Expression Profile and Ligand Screening of a Putative Odorant-Binding Protein, AcerOBP6, from the Asian Honeybee
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Sample Preparation
2.2. RNA Isolation and Quantitative Real-Time PCR
2.3. Cloning Full-Length of cDNA and Sequence Analysis
2.4. Recombinant Protein Expression and Purification
2.5. Fluorescence Competitive Binding Assay
2.6. dsRNA Synthesis and RNAi Verification
2.7. EAG Assays Based on RNAi
2.8. Statistical Analysis
3. Results
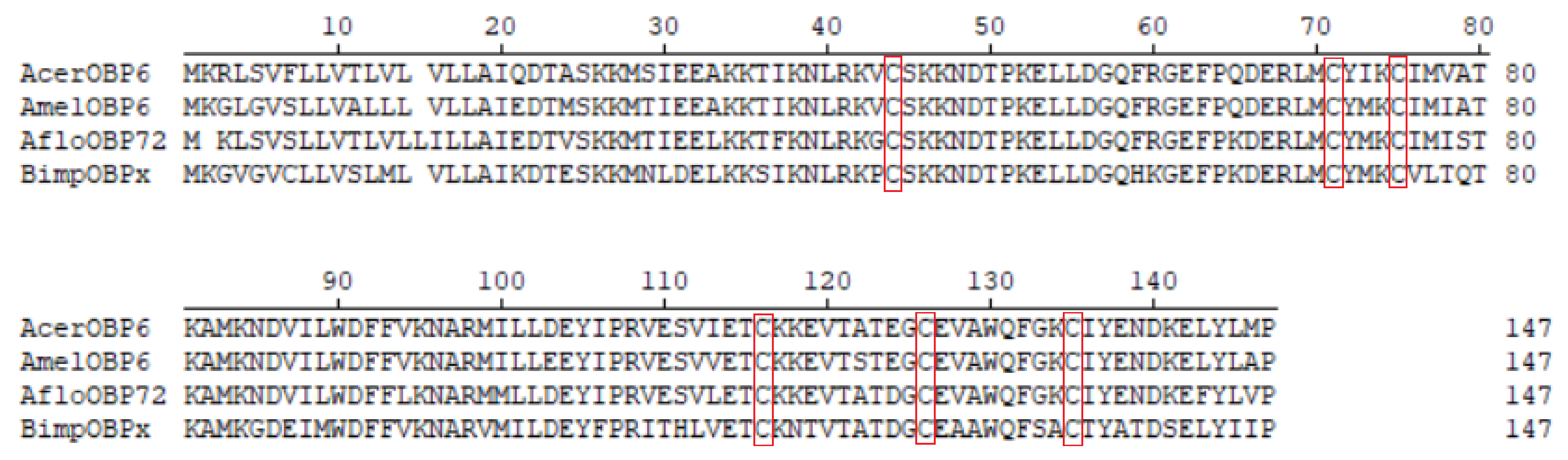
3.1. Sequence Characteristic of AcerOBP6
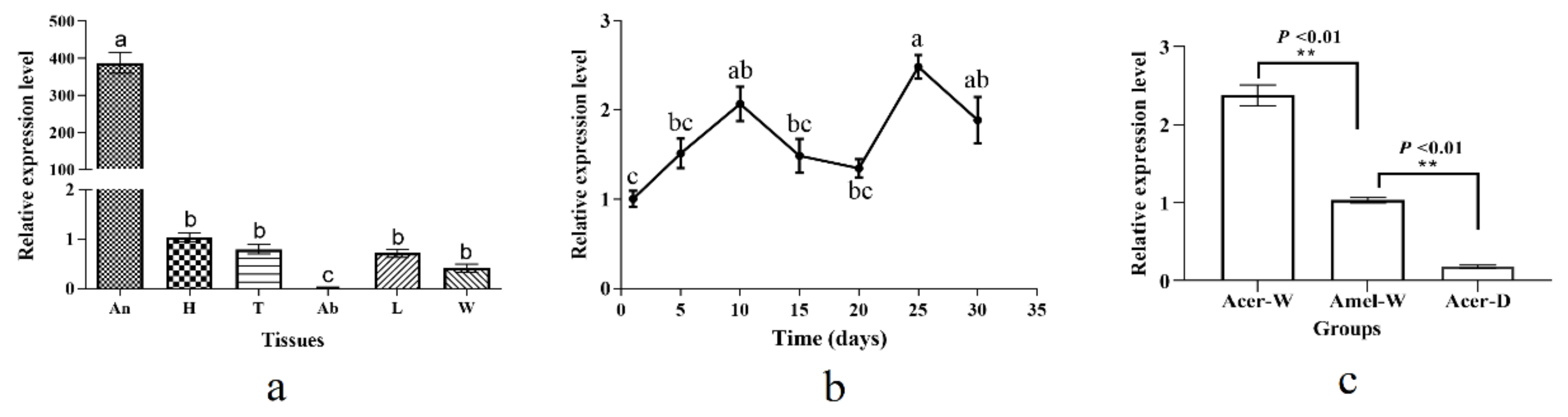
3.2. Expression Profiles of AcerOBP6
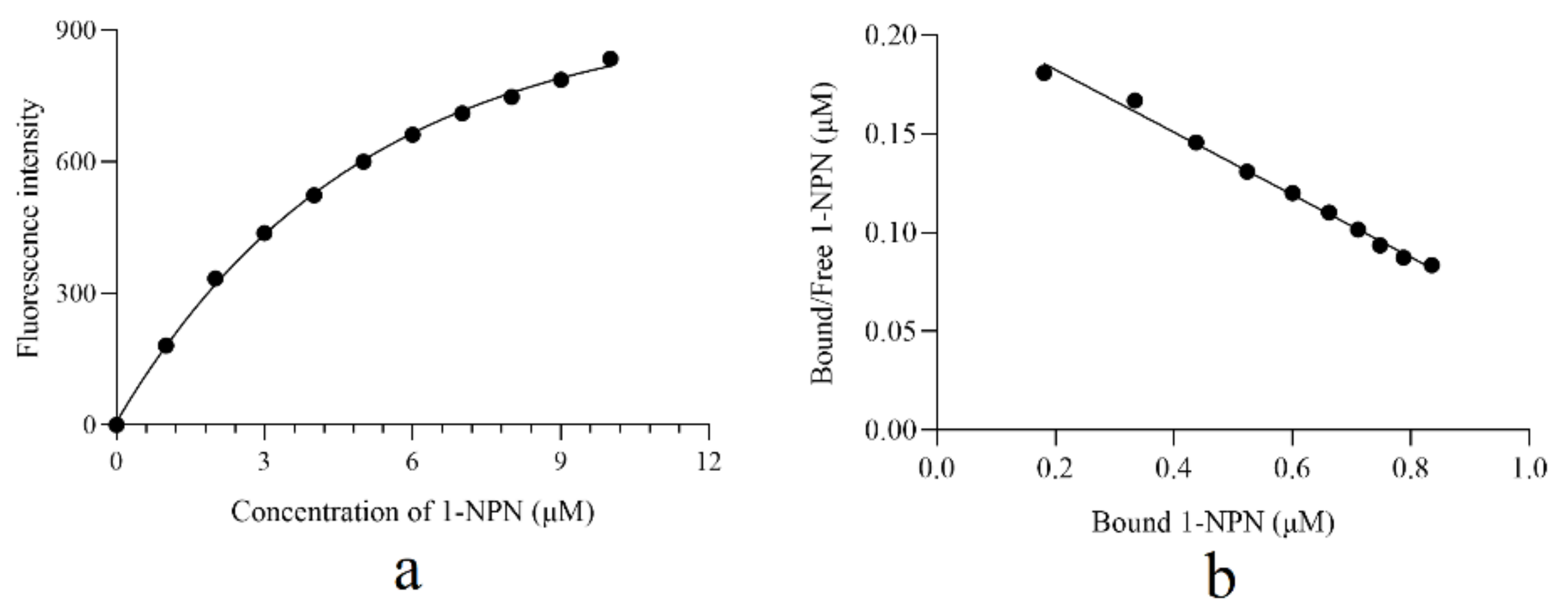
3.3. Ligand Binding Characteristic of AcerOBP6
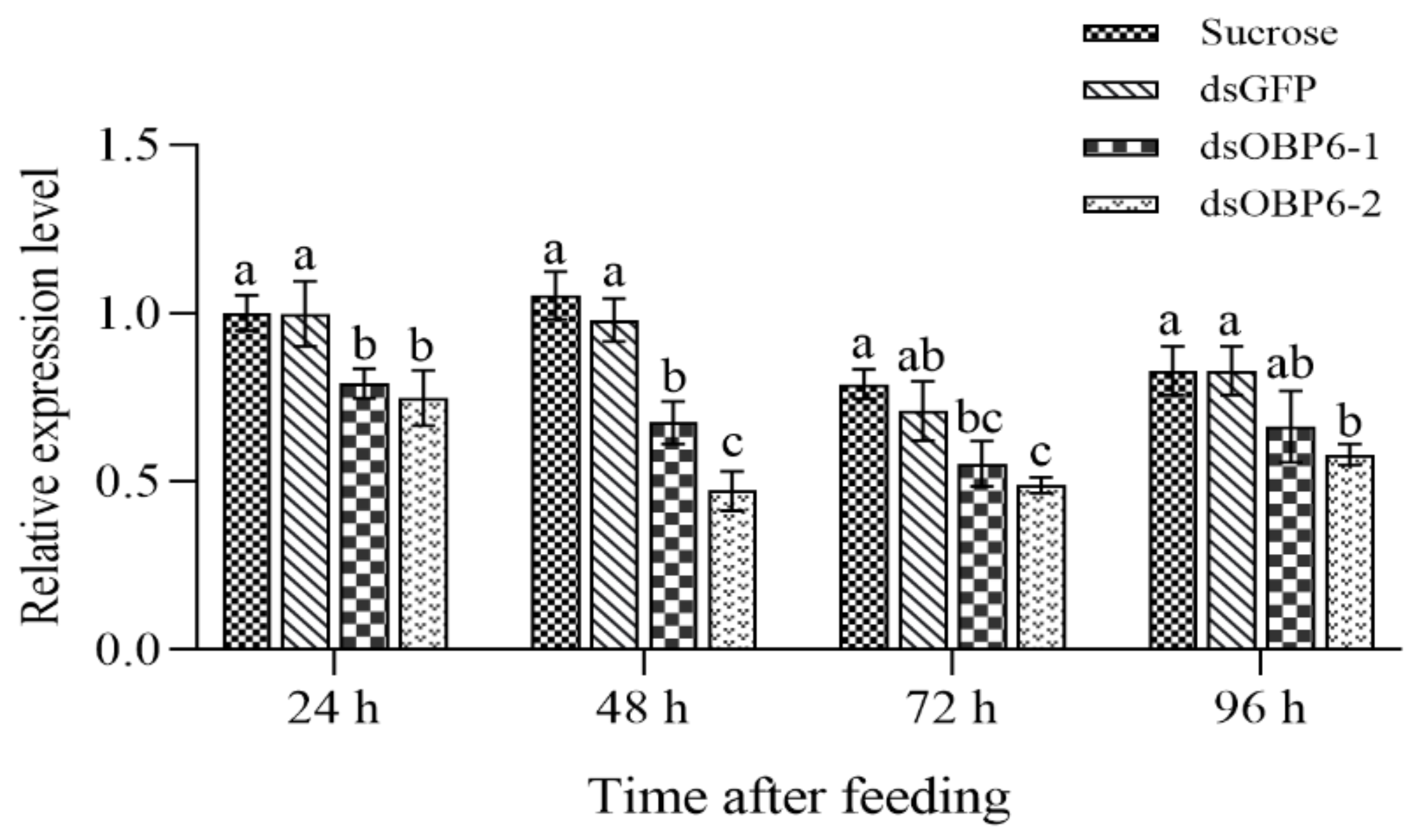
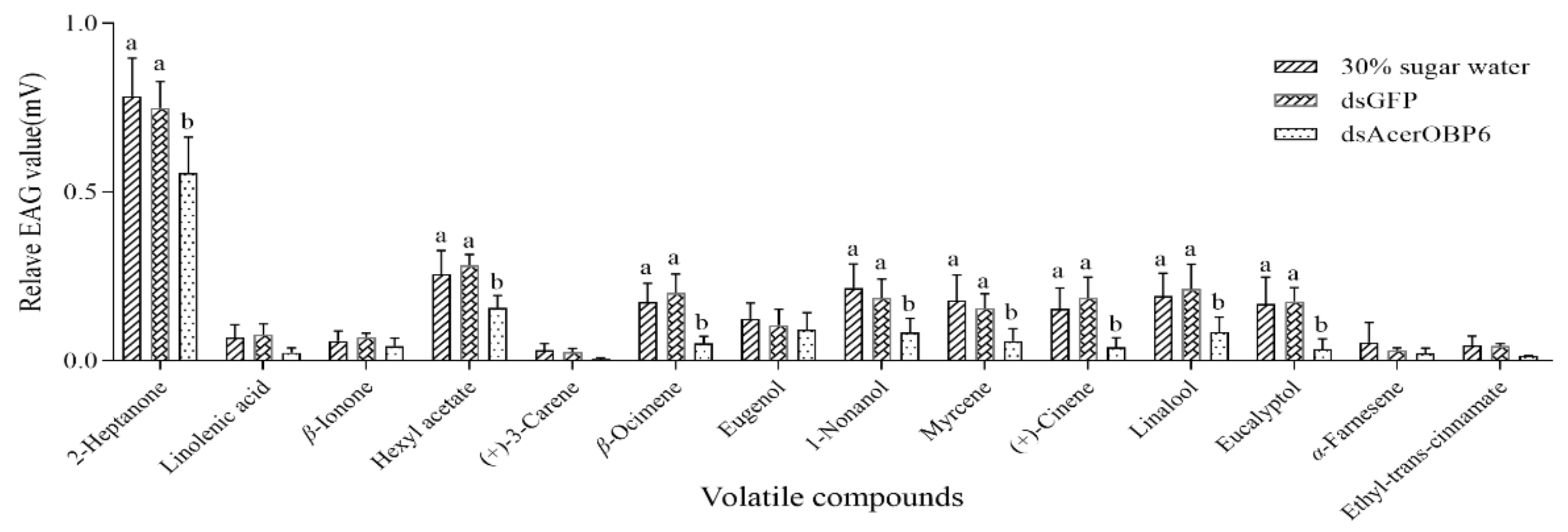
3.4. Further Results from RNAi and EAG Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ache, B.W.; Young, J.M. Olfaction: Diverse species, conserved principles. Neuron 2005, 48, 417–430. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wang, M.Q.; Zhang, G. Ultrastructural observations on antennal sensilla of Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). Microsc. Res. Tech. 2011, 74, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.C.; Wang, C.P.; Wei, J.Q. Advance in the research on insect peripheral olfactory system. Acta Agric. Univ. Jiangxiensis 2019, 41, 50–57. [Google Scholar]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Brito, N.F.; Moreira, M.F.; Melo, A.C. A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 2016, 95, 51–65. [Google Scholar] [CrossRef]
- Ban, L.P.; Scaloni, A.; D’Ambrosio, C.; Zhang, L.; Yan, Y.H.; Pelosi, P. Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell. Mol. Life Sci. 2003, 60, 390–400. [Google Scholar] [CrossRef]
