Modernizing the Toolkit for Arthropod Bloodmeal Identification
Abstract
Simple Summary
Abstract
1. Introduction
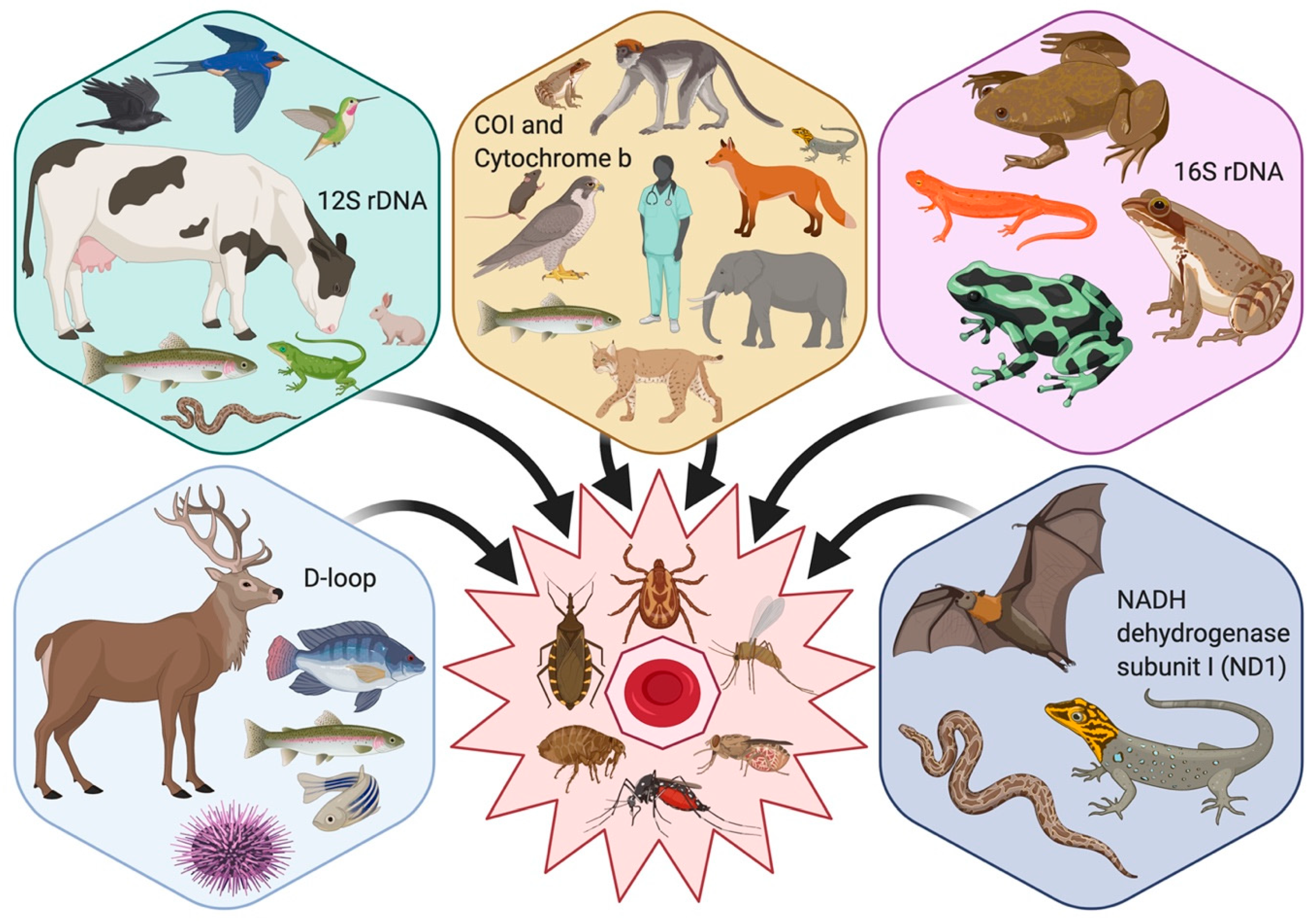
2. Diagnostic Markers: Mitochondrial DNA
2.1. Cytochrome c Oxidase Subunit I (COI)
2.2. Cytochrome b (Cyt b)
2.3. 12S and 16S Mitochondrial Ribosomal DNA (rDNA)
2.4. NADH Dehydrogenase Subunit I (ND1)
2.5. D-Loop
3. Diagnostic Markers: Nuclear Genes and Repetitive DNA Elements
3.1. Nuclear Ribosomal DNA
3.2. Prepronociceptin (PNOC)
3.3. Alu Transposable Elements
3.4. Other Nuclear Elements
4. Molecular Advances
4.1. Real-Time PCR and Quantitative PCR (qPCR)
4.2. High Resolution Melting Analysis (HRM)
4.3. Digital PCR (dPCR) and Droplet Digital PCR (ddPCR)
4.4. Next Generation Sequencing (NGS)
4.5. Microarray and Microsphere Assays
4.6. Mass Spectrometry
4.7. Stable Isotope Analysis (SIA)
5. Advances in Portability and Field-Forward Technologies
6. Xenosurveillance and Other Applications
7. Lessons Learned over the Past Decade
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Kent, R.J. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol. Ecol. Resour. 2009, 9, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Washino, R.K.; Tempelis, C.H. Mosquito host bloodmeal identification: Methodology and data analysis. Annu. Rev. Entomol. 1983, 28, 179–201. [Google Scholar] [CrossRef] [PubMed]
- Burkot, T.R.; Goodman, W.G.; DeFoliart, G.R. Identification of mosquito blood meals by enzyme-linked immunosorbent assay. Am. J. Trop. Med. Hyg. 1981, 30, 1336–1341. [Google Scholar] [CrossRef]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999, 1410, 103–123. [Google Scholar] [CrossRef]
- Natonek-Wisniewska, M.; Krzyscin, P. Evaluation of the suitability of mitochondrial DNA for species identification of microtraces and forensic traces. Acta. Biochim. Pol. 2017, 64, 705–708. [Google Scholar] [CrossRef]
- Beebe, N.W. DNA barcoding mosquitoes: Advice for potential prospectors. Parasitology 2018, 145, 622–633. [Google Scholar] [CrossRef]
- Saville, B.J.; Kohli, Y.; Anderson, J.B. mtDNA recombination in a natural population. Proc. Natl. Acad. Sci. USA 1998, 95, 1331–1335. [Google Scholar] [CrossRef]
- Hebert, P.D.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. Biol. Sci. 2003, 270 (Suppl. 1), S96–S99. [Google Scholar] [CrossRef]
- Hebert, P.D.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Paabo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef] [PubMed]
- Tobe, S.S.; Kitchener, A.C.; Linacre, A.M. Reconstructing mammalian phylogenies: A detailed comparison of the cytochrome B and cytochrome oxidase subunit I mitochondrial genes. PLoS ONE 2010, 5, e14156. [Google Scholar] [CrossRef] [PubMed]
- Humair, P.F.; Douet, V.; Moran Cadenas, F.; Schouls, L.M.; Van De Pol, I.; Gern, L. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J. Med. Entomol. 2007, 44, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Leger, E.; Liu, X.; Masseglia, S.; Noel, V.; Vourc’h, G.; Bonnet, S.; McCoy, K.D. Reliability of molecular host-identification methods for ticks: An experimental in vitro study with Ixodes ricinus. Parasit. Vectors 2015, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Collini, M.; Albonico, F.; Hauffe, H.C.; Mortarino, M. Identifying the last bloodmeal of questing sheep tick nymphs (Ixodes ricinus L.) using high resolution melting analysis. Vet. Parasitol. 2015, 210, 194–205. [Google Scholar] [CrossRef]
- Wodecka, B.; Rymaszewska, A.; Skotarczak, B. Host and pathogen DNA identification in blood meals of nymphal Ixodes ricinus ticks from forest parks and rural forests of Poland. Exp. Appl. Acarol. 2014, 62, 543–555. [Google Scholar] [CrossRef]
- Melton, T.; Holland, C. Routine forensic use of the mitochondrial 12S ribosomal RNA gene for species identification. J. Forensic. Sci. 2007, 52, 1305–1307. [Google Scholar] [CrossRef]
- Kitano, T.; Umetsu, K.; Tian, W.; Osawa, M. Two universal primer sets for species identification among vertebrates. Int. J. Legal Med. 2007, 121, 423–427. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Wang, Y.; Duan, J.; Shi, H.; Guo, J.; Wang, X.; Gao, T.; Ping, H.; Li, Z. Species identification of small fish in Xixuan Island coastal waters of Zhoushan using DNA barcoding. J. Appl. Icthyol. 2020, 36, 75–84. [Google Scholar] [CrossRef]
- Vences, M.; Thomas, M.; van der Meijden, A.; Chiari, Y.; Vieites, D.R. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Front. Zool. 2005, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.; Dietz, C.; Kiefer, A. Molecular species identification boosts bat diversity. Front. Zool. 2007, 4, 4. [Google Scholar] [CrossRef][Green Version]
- Ngamprasertwong, T.; Mackie, I.J.; Racey, P.A.; Piertney, S.B. Spatial distribution of mitochondrial and microsatellite DNA variation in Daubenton’s bat within Scotland. Mol. Ecol. 2008, 17, 3243–3258. [Google Scholar] [CrossRef] [PubMed]
- Karin, B.R.; Freitas, E.S.; Shonleben, S.; Grismer, L.L.; Bauer, A.M.; Das, I. Unrealized diversity in an urban rainforest: A new species of Lygosoma (Squamata: Scincidae) from western Sarawak, Malaysia (Borneo). Zootaxa 2018, 4370, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, I.; Hikida, T.; Hossman, M.Y.; Nishikawa, K. A new species of the genus Larutia (Squamata: Scincidae) from Gunung Penrissen, Sarawak, Borneo. Zootaxa 2019, 4661, 3–6. [Google Scholar] [CrossRef]
- Caviedes-Solis, I.W.; Nieto-Montes de Oca, A. A multilocus phylogeny of the genus Sarcohyla (Anura: Hylidae), and an investigation of species boundaries using statistical species delimitation. Mol. Phylogenet. Evol. 2018, 118, 184–193. [Google Scholar] [CrossRef]
- Rogers, S.D.; Peacock, M.M. The disappearing northern leopard frog (Lithobates pipiens): Conservation genetics and implications for remnant populations in western Nevada. Ecol. Evol. 2012, 2, 2040–2056. [Google Scholar] [CrossRef]
- Sbisa, E.; Tanzariello, F.; Reyes, A.; Pesole, G.; Saccone, C. Mammalian mitochondrial d-loop region structural analysis: Identification of new conserved sequences and their functional and evolutionary implications. Gene 1997, 205, 125–140. [Google Scholar] [CrossRef]
- Saccone, C.; Pesole, G.; Sbisa, E. The main regulatory region of mammalian mitochondrial DNA: Structure-function model and evolutionary pattern. J. Mol. Evol. 1991, 33, 83–91. [Google Scholar] [CrossRef]
- Purdue, J.R.; Smith, M.H.; Patton, J.C. Female philopatry and extreme spatial genetic heterogeneity in white-tailed deer. J. Mammal. 2000, 81, 179–185. [Google Scholar] [CrossRef]
- Hendrick, G.C.; Dolan, M.C.; McKay, T.; Sikkel, P.C. Host DNA integrity within blood meals of hematophagous larval gnathiid isopods (Crustacea, Isopoda, Gnathiidae). Parasit. Vectors 2019, 12, 316. [Google Scholar] [CrossRef] [PubMed]
- Miyake, T.; Aihara, N.; Maeda, K.; Shinzato, C.; Koyanagi, R.; Kobayashi, H.; Yamahira, K. Bloodmeal host identification with inferences to feeding habits of a fish-fed mosquito, Aedes baisasi. Sci. Rep. 2019, 9, 4002. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Knowlton, N.; Weigt, L.A. New dates and new rates for divergence across the isthmus of Panama. Proc. Royal Soc. B 1998, 265, 2257. [Google Scholar] [CrossRef]
