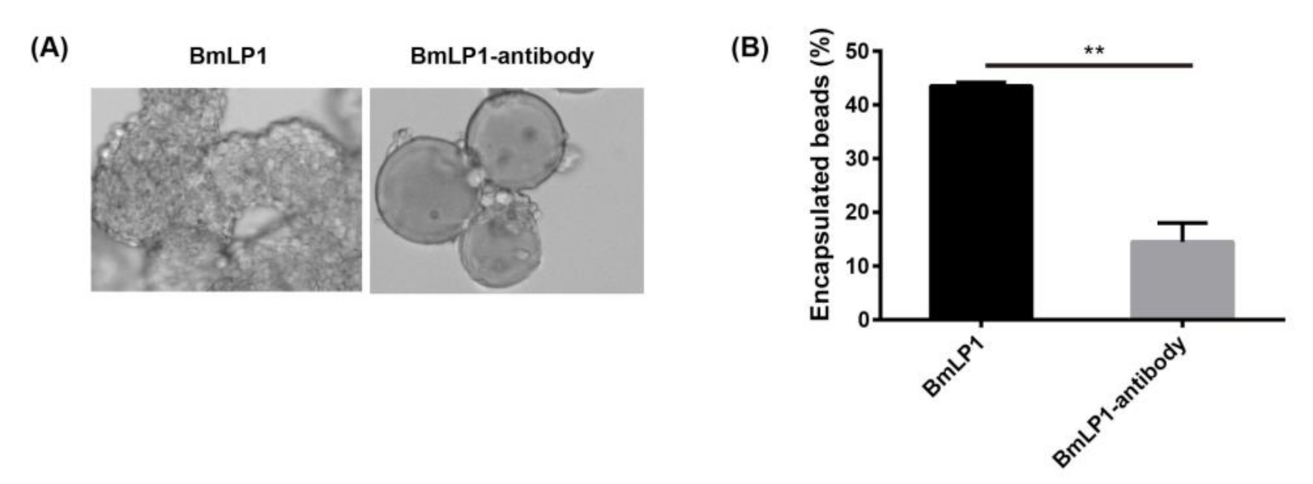
3.6. BmLP1 and BmLP4 Promotes Hemocyte Encapsulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gillott, C. The Circulatory System. In Entomology; Springer: Dordrecht, The Netherlands, 2005; pp. 515–536. [Google Scholar] [CrossRef]
- Wyatt, G.R.; Loughheed, T.C.; Wyatt, S.S. The chemistry of insect hemolymph. J. Gen. Physiol. 1956, 39, 853–868. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, G.R. The Biochemistry of Insect Hemolymph. Annu. Rev. Entomol. 1961, 6, 75–102. [Google Scholar] [CrossRef]
- Zhou, L.; Li, H.; Hao, F.; Li, N.; Liu, X.; Wang, G.; Wang, Y.; Tang, H. Developmental Changes for the Hemolymph Metabolome of Silkworm (Bombyx mori L.). J. Proteome Res. 2015, 14, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Tojo, S.; Nagata, M.; Kobayashi, M. Storage proteins in the silkworm, Bombyx mori. Insect Biochem. 1980, 10, 289–303. [Google Scholar] [CrossRef]
- Gamo, T. Low molecular weight lipo-proteins in the hemolymph of the silkworm, Bombyx mori: Inheritance, isolation and some properties. Insect Biochem. 1978, 8, 457–470. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Telfer, W.H.; Kunkel, J.G. The function and evolution of insect storage hexamers. Annu. Rev. Entomol. 1991, 36, 205–228. [Google Scholar] [CrossRef]
- Lee, D.J.; Martin, G.; Ferreras, F.M.; Matthews, P.G.D. Changes in hemolymph total CO2 content during the water-to-air respiratory transition of amphibiotic dragonflies. J. Exp. Biol. 2018, 221, jeb.181438. [Google Scholar] [CrossRef]
- Hernández-Martínez, S.; Rivera-Perez, C.; Nouzova, M.; Noriega, F.G. Coordinated changes in JH biosynthesis and JH hemolymph titers in Aedes aegypti mosquitoes. J. Insect Physiol. 2015, 72, 22–27. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Dong, Z.; Wang, D.; Wu, Y.; Song, Q.; Gu, P.; Zhao, P.; Xia, Q. Proteomics of larval hemolymph in Bombyx mori reveals various nutrient-storage and immunity-related proteins. Amino Acids 2014, 46, 1021–1031. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, Z.; Liu, S.; Yang, Q.; Zhao, P.; Xia, Q. Identification of novel members reveals the structural and functional divergence of lepidopteran-specific Lipoprotein_11 family. Funct. Integr. Genom. 2012, 12, 705–715. [Google Scholar] [CrossRef]
- Strand, M.R.; Hayakawa, Y.; Clark, K.D. Plasmatocyte spreading peptide (PSP1) and growth blocking peptide (GBP) are multifunctional homologs. J. Insect Physiol. 2000, 46, 817–824. [Google Scholar] [CrossRef]
- Bosquet, G.; Guillet, C.; Calvez, B.; Chavancy, G. The regulation of major haemolymph protein synthesis: Changes in mRNA content during the development of Bombyx mori larvae. Insect Biochem. 1989, 19, 29–39. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, P.; Liu, H.; Dong, Z.; Yang, Q.; Wang, D.; Xia, Q. The synthesis, transportation and degradation of BmLP3 and BmLP7, two highly homologous Bombyx mori 30K proteins. Insect Biochem. Mol. Biol. 2012, 42, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Pietrzyk, A.J.; Panjikar, S.; Bujacz, A.; Mueller-Dieckmann, J.; Lochynska, M.; Jaskolski, M.; Bujacz, G. High-resolution structure of Bombyx mori lipoprotein 7: Crystallographic determination of the identity of the protein and its potential role in detoxification. Acta Cryst. D Biol. Cryst. 