- Yang, M.X.; Tan, K.; Radloff, S.E.; Phiancharoen, M.; Hepburn, H.R. Comb construction in mixed-species colonies of honeybees, Apis cerana and Apis mellifera. J. Exp. Biol. 2010, 213 Pt 10, 1659–1664. [Google Scholar] [CrossRef] [Green Version]
- Diao, Q.Y.; Sun, L.X.; Zheng, H.J.; Zeng, Z.J.; Wang, S.Y.; Xu, S.F.; Zheng, H.Q.; Chen, Y.P.; Shi, Y.Y.; Wang, Y.Z.; et al. Genomic and transcriptomic analysis of the Asian honeybee Apis cerana provides novel insights into honeybee biology. Sci. Rep. 2018, 8, 822. [Google Scholar] [CrossRef] [PubMed]
- Forêt, S.; Maleszka, R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 2006, 16, 1404–1413. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Du, Y.; Gao, P.; Wang, S.; Pan, J.; Jiang, Y. Antennal transcriptome and differential expression analysis of five chemosensory gene families from the Asian honeybee Apis cerana cerana. PLoS ONE 2016, 11, e0165374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danty, E.; Briand, L.; Michard-Vanhee, C.; Perez, V.; Arnold, G.; Gaudemer, O.; Huet, D.; Huet, J.; Ouali, C.; Masson, C.; et al. Cloning and expression of a queen pheromone-binding protein in the honeybee: An olfactory-specific, developmentally regulated protein. J. Neurosci. 1999, 19, 7468–7475. [Google Scholar] [CrossRef] [Green Version]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.C.; Tegoni, M.; Cambillau, C. Structural basis of the honeybee PBP pheromone and pH-induced conformational change. J. Mol. Biol. 2008, 380, 158–169. [Google Scholar] [CrossRef]
- Li, H.L.; Gao, Q.K.; Cheng, J.A. Cloning and spatio-temporal expression of cDNA encoding pheromone binding protein ASPl in Apis cerana cerana (Hymenoptera Apidae). Acta Entomol. Sin. 2008, 51, 689–693. [Google Scholar]