- Doyle, J.J.; Gaut, B.S. Evolution of genes and taxa: A primer. Plant Mol. Biol. 2000, 42, 1–23. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef]
- Hajibabaei, M.; McKenna, C. DNA mini-barcodes. Methods Mol. Biol. 2012, 858, 339–353. [Google Scholar]
- Dean, M.D.; Ballard, J.W.O. Factors affecting mitochondrial DNA quality from museum preserved Drosophila simulans. Entomol. Exp. Appl. 2001, 98, 279–283. [Google Scholar] [CrossRef]
- Lee, P.L.; Prys-Jones, R.P. Extracting DNA from museum bird eggs, and whole genome amplification of archive DNA. Mol. Ecol. Resour. 2008, 8, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Tobe, S.S.; Kitchener, A.; Linacre, A. Cytochrome b or cytochrome c oxidase subunit I for mammalian species identification: An answer to the debate. Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 306–307. [Google Scholar] [CrossRef]
- Fornadel, C.M.; Norris, D.E. Increased endophily by the malaria vector Anopheles arabiensis in southern Zambia and identification of digested blood meals. Am. J. Trop. Med. Hyg. 2008, 79, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Nigro, L.; Sullivan, J.; Holsinger, K.; Martin, A.; Grapputo, A.; Franke, A.; McIntosh, C. Large differences in substitutional pattern and evolutionary rate of 12S ribosomal RNA genes. Mol. Biol. Evol. 1996, 13, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Sarri, C.; Stamatis, C.; Sarafidou, T.; Galara, I.; Godosopoulos, V.; Kolovos, M.; Liakou, C.; Tastsoglou, S.; Mamuris, Z. A new set of 16S rRNA universal primers for identification of animal species. Food Control 2014, 43, 35–41. [Google Scholar] [CrossRef]
- Xia, Y.; Gu, H.F.; Peng, R.; Chen, Q.; Zheng, Y.C.; Murphy, R.W.; Zeng, X.M. COI is better than 16S rRNA for DNA barcoding Asiatic salamanders (Amphibia: Caudata: Hynobiidae). Mol. Ecol. Resour. 2012, 12, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, D.M.; Steinman, H.A.; Witthuhn, R.C. Evaluation of the 16S and 12S rRNA genes as universal markers for the identification of commercial fish species in South Africa. Gene 2012, 491, 40–48. [Google Scholar] [CrossRef]
- Silva, P.D.; Jaramillo, C.; Bernal, X. Feeding site selection by frog-biting midges (Diptera: Corethrellidae) on anuran hosts. J. Insect Behav. 2013, 27, 302–316. [Google Scholar] [CrossRef]
- Slowik, T.J.; Lane, R.S. Feeding preferences of the immature stages of three western North American Ixodid ticks (Acari) for avian, reptilian, or rodent hosts. J. Med. Entomol. 2009, 46, 115–122. [Google Scholar] [CrossRef]
- Shi, R.; Xiong, X.; Huang, M.; Xu, W.; Li, Y.; Cao, M.; Xiong, X. High resolution melting (HRM) analysis of a 12S rRNA mini barcode as a novel approach for codfish species authentication in processed fish products. Eur. Food Res. Technol. 2020, 246, 891–899. [Google Scholar] [CrossRef]
- Britten, R.J.; Rowen, L.; Williams, J.; Cameron, R.A. Majority of divergence between closely related DNA samples is due to indels. Proc. Natl. Acad. Sci. USA 2003, 100, 4661–4665. [Google Scholar] [CrossRef] [PubMed]
- Coons, L.; Alberti, G. Acari: Ticks. In Microscopic Anatomy of Invertebrates; Chelicerata Arthropoda; Harrison, F., Foelix, R., Eds.; Wiley-Liss: New York, NY, USA, 1999; Volume 8B. [Google Scholar]
- Franta, Z.; Frantova, H.; Konvickova, J.; Horn, M.; Sojka, D.; Mares, M.; Kopacek, P. Dynamics of digestive proteolytic system during blood feeding of the hard tick Ixodes ricinus. Parasit. Vectors 2010, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Saccone, C.; De Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Cupp, E.W.; Zhang, D.; Yue, X.; Cupp, M.S.; Guyer, C.; Sprenger, T.R.; Unnasch, T.R. Identification of reptilian and amphibian blood meals from mosquitoes in an eastern equine encephalomyelitis virus focus in central Alabama. Am. J. Trop. Med. Hyg. 2004, 71, 272–276. [Google Scholar] [CrossRef]
- Toma, T.; Miyagi, I.; Tamashiro, M. Blood meal identification and feeding habits of Uranotaenia species collected in the Ryukyu archipelago. J. Am. Mosq. Control Assoc. 2014, 30, 215–218. [Google Scholar] [CrossRef]
- Nicholls, T.J.; Minczuk, M. In D-loop: 40 years of mitochondrial 7S DNA. Exp. Gerontol. 2014, 56, 175–181. [Google Scholar] [CrossRef]
- Xu, B.; Clayton, D.A. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: An implication for RNA-DNA hybrids serving as primers. EMBO J. 1996, 15, 3135–3143. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.; Raule, N.; Attardi, G. Discovery of a major D-loop replication origin reveals two modes of human mtDNA synthesis. Science 2004, 306, 2098–2101. [Google Scholar] [CrossRef]
- Kang, D.; Miyako, K.; Kai, Y.; Irie, T.; Takeshige, K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 1997, 272, 15275–15279. [Google Scholar] [CrossRef]
- Doda, J.N.; Wright, C.T.; Clayton, D.A. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl. Acad. Sci. USA 1981, 78, 6116–6120. [Google Scholar] [CrossRef]
- Hallberg, R.L. Mitochondrial DNA in Xenopus laevis oocytes. I. Displacement loop occurrence. Dev. Biol. 1974, 38, 346–355. [Google Scholar] [CrossRef]
- Annex, B.H.; Williams, R.S. Mitochondrial DNA structure and expression in specialized subtypes of mammalian striated muscle. Mol. Cell Biol. 1990, 10, 5671–5678. [Google Scholar] [CrossRef] [PubMed]
- Salim, D.; Gerton, J.L. Ribosomal DNA instability and genome adaptability. Chromosome Res. 2019, 27, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Dixon, M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991, 66, 411–453. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Powers, T.O.; Todd, T.C.; Burnell, A.M.; Murray, P.C.; Fleming, C.C.; Szalanski, A.L.; Adams, B.A.; Harris, T.S. The rDNA internal transcribed spacer region as a taxonomic marker for nematodes. J. Nematol. 1997, 29, 441–450. [Google Scholar]
- von der Schulenburg, J.H.; Hancock, J.M.; Pagnamenta, A.; Sloggett, J.J.; Majerus, M.E.; Hurst, G.D. Extreme length and length variation in the first ribosomal internal transcribed spacer of ladybird beetles (Coleoptera: Coccinellidae). Mol. Biol. Evol. 2001, 18, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Petes, T.D. Meiotic mapping of yeast ribosomal deoxyribonucleic acid on chromosome XII. J. Bacteriol. 1979, 138, 185–192. [Google Scholar] [CrossRef]
- Henderson, A.S.; Warburton, D.; Atwood, K.C. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. USA 1972, 69, 3394–3398. [Google Scholar] [CrossRef] [PubMed]
- Reeves, L.E.; Holderman, C.J.; Blosser, E.M.; Gillett-Kaufman, J.L.; Kawahara, A.Y.; Kaufman, P.E.; Burkett-Cadena, N.D. Identification of Uranotaenia sapphirina as a specialist of annelids broadens known mosquito host use patterns. Commun. Biol. 2018, 1, 92. [Google Scholar] [CrossRef] [PubMed]