2012, 68, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Pietrzyk, A.J.; Bujacz, A.; Mueller-Dieckmann, J.; Lochynska, M.; Jaskolski, M.; Bujacz, G. Two crystal structures of Bombyx mori lipoprotein 3-structural characterization of a new 30-kDa lipoprotein family member. PLoS ONE 2013, 8, e61303. [Google Scholar] [CrossRef] [PubMed]
- Pietrzyk, A.J.; Bujacz, A.; Lochynska, M.; Jaskolski, M.; Bujacz, G. Crystal structure of Bombyx mori lipoprotein 6: Comparative structural analysis of the 30-kDa lipoprotein family. PLoS ONE 2014, 9, e108761. [Google Scholar] [CrossRef]
- Yang, J.-P.; Ma, X.-X.; He, Y.-X.; Li, W.-F.; Kang, Y.; Bao, R.; Chen, Y.; Zhou, C.-Z. Crystal structure of the 30K protein from the silkworm Bombyx mori reveals a new member of the β-trefoil superfamily. J. Struct. Biol. 2011, 175, 97–103. [Google Scholar] [CrossRef]
- Zhan, M.Y.; Shahzad, T.; Yang, P.J.; Liu, S.; Yu, X.Q.; Rao, X.J. A single-CRD C-type lectin is important for bacterial clearance in the silkworm. Dev. Comp. Immunol. 2016, 65, 330–339. [Google Scholar] [CrossRef]
- Ujita, M.; Kimura, A.; Nishino, D.; Yokoyama, E.; Banno, Y.; Fujii, H.; Hara, A. Specific binding of silkworm Bombyx mori 30-kDa lipoproteins to carbohydrates containing glucose. Biosci. Biotechnol. Biochem. 2002, 66, 2264–2266. [Google Scholar] [CrossRef]
- Ujita, M.; Katsuno, Y.; Kawachi, I.; Ueno, Y.; Banno, Y.; Fujii, H.; Hara, A. Glucan-binding activity of silkworm 30-kDa apolipoprotein and its involvement in defense against fungal infection. J. Agric. Chem. Soc. Jpn. 2005, 69, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Sekimizu, K. Silkworm as an experimental animal for research on fungal infections. Microbiol. Immunol. 2019, 63, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Kurakado, S.; Sugita, T. Evaluating Candida albicans biofilm formation in silkworms. Med. Mycol. 2020, 58, myaa064. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.M.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.W.; Wilm, A.; Lopez, R.; et al. Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Patrice, G.; Xavier, R.; Emmanuel, C. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003, 31, 3320–3323. [Google Scholar] [CrossRef]
- Gillespie, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef]
- Wang, H.; Song, L.; Li, C.; Zhao, J.; Zhang, H.; Ni, D.; Xu, W. Cloning and characterization of a novel C-type lectin from Zhikong scallop Chlamys farreri. Mol. Immunol. 2007, 44, 722–731. [Google Scholar] [CrossRef]
- Ao, J.; Ling, E.; Yu, X.Q. Drosophila C-type lectins enhance cellular encapsulation. Mol. Immunol. 2007, 44, 2541–2548. [Google Scholar] [CrossRef]
- Yang, J.; Wang, L.; Zhang, H.; Qiu, L.; Wang, H.; Song, L. C-type lectin in Chlamys farreri (CfLec-1) mediating immune recognition and opsonization. PLoS ONE 2011, 6, e17089. [Google Scholar] [CrossRef]