- Weng, C.; Fu, Y.; Jiang, H.; Zhuang, S.; Li, H. Binding interaction between a queen pheromone component HOB and pheromone binding protein ASP1 of Apis cerana. Int. J. Biol. Macromol. 2015, 72, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Danty, E.; Michard Vanhee, C.; Huet, J.C.; Genecque, E.; Pernollet, J.C.; Masson, C. Biochemical characterization, molecular cloning and localization of a putative odorant-binding protein in the honeybee Apis mellifera L. (Hymenoptem Apidea). FEBS. Lett. 1997, 414, 595–598. [Google Scholar] [CrossRef] [Green Version]
- Briand, L.; Nespoulous, C.; Huet, J.C.; Takahashi, M.; Pernollet, J.C. Ligand binding and physico-chemical properties of ASP2, a recombinant odorant-binding protein from honeybee (Apis mellifera L.). Eur. J. Biochem. 2001, 268, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Song, X.M.; Wu, F.; Qiu, Y.L.; Fu, X.B.; Zhang, L.Y.; Tan, J. Chemical structure of semiochemicals and key binding sites together determine the olfactory functional modes of odorant-binding protein 2 in Eastern honeybee, Apis cerana. Int. J. Biol. Macromol. 2020, 145, 876–884. [Google Scholar] [CrossRef]
- Iovinella, I.; Dani, F.R.; Niccolini, A.; Sagona, S.; Michelucci, E.; Gazzano, A.; Turillazzi, S.; Felicioli, A.; Pelosi, P. Differential expression of odorant-binding proteins in the mandibular glands of the honeybee according to caste and age. J. Proteome Res. 2011, 10, 3439–3449. [Google Scholar] [CrossRef]
- Song, X.M.; Zhang, L.Y.; Fu, X.B.; Wu, F.; Tan, J.; Li, H.L. Various Bee Pheromones Binding Affinity, Exclusive Chemosensillar Localization, and Key Amino Acid Sites Reveal the Distinctive Characteristics of Odorant-Binding Protein 11 in the Eastern Honeybee, Apis cerana. Front. Physiol. 2018, 9, 422. [Google Scholar] [CrossRef]