- Mollereau, C.; Simons, M.J.; Soularue, P.; Liners, F.; Vassart, G.; Meunier, J.C.; Parmentier, M. Structure, tissue distribution, and chromosomal localization of the prepronociceptin gene. Proc. Natl. Acad. Sci. USA 1996, 93, 8666–8670. [Google Scholar] [CrossRef] [PubMed]
- Slama, D.; Haouas, N.; Mezhoud, H.; Babba, H.; Chaker, E. Blood meal analysis of Culicoides (Diptera: Ceratopogonidae) in central Tunisia. PLoS ONE 2015, 10, e0120528. [Google Scholar] [CrossRef] [PubMed]
- Chargui, N.; Slama, D.; Haouas, N.; Rmadi, L.; Babba, H. Transmission cycle analysis in a Leishmania infantum focus: Infection rates and blood meal origins in sand flies (Diptera: Psychodidae). J. Vector Ecol. 2018, 43, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Hadj-Henni, L.; De Meulemeester, T.; Depaquit, J.; Noel, P.; Germain, A.; Helder, R.; Augot, D. Comparison of vertebrate cytochrome b and prepronociceptin for blood meal analyses in Culicoides. Front. Vet. Sci. 2015, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef]
- Siripattanapipong, S.; Leelayoova, S.; Ninsaeng, U.; Mungthin, M. Detection of DNA of Leishmania siamensis in Sergentomyia (Neophlebotomus) iyengari (Diptera: Psychodidae) and molecular identification of blood meals of sand flies in an affected area, Southern Thailand. J. Med. Entomol. 2018, 55, 1277–1283. [Google Scholar] [CrossRef]
- Mejia, A.; Matamoros, G.; Fontecha, G.; Sosa-Ochoa, W. Bionomic aspects of Lutzomyia evansi and Lutzomyia longipalpis, proven vectors of Leishmania infantum in an endemic area of non-ulcerative cutaneous leishmaniasis in Honduras. Parasit. Vectors 2018, 11, 15. [Google Scholar] [CrossRef]
- Haouas, N.; Pesson, B.; Boudabous, R.; Dedet, J.P.; Babba, H.; Ravel, C. Development of a molecular tool for the identification of Leishmania reservoir hosts by blood meal analysis in the insect vectors. Am. J. Trop. Med. Hyg. 2007, 77, 1054–1059. [Google Scholar] [CrossRef]
- Breviario, D. Is there any alternative to canonical DNA barcoding of multicellular eukaryotic species? A case for the tubulin gene family. Int. J. Mol. Sci. 2017, 18, 827. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Fazekas, A.J.; Steeves, R.A.; Janovec, J. Testing candidate plant barcode regions in the Myristicaceae. Mol. Ecol. Resour. 2008, 8, 480–490. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Groenewald, J.Z.; de Jesus Yanez-Morales, M.; Crous, P.W. DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Persoonia 2012, 29, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Paracchini, V.; Petrillo, M.; Lievens, A.; Puertas Gallardo, A.; Martinsohn, J.T.; Hofherr, J.; Maquet, A.; Silva, A.P.B.; Kagkli, D.M.; Querci, M.; et al. Novel nuclear barcode regions for the identification of flatfish species. Food Control 2017, 79, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Paetkau, D.; Strobeck, C. Microsatellite analysis of genetic variation in black bear populations. Mol. Ecol. 1994, 3, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Keven, J.B.; Walker, E.D.; Venta, P.J. A microsatellite multiplex assay for profiling pig DNA in mosquito bloodmeals. J. Med. Entomol. 2019, 56, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Molaei, G.; Huang, S.; Andreadis, T.G. Vector-host interactions of Culex pipiens complex in northeastern and southwestern USA. J. Am. Mosq. Control Assoc. 2012, 28, 127–136. [Google Scholar] [CrossRef]
- Levin, I.I.; Parker, P.G. Infection with Haemoproteus iwa affects vector movement in a hippoboscid fly-frigatebird system. Mol. Ecol. 2014, 23, 947–953. [Google Scholar] [CrossRef]
- Martínez de la Puente, J.; Méndez, M.; Ruiz, S.; Godoy, J.A.; Soriguer, R.C.; Figuerola, J. Individual identification of endangered species using mosquito blood meals: A proof-of-concept study in Iberian lynx. Parasitol. Res. 2015, 114, 1607–1610. [Google Scholar] [CrossRef]
- Alcaide, M.; Rico, C.; Ruiz, S.; Soriguer, R.; Munoz, J.; Figuerola, J. Disentangling vector-borne transmission networks: A universal DNA barcoding method to identify vertebrate hosts from arthropod bloodmeals. PLoS ONE 2009, 4, e7092. [Google Scholar] [CrossRef]
- Nagy, Z.T.; Sonet, G.; Glaw, F.; Vences, M. First large-scale DNA barcoding assessment of reptiles in the biodiversity hotspot of Madagascar, based on newly designed COI primers. PLoS ONE 2012, 7, e34506. [Google Scholar] [CrossRef]
- Reeves, L.E.; Gillett-Kaufman, J.L.; Kawahara, A.Y.; Kaufman, P.E. Barcoding blood meals: New vertebrate-specific primer sets for assigning taxonomic identities to host DNA from mosquito blood meals. PLoS Negl. Trop. Dis. 2018, 12, e0006767. [Google Scholar] [CrossRef]
- Abbasi, I.; Cunio, R.; Warburg, A. Identification of blood meals imbibed by phlebotomine sand flies using cytochrome b PCR and reverse line blotting. Vector Borne Zoonotic Dis. 2009, 9, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Berdjane-Brouk, Z.; Kone, A.K.; Djimde, A.A.; Charrel, R.N.; Ravel, C.; Delaunay, P.; del Giudice, P.; Diarra, A.Z.; Doumbo, S.; Goita, S.; et al. First detection of Leishmania major DNA in Sergentomyia (Spelaeomyia) darlingi from cutaneous leishmaniasis foci in Mali. PLoS ONE 2012, 7, e28266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sales, K.G.; Costa, P.L.; de Morais, R.C.; Otranto, D.; Brandao-Filho, S.P.; Cavalcanti Mde, P.; Dantas-Torres, F. Identification of phlebotomine sand fly blood meals by real-time PCR. Parasit. Vectors 2015, 8, 230. [Google Scholar] [CrossRef]
- Garros, C.; Gardes, L.; Allene, X.; Rakotoarivony, I.; Viennet, E.; Rossi, S.; Balenghien, T. Adaptation of a species-specific multiplex PCR assay for the identification of blood meal source in Culicoides (Ceratopogonidae: Diptera): Applications on Palaearctic biting midge species, vectors of Orbiviruses. Infect. Genet. Evol. 2011, 11, 1103–1110. [Google Scholar] [CrossRef]
- Valinsky, L.; Ettinger, G.; Bar-Gal, G.K.; Orshan, L. Molecular identification of bloodmeals from sand flies and mosquitoes collected in Israel. J. Med. Entomol. 2014, 51, 678–685. [Google Scholar] [CrossRef]
- Sawabe, K.; Isawa, H.; Hoshino, K.; Sasaki, T.; Roychoudhury, S.; Higa, Y.; Kasai, S.; Tsuda, Y.; Nishiumi, I.; Hisai, N.; et al. Host-feeding habits of Culex pipiens and Aedes albopictus (Diptera: Culicidae) collected at the urban and suburban residential areas of Japan. J. Med. Entomol. 2010, 47, 442–450. [Google Scholar] [CrossRef]
- Curler, G.R.; Moulton, J.K.; Madriz, R.I. Redescription of Aposycorax chilensis (Tonnoir) (Diptera, Psychodidae, Sycoracinae) with the first identification of a blood meal host for the species. Zootaxa 2015, 4048, 114–126. [Google Scholar] [CrossRef]
- Omondi, D.; Masiga, D.K.; Ajamma, Y.U.; Fielding, B.C.; Njoroge, L.; Villinger, J. Unraveling host-vector-arbovirus interactions by two-gene high resolution melting mosquito bloodmeal analysis in a Kenyan wildlife-livestock interface. PLoS ONE 2015, 10, e0134375. [Google Scholar] [CrossRef]
- Anaguano, D.F.; Ponce, P.; Baldeon, M.E.; Santander, S.; Cevallos, V. Blood-meal identification in phlebotomine sand flies (Diptera: Psychodidae) from Valle Hermoso, a high prevalence zone for cutaneous leishmaniasis in Ecuador. Acta Trop. 2015, 152, 116–120. [Google Scholar] [CrossRef]