- Ling, E.; Yu, X. Hemocytes from the tobacco hornworm Manduca sexta have distinct functions in phagocytosis of foreign particles and self dead cells. Dev. Comp. Immunol. 2006, 30, 301–309. [Google Scholar] [CrossRef]
- Shu, M.; Mang, D.; Fu, G.S.; Tanaka, S.; Endo, H.; Kikuta, S.; Sato, R. Mechanisms of nodule-specific melanization in the hemocoel of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2016, 70, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Ohta, M.; Suzuki, A.; Takase, H.; Yoshizawa, Y.; Kitami, M.; Sato, R. Localization of the serine protease homolog BmSPH-1 in nodules of E. coli-injected Bombyx mori larvae and functional analysis of its role in nodule melanization. Dev. Comp. Immunol. 2011, 35, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; You, M.; Rao, X.J.; Yu, X.Q. Insect C-type lectins in innate immunity. Dev. Comp. Immunol. 2018, 83, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, J.H.; Park, H.H.; Rhee, W.J.; Choi, S.S.; Park, T.H. A protein delivery system using 30Kc19 cell-penetrating protein originating from silkworm. Biomaterials 2012, 33, 9127–9134. [Google Scholar] [CrossRef]
- Park, H.H.; Sohn, Y.; Yeo, J.W.; Park, J.H.; Lee, H.J.; Ryu, J.; Rhee, W.J.; Park, T.H. Dimerization of 30Kc19 protein in the presence of amphiphilic moiety and importance of Cys-57 during cell penetration. Biotechnol. J. 2014, 9, 1582–1593. [Google Scholar] [CrossRef][Green Version]
- Richard, J.P.; Melikov, K.; Vives, E.; Ramos, C.; Verbeure, B.; Gait, M.J.; Chernomordik, L.V.; Lebleu, B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003, 278, 585–590. [Google Scholar] [CrossRef]
- Jan Akhunzada, M.; Chandramouli, B.; Bhattacharjee, N.; Macchi, S.; Cardarelli, F.; Brancato, G. The role of Tat peptide self-aggregation in membrane pore stabilization: Insights from a computational study. Phys. Chem. Chem. Phys. 2017, 19, 27603–27610. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, L.; Zhang, Y.; Dong, Z.; Guo, P.; Zhao, D.; Li, H.; Hu, H.; Zhou, X.; Chen, H.; Zhao, P. Five Silkworm 30K Proteins Are Involved in the Cellular Immunity against Fungi. Insects 2021, 12, 107. https://doi.org/10.3390/insects12020107
Ye L, Zhang Y, Dong Z, Guo P, Zhao D, Li H, Hu H, Zhou X, Chen H, Zhao P. Five Silkworm 30K Proteins Are Involved in the Cellular Immunity against Fungi. Insects. 2021; 12(2):107. https://doi.org/10.3390/insects12020107
Chicago/Turabian StyleYe, Lin, Yan Zhang, Zhaoming Dong, Pengchao Guo, Dongchao Zhao, Haoyun Li, Hang Hu, Xiaofang Zhou, Haiqin Chen, and Ping Zhao. 2021. "Five Silkworm 30K Proteins Are Involved in the Cellular Immunity against Fungi" Insects 12, no. 2: 107. https://doi.org/10.3390/insects12020107
APA StyleYe, L., Zhang, Y., Dong, Z., Guo, P., Zhao, D., Li, H., Hu, H., Zhou, X., Chen, H., & Zhao, P. (2021). Five Silkworm 30K Proteins Are Involved in the Cellular Immunity against Fungi. Insects, 12(2), 107. https://doi.org/10.3390/insects12020107