- Guo, D.Z.; Hao, C.H.; Cui, X.P.; Wang, Y.; Liu, Z.G.; Xu, B.H.; Guo, X.Q. Molecular and functional characterization of the novel odorant-binding protein gene AccOBP10 from Apis cerana cerana. J. Biochem. 2021, 169, 215–225. [Google Scholar] [CrossRef]
- Du, Y.L.; Xu, K.; Zhao, H.T.; Jiang, Y.S.; Li, H.Q. Identification and functional characterization of AcerOBP15 from Apis cerana cerana (Hymenoptera: Apidae). Apidologie 2021, 52, 668–683. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, J.J.; Vieira, F.G.; He, X.L.; Smadja, C.; Liu, R.; Rozas, J.; Field, L.M. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2010, 19 (Suppl. S2), 113–122. [Google Scholar] [CrossRef] [Green Version]
- Hull, J.J.; Peretra, O.P.; Snodgrass, L. Cloning and expression profiling of odorant-binding proteins in the tarnished plant bug, Lygus lineolaris. Insect Mol. Biol. 2014, 23, 78–97. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iovinella, I.; Cappa, F.; Cini, A.; Petrocelli, I.; Cervo, R.; Turillazzi, S.; Dani, F.R. Antennal protein profile in honeybees: Caste and task matter more than age. Front. Physiol. 2018, 9, 748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, Y.; Ishibashi, J.; Leal, W.S. Fatty acid solubilizer from the oral disk of the blowfly. PLoS ONE 2013, 8, e51779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- William, B.W.; Amit, R.; Peter, A.; Fredrik, S.; Bill, S.H.; Mattias, C.L. Transcriptome Analysis of Gene Families Involved in Chemosensory Function in Spodoptera littoralis (Lepidoptera: Noctuidae). BMC Genom. 2019, 20, 428. [Google Scholar]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. Camb. Philos. Soc. 2018, 93, 184–200. [Google Scholar] [CrossRef] [Green Version]