- Jaouadi, K.; Bettaieb, J.; Bennour, A.; Salem, S.; Ghawar, W.; Rjeibi, M.R.; Khabouchi, N.; Gonzalez, J.P.; Diouani, M.F.; Ben Salah, A. Blood meal analysis of phlebotomine sandflies (Diptera: Psychodidae: Phlebotominae) for Leishmania spp. identification and vertebrate blood origin, central Tunisia, 2015-2016. Am. J. Trop. Med. Hyg. 2018, 98, 146–149. [Google Scholar] [CrossRef]
- Baum, M.; de Castro, E.A.; Pinto, M.C.; Goulart, T.M.; Baura, W.; Klisiowicz Ddo, R.; Vieira da Costa-Ribeiro, M.C. Molecular detection of the blood meal source of sand flies (Diptera: Psychodidae) in a transmission area of American cutaneous leishmaniasis, Parana State, Brazil. Acta Trop. 2015, 143, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Eizirik, E.; Johnson, W.E.; Zhang, Y.P.; Ryder, O.A.; O’Brien, S.J. Molecular phylogenetics and the origins of placental mammals. Nature 2001, 409, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shi, G.; Cheng, P.; Liu, L.; Gong, M. Host preferences and feeding patterns of Anopheles sinensis Wiedemann in three sites of Shandong province, China. J. Vector Borne Dis. 2017, 54, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Mucci, L.F.; Junior, R.P.; de Paula, M.B.; Scandar, S.A.; Pacchioni, M.L.; Fernandes, A.; Consales, C.A. Feeding habits of mosquitoes (Diptera: Culicidae) in an area of sylvatic transmission of yellow fever in the state of Sao Paulo, Brazil. J. Venom Anim. Toxins Incl. Trop. Dis. 2015, 21, 6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tanure, A.; Peixoto, J.C.; Afonso, M.M.; Duarte, R.; Pinheiro Ada, C.; Coelho, S.V.; Barata, R.A. Identification of sandflies (Diptera: Psychodidae: Phlebotominae) blood meals in an endemic leishmaniasis area in Brazil. Rev. Inst. Med. Trop. Sao Paulo 2015, 57, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Maleki-Ravasan, N.; Oshaghi, M.; Javadian, E.; Rassi, Y.; Sadraei, J.; Mohtarami, F. Blood meal identification in field-captured sand flies: Comparison of PCR-RFLP and ELISA assays. Iran J. Arthropod Borne Dis. 2009, 3, 8–18. [Google Scholar] [PubMed]
- Fonteles, R.S.; Pereira Filho, A.A.; Moraes, J.L.P.; Pereira, S.R.F.; Rodrigues, B.L.; Rebelo, J.M.M. Detection of Leishmania DNA and blood meal identification in sand flies (Diptera: Psychodidae) from Lencois Maranhenses National Park region, Brazil. J. Med. Entomol. 2018, 55, 445–451. [Google Scholar] [CrossRef]
- Gonzalez, E.; Gallego, M.; Molina, R.; Abras, A.; Alcover, M.M.; Ballart, C.; Fernandez, A.; Jimenez, M. Identification of blood meals in field captured sand flies by a PCR-RFLP approach based on cytochrome b gene. Acta Trop. 2015, 152, 96–102. [Google Scholar] [CrossRef]
- Das, S.; Henning, T.C.; Simubali, L.; Hamapumbu, H.; Nzira, L.; Mamini, E.; Makuwaza, A.; Muleba, M.; Norris, D.E.; Stevenson, J.C.; et al. Underestimation of foraging behaviour by standard field methods in malaria vector mosquitoes in southern Africa. Malar. J. 2015, 14, 12. [Google Scholar] [CrossRef][Green Version]
- Soares, V.Y.; Silva, J.C.; Silva, K.R.; Pires e Cruz Mdo, S.; Santos, M.P.; Ribolla, P.E.; Alonso, D.P.; Coelho, L.F.; Costa, D.L.; Costa, C.H. Identification of blood meal sources of Lutzomyia longipalpis using polymerase chain reaction-restriction fragment length polymorphism analysis of the cytochrome B gene. Mem. Inst. Oswaldo Cruz 2014, 109, 379–383. [Google Scholar] [CrossRef][Green Version]
- Wodecka, B.; Skotarczak, B. Identification of host blood-meal sources and Borrelia in field-collected Ixodes ricinus ticks in north-western Poland. Ann. Agric. Environ. Med. 2016, 23, 59–63. [Google Scholar] [CrossRef]
- Azizi, K.; Askari, M.B.; Kalantari, M.; Moemenbellah-Fard, M.D. Molecular detection of Leishmania parasites and host blood meal identification in wild sand flies from a new endemic rural region, south of Iran. Pathog. Glob. Health 2016, 110, 303–309. [Google Scholar] [CrossRef]
- Honig, V.; Carolan, H.E.; Vavruskova, Z.; Massire, C.; Mosel, M.R.; Crowder, C.D.; Rounds, M.A.; Ecker, D.J.; Ruzek, D.; Grubhoffer, L.; et al. Broad-range survey of vector-borne pathogens and tick host identification of Ixodes ricinus from Southern Czech Republic. FEMS Microbiol. Ecol. 2017, 93, fix129. [Google Scholar] [CrossRef] [PubMed]
- Samuel, P.P.; Thenmozhi, V.; Nagaraj, J.; Kumar, T.D.; Tyagi, B.K. Dengue vectors prevalence and the related risk factors involved in the transmission of dengue in Thiruvananthapuram district, Kerala, South India. J. Vector Borne Dis. 2014, 51, 313–319. [Google Scholar] [PubMed]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Lanciotti, R.S.; Kerst, A.J.; Nasci, R.S.; Godsey, M.S.; Mitchell, C.J.; Savage, H.M.; Komar, N.; Panella, N.A.; Allen, B.C.; Volpe, K.E.; et al. Rapid detection of west nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J. Clin. Microbiol. 2000, 38, 4066–4071. [Google Scholar] [CrossRef] [PubMed]
- Mukabana, W.R.; Takken, W.; Knols, B.G. Analysis of arthropod bloodmeals using molecular genetic markers. Trends Parasitol. 2002, 18, 505–509. [Google Scholar] [CrossRef]
- van den Hurk, A.F.; Smith, I.L.; Smith, G.A. Development and evaluation of real-time polymerase chain reaction assays to identify mosquito (Diptera: Culicidae) bloodmeals originating from native Australian mammals. J. Med. Entomol. 2007, 44, 85–92. [Google Scholar] [CrossRef]
- Wittwer, C.T.; Reed, G.H.; Gundry, C.N.; Vandersteen, J.G.; Pryor, R.J. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003, 49, 853–860. [Google Scholar] [CrossRef]
- Vossen, R.H.; Aten, E.; Roos, A.; den Dunnen, J.T. High-resolution melting analysis (HRMA): More than just sequence variant screening. Hum. Mutat. 2009, 30, 860–866. [Google Scholar] [CrossRef]
- Wittwer, C.T. High-resolution DNA melting analysis: Advancements and limitations. Hum. Mutat. 2009, 30, 857–859. [Google Scholar] [CrossRef] [PubMed]
- Ouso, D.O.; Otiende, M.Y.; Jeneby, M.M.; Oundo, J.W.; Bargul, J.L.; Miller, S.E.; Wambua, L.; Villinger, J. Three-gene PCR and high-resolution melting analysis for differentiating vertebrate species mitochondrial DNA for biodiversity research and complementing forensic surveillance. Sci. Rep. 2020, 10, 4741. [Google Scholar] [CrossRef] [PubMed]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef]
- Saunderson, E.A.; Baker, A.M.; Williams, M.; Curtius, K.; Jones, J.L.; Graham, T.A.; Ficz, G. A novel use of random priming-based single-strand library preparation for whole genome sequencing of formalin-fixed paraffin-embedded tissue samples. NAR Genom. Bioinform. 2020, 2, lqz017. [Google Scholar] [CrossRef]
- Logue, K.; Keven, J.B.; Cannon, M.V.; Reimer, L.; Siba, P.; Walker, E.D.; Zimmerman, P.A.; Serre, D. Unbiased Characterization of Anopheles Mosquito Blood Meals by Targeted High-Throughput Sequencing. PLoS Negl. Trop. Dis. 2016, 10, e0004512. [Google Scholar] [CrossRef]