- Pelosi, P.; Mastrogiacomo, R.; Iovinella, I.; Tuccori, E.; Persaud, K.C. Structure and biotechnological applications of odorant binding proteins. Appl. Microbiol. Biot. 2014, 98, 61–70. [Google Scholar] [CrossRef]
- Huang, G.Z.; Liu, J.T.; Zhou, J.J.; Wang, Q.; Dong, J.Z.; Zhang, Y.J.; Li, X.C.; Li, J.; Gu, S.H. Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect Biochem. Mol. 2018, 7, 98. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Yan, Q.; Li, L.L.; Xu, J.W.; Mang, D.; Wang, X.L.; Hoh, H.H.; Ye, J.; Ju, Q.; Ma, Y.; et al. Different binding properties of two general-odorant binding proteins in Athetis lepigone with sex pheromones, host plant volatiles and insecticides. Pestic. Biochem. Physiol. 2020, 164, 173–182. [Google Scholar] [CrossRef]
- Weng, C.; Zhang, L.Y.; Zhao, L.; Fu, Y.X.; Luo, C.; Li, H.L. Prokaryotic expression and ligand binding characteristics of pheromone binding protein ASP1 in the Chinses honeybee (Apis cerana cerana). Acta Entomol. Sin. 2013, 56, 1110–1116. [Google Scholar]
- Wu, F.; Huang, J.J.; Tan, J.; Tang, M.Z.; Li, H.L. Molecular cloning, prokaryotic expression and lignand-binding characterization of a novel pheromone binding protein OBP10 in Apis cerana cerana (Hymenoptera: Apidae). Acta Entomol. Sin. 2016, 59, 25–32. [Google Scholar]
- Sun, S.F.; Zeng, F.F.; Yi, S.C.; Wang, M.Q. Molecular screening of behaviorally active compounds with CmedOBP14 from the rice leaf folder Cnaphalocrocis medinalis. J. Chem. Ecol. 2019, 45, 849–857. [Google Scholar] [CrossRef]
- Manning, R. Fatty acids in pollen: A review of their importance for honeybees. Bee World 2001, 82, 60–75. [Google Scholar] [CrossRef]