- Francoso, E.; Zuntini, A.R.; Ricardo, P.C.; Silva, J.P.N.; Brito, R.; Oldroyd, B.P.; Arias, M.C. Conserved numts mask a highly divergent mitochondrial-COI gene in a species complex of Australian stingless bees Tetragonula (Hymenoptera: Apidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2019, 30, 806–817. [Google Scholar] [CrossRef]
- Barzon, L.; Lavezzo, E.; Militello, V.; Toppo, S.; Palu, G. Applications of next-generation sequencing technologies to diagnostic virology. Int. J. Mol. Sci. 2011, 12, 7861–7884. [Google Scholar] [CrossRef]
- Thiemann, T.C.; Brault, A.C.; Ernest, H.B.; Reisen, W.K. Development of a high-throughput microsphere-based molecular assay to identify 15 common bloodmeal hosts of Culex mosquitoes. Mol. Ecol. Resour. 2012, 12, 238–246. [Google Scholar] [CrossRef]
- Thiemann, T.C.; Wheeler, S.S.; Barker, C.M.; Reisen, W.K. Mosquito host selection varies seasonally with host availability and mosquito density. PLoS Negl. Trop. Dis. 2011, 5, e1452. [Google Scholar] [CrossRef]
- Thiemann, T.C.; Reisen, W.K. Evaluating sampling method bias in Culex tarsalis and Culex quinquefasciatus (Diptera: Culicidae) bloodmeal identification studies. J. Med. Entomol. 2012, 49, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Petz, L.N.; Melanson, V.R.; McMenamy, S.S.; Turell, M.J.; Long, L.S.; Pisarcik, S.E.; Kengluecha, A.; Jaichapor, B.; O’Guinn, M.L.; et al. Evaluation of a field-portable DNA microarray platform and nucleic acid amplification strategies for the detection of arboviruses, arthropods, and bloodmeals. Am. J. Trop. Med. Hyg. 2013, 88, 245–253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dunbar, S.A. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta. 2006, 363, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Önder, Ö.; Shao, W.; Lam, H.; Brisson, D. Tracking the sources of blood meals of parasitic arthropods using shotgun proteomics and unidentified tandem mass spectral libraries. Nat. Protoc. 2014, 9, 842–850. [Google Scholar] [CrossRef]
- Dasari, S.; Chambers, M.C.; Martinez, M.A.; Carpenter, K.L.; Ham, A.J.; Vega-Montoto, L.J.; Tabb, D.L. Pepitome: Evaluating improved spectral library search for identification complementarity and quality assessment. J. Proteome Res. 2012, 11, 1686–1695. [Google Scholar] [CrossRef]
- Lam, H.; Aebersold, R. Building and searching tandem mass (MS/MS) spectral libraries for peptide identification in proteomics. Methods 2011, 54, 424–431. [Google Scholar] [CrossRef]
- Lam, H.; Deutsch, E.W.; Eddes, J.S.; Eng, J.K.; Stein, S.E.; Aebersold, R. Building consensus spectral libraries for peptide identification in proteomics. Nat. Methods 2008, 5, 873–875. [Google Scholar] [CrossRef]
- Craig, R.; Cortens, J.C.; Fenyo, D.; Beavis, R.C. Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 2006, 5, 1843–1849. [Google Scholar] [CrossRef]
- Frewen, B.E.; Merrihew, G.E.; Wu, C.C.; Noble, W.S.; MacCoss, M.J. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 2006, 78, 5678–5684. [Google Scholar] [CrossRef]
- Keller, J.I.; Ballif, B.A.; St Clair, R.M.; Vincent, J.J.; Monroy, M.C.; Stevens, L. Chagas disease vector blood meal sources identified by protein mass spectrometry. PLoS ONE 2017, 12, e0189647. [Google Scholar] [CrossRef]
- Niare, S.; Tandina, F.; Davoust, B.; Doumbo, O.; Raoult, D.; Parola, P.; Almeras, L. Accurate identification of Anopheles gambiae Giles trophic preferences by MALDI-TOF MS. Infect. Genet. Evol. 2018, 63, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Tandina, F.; Laroche, M.; Davoust, B.; Ogobara, K.D.; Parola, P. Blood meal identification in the cryptic species Anopheles gambiae and Anopheles coluzzii using MALDI-TOF MS. Parasite 2018, 25, 40. [Google Scholar] [CrossRef] [PubMed]
- Niare, S.; Berenger, J.M.; Dieme, C.; Doumbo, O.; Raoult, D.; Parola, P.; Almeras, L. Identification of blood meal sources in the main African malaria mosquito vector by MALDI-TOF MS. Malar. J. 2016, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Rasgon, J.L. Stable isotope analysis can potentially identify completely-digested bloodmeals in mosquitoes. PLoS ONE 2008, 3, e2198. [Google Scholar] [CrossRef]
- LoGiudice, K.; Kurchena, K.; Christopher, K.; Scott, N. Exploration of stable isotope analysis for tick host identification. Ticks Tick Borne Dis. 2018, 9, 151–154. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Caraguel, C.G.; Stryhn, H.; Gagne, N.; Dohoo, I.R.; Hammell, K.L. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: Analytical and epidemiologic approaches. J. Vet. Diagn. Invest. 2011, 23, 2–15. [Google Scholar] [CrossRef]
- Keven, J.B.; Artzberger, G.; Gillies, M.L.; Mbewe, R.B.; Walker, E.D. Probe-based multiplex qPCR identifies blood-meal hosts in Anopheles mosquitoes from Papua New Guinea. Parasit. Vectors 2020, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Sales, K.; Miranda, D.E.O.; Paiva, M.H.S.; Figueredo, L.A.; Otranto, D.; Dantas-Torres, F. Fast multiplex real-time PCR assay for simultaneous detection of dog and human blood and Leishmania parasites in sand flies. Parasit. Vectors 2020, 13, 131. [Google Scholar] [CrossRef]
- Woods, M.E.; Montenieri, J.A.; Eisen, R.J.; Zeidner, N.S.; Borchert, J.N.; Laudisoit, A.; Babi, N.; Atiku, L.A.; Enscore, R.E.; Gage, K.L. Identification of flea blood meals using multiplexed real-time polymerase chain reaction targeting mitochondrial gene fragments. Am. J. Trop. Med. Hyg. 2009, 80, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Van Der Saag, M.; Gu, X.; Ward, M.P.; Kirkland, P.D. Development and evaluation of real-time PCR assays for bloodmeal identification in Culicoides midges. Med. Vet. Entomol. 2016, 30, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Gunathilaka, N.; Denipitiya, T.; Hapugoda, M.; Abeyewickreme, W.; Wickremasinghe, R. Determination of the foraging behaviour and blood meal source of malaria vector mosquitoes in Trincomalee District of Sri Lanka using a multiplex real time polymerase chain reaction assay. Malar. J. 2016, 15, 242. [Google Scholar] [CrossRef] [PubMed]
- Orsborne, J.; Furuya-Kanamori, L.; Jeffries, C.L.; Kristan, M.; Mohammed, A.R.; Afrane, Y.A.; O’Reilly, K.; Massad, E.; Drakeley, C.; Walker, T.; et al. Investigating the blood-host plasticity and dispersal of Anopheles coluzzii using a novel field-based methodology. Parasit. Vectors 2019, 12, 143. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T.; Herrmann, M.G.; Moss, A.A.; Rasmussen, R.P. Continuous fluorescence monitoring of rapid cycle DNA amplification. 1997. Biotechniques 2013, 54, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Peña, V.H.; Fernandez, G.J.; Gomez-Palacio, A.M.; Mejia-Jaramillo, A.M.; Cantillo, O.; Triana-Chavez, O. High-resolution melting (HRM) of the cytochrome B gene: A powerful approach to identify blood-meal sources in Chagas disease Vectors. PLoS Negl. Trop. Dis. 2012, 6, e1530. [Google Scholar] [CrossRef]
- Graham, C.B.; Borchert, J.N.; Black, W.C.t.; Atiku, L.A.; Mpanga, J.T.; Boegler, K.A.; Moore, S.M.; Gage, K.L.; Eisen, R.J. Blood meal identification in off-host cat fleas (Ctenocephalides felis) from a plague-endemic region of Uganda. Am. J. Trop. Med. Hyg. 2013, 88, 381–389. [Google Scholar] [CrossRef][Green Version]