- Arien, Y.; Dag, A.; Shafir, S. Omega-6:3 Ratio More Than Absolute Lipid Level in Diet Affects Associative Learning in Honeybees. Front. Psychol. 2018, 9, 1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adal, A.M.; Sarker, L.S.; Lemke, A.D.; Mahmoud, S.S. Isolation and functional characterization of a methyl jasmonate-responsive 3-Carene synthase from Lavandula x intermedia. Plant Mol. Biol. 2017, 93, 641–657. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, N.M.; Qiu, B.-L.; Hordijk, C.A.; Vet, L.E.M.; Jansen, J.J. Identification of Biologically Relevant Compounds in Aboveground and Belowground Induced Volatile Blends. J. Chem. Ecol. 2010, 36, 1006–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bratt, K.; Sunnerheim, K.; Nordenhem, H.; Nordlander, G.; Langström, B. Pine weevil (Hylobius abietis) antifeedants from lodgepole pine (Pinus contorta). J. Chem. Ecol. 2001, 27, 2253–2262. [Google Scholar] [CrossRef] [PubMed]
- Aceña, L.; Vera, L.; Guasch, J.; Busto, O.; Mestres, M. Chemical characterization of commercial Sherry vinegar aroma by headspace solid-phase microextraction and gas chromatography-olfactometry. J. Agric. Food. Chem. 2011, 59, 4062–4070. [Google Scholar] [CrossRef]
- Vogt, R.G.; Prestwich, G.D.; Lerner, M.R. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J. Neurobiol. 1991, 22, 74–84. [Google Scholar] [CrossRef]
- Swevers, L.; Huvenne, H.; Menschaert, G.; Kontogiannatos, D.; Kourti, A.; Pauchet, Y.; Ffrench-Constant, R.; Smagghe, G. Colorado potato beetle (Coleoptera) gut transcriptome analysis: Expression of RNA interference-related genes. Insect Mol. Biol. 2013, 22, 668–684. [Google Scholar] [CrossRef]
- San Miguel, K.; Scott, J.G. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest. Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef]