- Graham, C.B.; Black, W.C.; Boegler, K.A.; Montenieri, J.A.; Holmes, J.L.; Gage, K.L.; Eisen, R.J. Combining real-time polymerase chain reaction using SYBR Green I detection and sequencing to identify vertebrate bloodmeals in fleas. J. Med. Entomol. 2012, 49, 1442–1452. [Google Scholar] [CrossRef]
- Ibanez-Cervantes, G.; Martinez-Ibarra, A.; Nogueda-Torres, B.; Lopez-Orduna, E.; Alonso, A.L.; Perea, C.; Maldonado, T.; Hernandez, J.M.; Leon-Avila, G. Identification by Q-PCR of Trypanosoma cruzi lineage and determination of blood meal sources in triatomine gut samples in Mexico. Parasitol. Int. 2013, 62, 36–43. [Google Scholar] [CrossRef]
- Peña-Garcia, V.H.; Gomez-Palacio, A.M.; Triana-Chavez, O.; Mejia-Jaramillo, A.M. Eco-epidemiology of Chagas disease in an endemic area of Colombia: Risk factor estimation, Trypanosoma cruzi characterization and identification of blood-meal sources in bugs. Am. J. Trop. Med. Hyg. 2014, 91, 1116–1124. [Google Scholar] [CrossRef]
- Collini, M.; Albonico, F.; Rosa, R.; Tagliapietra, V.; Arnoldi, D.; Conterno, L.; Rossi, C.; Mortarino, M.; Rizzoli, A.; Hauffe, H.C. Identification of Ixodes ricinus blood meals using an automated protocol with high resolution melting analysis (HRMA) reveals the importance of domestic dogs as larval tick hosts in Italian alpine forests. Parasit. Vectors 2016, 9, 638. [Google Scholar] [CrossRef]
- Dube, S.; Qin, J.; Ramakrishnan, R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE 2008, 3, e2876. [Google Scholar] [CrossRef] [PubMed]
- Whale, A.S.; Cowen, S.; Foy, C.A.; Huggett, J.F. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS ONE 2013, 8, e58177. [Google Scholar] [CrossRef] [PubMed]
- Sykes, P.J.; Neoh, S.H.; Brisco, M.J.; Hughes, E.; Condon, J.; Morley, A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992, 13, 444–449. [Google Scholar] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Duffy, D.C.; McDonald, J.C.; Schueller, O.J.; Whitesides, G.M. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 1998, 70, 4974–4984. [Google Scholar] [CrossRef]
- Du, W.; Li, L.; Nichols, K.P.; Ismagilov, R.F. SlipChip. Lab Chip 2009, 9, 2286–2292. [Google Scholar] [CrossRef]
- Holtze, C.; Rowat, A.C.; Agresti, J.J.; Hutchison, J.B.; Angile, F.E.; Schmitz, C.H.; Koster, S.; Duan, H.; Humphry, K.J.; Scanga, R.A.; et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip 2008, 8, 1632–1639. [Google Scholar] [CrossRef]
- Thorsen, T.; Roberts, R.W.; Arnold, F.H.; Quake, S.R. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 2001, 86, 4163–4166. [Google Scholar] [CrossRef]
- Anna, S.L.; Bontoux, N.; Stone, H.A. Formation of dispersions using “flow focusing” in microchannels. App. Phys. Lett. 2003, 82, 364–366. [Google Scholar] [CrossRef]
- Sugiura, S.; Nakajima, M.; Iwamoto, S.; Seki, M. Interfacial tension driven monodispersed droplet formation from microfabricated channel array. Langmuir 2001, 17, 5562–5566. [Google Scholar] [CrossRef]
- Batovska, J.; Mee, P.T.; Lynch, S.E.; Sawbridge, T.I.; Rodoni, B.C. Sensitivity and specificity of metatranscriptomics as an arbovirus surveillance tool. Sci. Rep. 2019, 9, 19398. [Google Scholar] [CrossRef] [PubMed]
- Almeida, P.R.; Eisen, A.K.A.; Demoliner, M.; Spilki, F.R. RT-dPCR in mosquito samples for ZIKV detection: Effects of RNA extraction and reverse transcription in target concentration. Viruses 2020, 12, 827. [Google Scholar] [CrossRef] [PubMed]
- Koepfli, C.; Nguitragool, W.; Hofmann, N.E.; Robinson, L.J.; Ome-Kaius, M.; Sattabongkot, J.; Felger, I.; Mueller, I. Sensitive and accurate quantification of human malaria parasites using droplet digital PCR (ddPCR). Sci. Rep. 2016, 6, 39183. [Google Scholar] [CrossRef] [PubMed]
- Srisutham, S.; Saralamba, N.; Malleret, B.; Renia, L.; Dondorp, A.M.; Imwong, M. Four human Plasmodium species quantification using droplet digital PCR. PLoS ONE 2017, 12, e0175771. [Google Scholar] [CrossRef] [PubMed]
- Pomari, E.; Piubelli, C.; Perandin, F.; Bisoffi, Z. Digital PCR: A new technology for diagnosis of parasitic infections. Clin. Microbiol. Infect. 2019, 25, 1510–1516. [Google Scholar] [CrossRef]
- King, J.L.; Smith, A.D.; Mitchell, E.A.; Allen, M.S. Validation of droplet digital PCR for the detection and absolute quantification of Borrelia DNA in Ixodes scapularis ticks. Parasitology 2017, 144, 359–367. [Google Scholar] [CrossRef]
- Kumar, K.; Arshad, S.S.; Toung, O.P.; Abba, Y.; Selvarajah, G.T.; Abu, J.; Yasmin, A.R.; Ong, B.L.; Bande, F. The distribution of important sero-complexes of flaviviruses in Malaysia. Trop. Anim. Health Prod. 2019, 51, 495–506. [Google Scholar] [CrossRef]
- Rice, L.M.; Robb, L.L.; Hartman, D.A.; Anderson, J.R.; Kading, R.C. Application of the droplet digital polymerase chain reaction (ddPCR) platform for detection and quantification of vertebrate host DNA in engorged mosquitoes. J. Med. Entomol. 2019, 56, 1150–1153. [Google Scholar] [CrossRef]
- Brinkmann, A.; Nitsche, A.; Kohl, C. Viral metagenomics on blood-feeding arthropods as a tool for human disease surveillance. Int. J. Mol. Sci. 2016, 17, 1743. [Google Scholar] [CrossRef]
- Levy, S.E.; Myers, R.M. Advancements in next-generation sequencing. Annu. Rev. Genomics Hum. Genet. 2016, 17, 95–115. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of next-generation sequencing technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef] [PubMed]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A next generation sequencing platform for genomic analysis. Methods Mol. Biol. 2018, 1706, 223–232. [Google Scholar] [PubMed]
- Van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Richards, S. Arthropod genome sequencing and assembly strategies. Methods Mol. Biol. 2019, 1858, 1–14. [Google Scholar] [PubMed]
- Batovska, J.; Lynch, S.E.; Cogan, N.O.I.; Brown, K.; Darbro, J.M.; Kho, E.A.; Blacket, M.J. Effective mosquito and arbovirus surveillance using metabarcoding. Mol. Ecol. Resour. 2018, 18, 32–40. [Google Scholar] [CrossRef]
- Kieran, T.J.; Gottdenker, N.L.; Varian, C.P.; Saldana, A.; Means, N.; Owens, D.; Calzada, J.E.; Glenn, T.C. Blood meal source characterization using Illumina sequencing in the Chagas disease vector Rhodnius pallescens (Hemiptera: Reduviidae) in Panama. J. Med. Entomol. 2017, 54, 1786–1789. [Google Scholar] [CrossRef]
- Campana, M.G.; Hawkins, M.T.; Henson, L.H.; Stewardson, K.; Young, H.S.; Card, L.R.; Lock, J.; Agwanda, B.; Brinkerhoff, J.; Gaff, H.D.; et al. Simultaneous identification of host, ectoparasite and pathogen DNA via in-solution capture. Mol. Ecol. Resour. 2016, 16, 1224–1239. [Google Scholar] [CrossRef]
- Page, G.P.; Zakharkin, S.O.; Kim, K.; Mehta, T.; Chen, L.; Zhang, K. Microarray Analysis; Humana Press: Totowa, NJ, USA, 2007; pp. 409–430. [Google Scholar]
- Wickramasekara, S.; Bunikis, J.; Wysocki, V.; Barbour, A.G. Identification of residual blood proteins in ticks by mass spectrometry proteomics. Emerg. Infect. Dis. 2008, 14, 1273–1275. [Google Scholar] [CrossRef]
- Song, Y.; Laskay, U.A.; Vilcins, I.M.; Barbour, A.G.; Wysocki, V.H. Top-down-assisted bottom-up method for homologous protein sequencing: Hemoglobin from 33 bird species. J. Am. Soc. Mass Spectrom. 2015, 26, 1875–1884. [Google Scholar] [CrossRef][Green Version]
- Steen, H.; Mann, M. The ABC’s (and XYZ’s) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 2004, 5, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Greenwalt, D.E.; Goreva, Y.S.; Siljestrom, S.M.; Rose, T.; Harbach, R.E. Hemoglobin-derived porphyrins preserved in a Middle Eocene blood-engorged mosquito. Proc. Natl. Acad. Sci. USA 2013, 110, 18496–18500. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.A.; Rush, J.; Stemman, O.; Kirschner, M.W.; Gygi, S.P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA 2003, 100, 6940–6945. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, C.; Hoffmann, F.G.; Lanier, H.C.; Wolf, C.J.; Cheviron, Z.A.; Spangler, M.L.; Weber, R.E.; Fago, A.; Storz, J.F. Intraspecific polymorphism, interspecific divergence, and the origins of function-altering mutations in deer mouse hemoglobin. Mol. Biol. Evol. 2015, 32, 978–997. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, C.; Schaffner, F.; Ziegler, D.; Pfluger, V.; Mathis, A. Identification of field-caught Culicoides biting midges using matrix-assisted laser desorption/ionization time of flight mass spectrometry. Parasitology 2012, 139, 248–258. [Google Scholar] [CrossRef]
- Yssouf, A.; Flaudrops, C.; Drali, R.; Kernif, T.; Socolovschi, C.; Berenger, J.M.; Raoult, D.; Parola, P. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid identification of tick vectors. J. Clin. Microbiol. 2013, 51, 522–528. [Google Scholar] [CrossRef]
- Sougoufara, S.; Sokhna, C.; Diagne, N.; Doucoure, S.; Sembene, P.M.; Harry, M. The implementation of long-lasting insecticidal bed nets has differential effects on the genetic structure of the African malaria vectors in the Anopheles gambiae complex in Dielmo, Senegal. Malar. J. 2017, 16, 337. [Google Scholar] [CrossRef]
- Tandina, F.; Niare, S.; Laroche, M.; Kone, A.K.; Diarra, A.Z.; Ongoiba, A.; Berenger, J.M.; Doumbo, O.K.; Raoult, D.; Parola, P. Using MALDI-TOF MS to identify mosquitoes collected in Mali and their blood meals. Parasitology 2018, 145, 1170–1182. [Google Scholar] [CrossRef]
- Diarra, A.Z.; Laroche, M.; Berger, F.; Parola, P. Use of MALDI-TOF MS for the identification of Chad mosquitoes and the origin of their blood meal. Am. J. Trop. Med. Hyg. 2019, 100, 47–53. [Google Scholar] [CrossRef]
- Laskay, U.A.; Burg, J.; Kaleta, E.J.; Vilcins, I.M.; Telford Iii, S.R.; Barbour, A.G.; Wysocki, V.H. Development of a host blood meal database: De novo sequencing of hemoglobin from nine small mammals using mass spectrometry. Biol. Chem. 2012, 393, 195–201. [Google Scholar] [CrossRef]
- Crawford, K.; McDonald, R.A.; Bearhop, S. Applications of stable isotope techniques to the ecology of mammals. Mamm. Rev. 2008, 38, 87–107. [Google Scholar] [CrossRef]
- Martínez del Rio, C.; Carleton, S.A. How fast and how faithful: The dynamics of isotopic incorporation into animal tissues. J. Mammal. 2012, 93, 353–359. [Google Scholar] [CrossRef]
- Wada, E.; Mizutani, H.; Minagawa, M. The use of stable isotopes for food web analysis. Crit. Rev. Food Sci. Nutr. 1991, 30, 361–371. [Google Scholar] [CrossRef]
- Grupe, G.; Kruger, H.H. Feeding ecology of the stone and pine marten revealed by element analysis of their skeletons. Sci. Total Environ. 1990, 90, 227–240. [Google Scholar] [CrossRef]
- Gomez-Diaz, E.; Figuerola, J. New perspectives in tracing vector-borne interaction networks. Trends Parasitol. 2010, 26, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, O.; Dautel, H.; Newton, J.; Gray, J.S. Natural isotope signatures of host blood are replicated in moulted ticks. Ticks Tick Borne Dis. 2011, 2, 225–227. [Google Scholar] [CrossRef]
- Hamer, S.A.; Weghorst, A.C.; Auckland, L.D.; Roark, E.B.; Strey, O.F.; Teel, P.D.; Hamer, G.L. Comparison of DNA and carbon and nitrogen stable isotope-based techniques for identification of prior vertebrate hosts of ticks. J. Med. Entomol. 2015, 52, 1043–1049. [Google Scholar] [CrossRef]
- Stapp, P.; Salkeld, D.J. Inferring host-parasite relationships using stable isotopes: Implications for disease transmission and host specificity. Ecology 2009, 90, 3268–3273. [Google Scholar] [CrossRef]
- Njabo, K.; Smith, T.; Yohannes, E. Feeding habits of culicine mosquitoes in the Cameroon lowland forests based on stable isotopes and blood meal analyses. J. Parasitol. Vector Biol. 2013, 5, 6–12. [Google Scholar]
- Cormie, A.B.; Schwarcz, H.P. Stable isotopes of nitrogen and carbon of North American white-tailed deer and implications for paleodietary and other food web studies. Palaeogeogr. Palaeoclimatol. Palaeoecol. 1994, 107, 227–241. [Google Scholar] [CrossRef]
- Lavin, S.; Deelen, T.; Brown, P.; Warner, R.; Ambrose, S. Prey use by red foxes (Vulpes vulpes) in urban and rural areas of Illinois. Can. J. Zool. 2011, 81, 1070–1082. [Google Scholar] [CrossRef]
- O’Guinn, M.L.; Lee, J.S.; Kondig, J.P.; Fernandez, R.; Carbajal, F. Field detection of eastern equine encephalitis virus in the Amazon Basin region of Peru using reverse transcription-polymerase chain reaction adapted for field identification of arthropod-borne pathogens. Am. J. Trop. Med. Hyg. 2004, 70, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Giordano, F.; Aigrain, L.; Quail, M.A.; Coupland, P.; Bonfield, J.K.; Davies, R.M.; Tischler, G.; Jackson, D.K.; Keane, T.M.; Li, J.; et al. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci. Rep. 2017, 7, 3935. [Google Scholar] [CrossRef] [PubMed]
- Mongan, A.E.; Tuda, J.S.B.; Runtuwene, L.R. Portable sequencer in the fight against infectious disease. J. Hum. Genet. 2020, 65, 35–40. [Google Scholar] [CrossRef]
- Faria, N.R.; Sabino, E.C.; Nunes, M.R.; Alcantara, L.C.; Loman, N.J.; Pybus, O.G. Mobile real-time surveillance of Zika virus in Brazil. Genome Med. 2016, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Castro-Wallace, S.L.; Chiu, C.Y.; John, K.K.; Stahl, S.E.; Rubins, K.H.; McIntyre, A.B.R.; Dworkin, J.P.; Lupisella, M.L.; Smith, D.J.; Botkin, D.J.; et al. Nanopore DNA sequencing and genome assembly on the International Space Station. Sci. Rep. 2017, 7, 18022. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A.; Campos, B.; Stone, J.; Blosser, E.M.; Burkett-Cadena, N.; Jacobs, J.L. Unbiased strain-typing of arbovirus directly from mosquitoes using nanopore sequencing: A field-forward biosurveillance protocol. Sci. Rep. 2018, 8, 5417. [Google Scholar] [CrossRef] [PubMed]
- Seah, A.; Lim, M.C.W.; McAloose, D.; Prost, S.; Seimon, T.A. MinION-based DNA barcoding of preserved and non-invasively collected wildlife samples. Genes 2020, 11, 445. [Google Scholar] [CrossRef] [PubMed]
- Biomeme. Franklin by Biomeme. Available online: https://info.biomeme.com/mobile-qpcr-thermocyclers (accessed on 18 August 2020).