- Li, F.Q.; Li, D.; Dewer, Y.; Qu, C.; Yang, Z.; Tian, J.H.; Luo, C. Discrimination of oviposition deterrent volatile β-ionone by odorant-binding proteins 1 and 4 in the whitefly Bemisia tabaci. Biomolecules 2019, 9, 563. [Google Scholar] [CrossRef] [Green Version]
- Shearer, D.A.; Boch, R. 2-heptanone in the mandibular gland secretion of the honeybee. Nature 1965, 206, 530. [Google Scholar] [CrossRef]
- Reith, J.P.; Wilson, W.T.; Levin, M.D. Repelling honey bees from insecticide treated flowers with 2-heptanone. J. Apicult. Res. 1986, 25, 78–84. [Google Scholar] [CrossRef]
- Vallet, A.; Cassier, P.; Lensky, Y. Ontogeny of the fine structure of the mandibular glands of the honeybee (Apis mellifera L.) workers and the pheromonal activity of 2-heptanone. J. Insect Physiol. 1991, 37, 789–804. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Peng, Z.; Huang, L.; Zhao, S.; Liu, M. Expression Profile and Ligand Screening of a Putative Odorant-Binding Protein, AcerOBP6, from the Asian Honeybee. Insects 2021, 12, 955. https://doi.org/10.3390/insects12110955
Zhao H, Peng Z, Huang L, Zhao S, Liu M. Expression Profile and Ligand Screening of a Putative Odorant-Binding Protein, AcerOBP6, from the Asian Honeybee. Insects. 2021; 12(11):955. https://doi.org/10.3390/insects12110955
Chicago/Turabian StyleZhao, Huiting, Zhu Peng, Li Huang, Shuguo Zhao, and Miaomiao Liu. 2021. "Expression Profile and Ligand Screening of a Putative Odorant-Binding Protein, AcerOBP6, from the Asian Honeybee" Insects 12, no. 11: 955. https://doi.org/10.3390/insects12110955
APA StyleZhao, H., Peng, Z., Huang, L., Zhao, S., & Liu, M. (2021). Expression Profile and Ligand Screening of a Putative Odorant-Binding Protein, AcerOBP6, from the Asian Honeybee. Insects, 12(11), 955. https://doi.org/10.3390/insects12110955