- Tomaszewicz Brown, A.; McAloose, D.; Calle, P.P.; Auer, A.; Posautz, A.; Slavinski, S.; Brennan, R.; Walzer, C.; Seimon, T.A. Development and validation of a portable, point-of-care canine distemper virus qPCR test. PLoS ONE 2020, 15, e0232044. [Google Scholar] [CrossRef]
- Hole, K.; Nfon, C. Foot-and-mouth disease virus detection on a handheld real-time polymerase chain reaction platform. Transbound Emerg. Dis. 2019, 66, 1789–1795. [Google Scholar] [CrossRef]
- Rutkowski, N.; Dong, Y.; Dimopoulos, G. Field-deployable molecular diagnostic platform for arbovirus detection in Aedes aegypti. Parasit. Vectors 2020, 13, 489. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, F.; Perrone, A.; Della Noce, I.; Colombo, L.; Lo Priore, S.; Romano, S. Application of a portable instrument for rapid and reliable detection of SARS-CoV-2 infection in any environment. Immunol. Rev. 2020, 295 (Suppl. s1), 4–10. [Google Scholar] [CrossRef] [PubMed]
- Pashchenko, O.; Shelby, T.; Banerjee, T.; Santra, S. A comparison of optical, electrochemical, magnetic, and colorimetric point-of-care biosensors for infectious disease diagnosis. ACS Infect. Dis. 2018, 4, 1162–1178. [Google Scholar] [CrossRef] [PubMed]
- Lemuth, K.; Rupp, S. Microarrays as research tools and diagnostic devices. In RNA and DNA Diagnostics; Springer: Cham, Switzerland, 2015; pp. 259–280. [Google Scholar]
- Toner, M.; Irimia, D. Blood-on-a-chip. Annu. Rev. Biomed. Eng. 2005, 7, 77–103. [Google Scholar] [CrossRef]
- Liu, C.; Mauk, M.G.; Hart, R.; Bonizzoni, M.; Yan, G.; Bau, H.H. A low-cost microfluidic chip for rapid genotyping of malaria-transmitting mosquitoes. PLoS ONE 2012, 7, e42222. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Sharma, S.; Krajacich, B.J.; Fakoli, L.S., III.; Bolay, F.K.; Diclaro, J.W., II.; Johnson, W.E.; Ebel, G.D.; Foy, B.D.; Brackney, D.E. Xenosurveillance: A novel mosquito-based approach for examining the human-pathogen landscape. PLoS Negl. Trop. Dis. 2015, 9, e0003628. [Google Scholar] [CrossRef]
- Fauver, J.R.; Gendernalik, A.; Weger-Lucarelli, J.; Grubaugh, N.D.; Brackney, D.E.; Foy, B.D.; Ebel, G.D. The use of xenosurveillance to detect human bacteria, parasites, and viruses in mosquito bloodmeals. Am. J. Trop. Med. Hyg. 2017, 97, 324–329. [Google Scholar] [CrossRef]
- Kocher, A.; de Thoisy, B.; Catzeflis, F.; Valière, S.; Bañuls, A.L.; Murienne, J. iDNA screening: Disease vectors as vertebrate samplers. Mol. Ecol. 2017, 26, 6478–6486. [Google Scholar] [CrossRef]
- Komar, N.; Panella, N.A.; Young, G.R.; Basile, A.J. Methods for detection of West Nile virus antibodies in mosquito blood meals. J. Am. Mosq. Control Assoc. 2015, 31, 1–6. [Google Scholar] [CrossRef]
- Greenberg, J.A.; DiMenna, M.A.; Hanelt, B.; Hofkin, B.V. Analysis of post-blood meal flight distances in mosquitoes utilizing zoo animal blood meals. J. Vector. Ecol. 2012, 37, 83–89. [Google Scholar] [CrossRef]
- Martínez-de la Puente, J.; Soriguer, R.; Senar, J.C.; Figuerola, J.; Bueno-Mari, R.; Montalvo, T. Mosquitoes in an urban zoo: Identification of blood meals, flight distances of engorged females, and avian malaria infection. Front. Vet. Sci. 2020, 7, 460. [Google Scholar] [CrossRef] [PubMed]
- Michael, E.; Ramaiah, K.D.; Hoti, S.L.; Barker, G.; Paul, M.R.; Yuvaraj, J.; Das, P.K.; Grenfell, B.T.; Bundy, D.A. Quantifying mosquito biting patterns on humans by DNA fingerprinting of bloodmeals. Am. J. Trop. Med. Hyg. 2001, 65, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Soremekun, S.; Maxwell, C.; Zuwakuu, M.; Chen, C.; Michael, E.; Curtis, C. Measuring the efficacy of insecticide treated bednets: The use of DNA fingerprinting to increase the accuracy of personal protection estimates in Tanzania. Trop. Med. Int. Health 2004, 9, 664–672. [Google Scholar] [CrossRef]
- Tedrow, R.E.; Zimmerman, P.A.; Abbott, K.C. Multiple blood feeding: A force multiplier for transmission. Trends Parasitol. 2019, 35, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.W.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- Allio, R.; Donega, S.; Galtier, N.; Nabholz, B. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: Implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 2017, 34, 2762–2772. [Google Scholar] [CrossRef]
- Meyer, C.P.; Paulay, G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005, 3, e422. [Google Scholar] [CrossRef]
- Avise, J.C.; Walker, D. Species realities and numbers in sexual vertebrates: Perspectives from an asexually transmitted genome. Proc. Natl. Acad. Sci. USA 1999, 96, 992–995. [Google Scholar] [CrossRef]
- Meier, R.; Zhang, G.; Ali, F. The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Syst. Biol. 2008, 57, 809–813. [Google Scholar] [CrossRef]
- Mallet, J.; Willmott, K. Taxonomy: Renaissance or Tower of Babel? Trends Ecol. Evol. 2003, 18, 57–59. [Google Scholar] [CrossRef]
- Moritz, C.; Cicero, C. DNA barcoding: Promise and pitfalls. PLoS Biol. 2004, 2, e354. [Google Scholar] [CrossRef] [PubMed]
- Bensasson, D.; Zhang, D.; Hartl, D.L.; Hewitt, G.M. Mitochondrial pseudogenes: Evolution’s misplaced witnesses. Trends Ecol. Evol. 2001, 16, 314–321. [Google Scholar] [CrossRef]
- da Cruz, M.O.R.; Weksler, M.; Bonvicino, C.R.; Bezerra, A.M.R.; Prosdocimi, F.; Furtado, C.; Geise, L.; Catzeflis, F.; de Thoisy, B.; de Oliveira, L.F.B.; et al. DNA barcoding of the rodent genus Oligoryzomys (Cricetidae: Sigmodontinae): Mitogenomic-anchored database and identification of nuclear mitochondrial translocations (Numts). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2019, 30, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Borland, E.M.; Hartman, D.A.; Hopken, M.W.; Piaggio, A.J.; Kading, R.C. Technical limitations associated with molecular barcoding of arthropod bloodmeals taken from North American deer species. J. Med. Entomol. 2020, 57, 2002–2006. [Google Scholar] [CrossRef]
- Hickerson, M.J.; Meyer, C.P.; Moritz, C. DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 2006, 55, 729–739. [Google Scholar] [CrossRef]
- Iyiola, O.A.; Nneji, L.M.; Mustapha, M.K.; Nzeh, C.G.; Oladipo, S.O.; Nneji, I.C.; Okeyoyin, A.O.; Nwani, C.D.; Ugwumba, O.A.; Ugwumba, A.A.A.; et al. DNA barcoding of economically important freshwater fish species from north-central Nigeria uncovers cryptic diversity. Ecol. Evol. 2018, 8, 6932–6951. [Google Scholar] [CrossRef]
- Decru, E.; Moelants, T.; De Gelas, K.; Vreven, E.; Verheyen, E.; Snoeks, J. Taxonomic challenges in freshwater fishes: A mismatch between morphology and DNA barcoding in fish of the north-eastern part of the Congo basin. Mol. Ecol. Resour. 2016, 16, 342–352. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2019, 47, D94–D99. [Google Scholar] [CrossRef]
- Shen, Y.Y.; Chen, X.; Murphy, R.W. Assessing DNA barcoding as a tool for species identification and data quality control. PLoS ONE 2013, 8, e57125. [Google Scholar] [CrossRef]
- Li, X.; Shen, X.; Chen, X.; Xiang, D.; Murphy, R.W.; Shen, Y. Detection of potential problematic Cyt b gene sequences of fishes in GenBank. Front. Genet. 2018, 9, 30. [Google Scholar] [CrossRef]
- Mioduchowska, M.; Czyz, M.J.; Goldyn, B.; Kur, J.; Sell, J. Instances of erroneous DNA barcoding of metazoan invertebrates: Are universal cox1 gene primers too “universal”? PLoS ONE 2018, 13, e0199609. [Google Scholar] [CrossRef] [PubMed]
- Elmeer, K.; Almalki, A.; Mohran, K.A.; Al-Qahtani, K.N.; Almarri, M. DNA barcoding of Oryx leucoryx using the mitochondrial cytochrome c oxidase gene. Genet. Mol. Res. 2012, 11, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Leray, M.; Knowlton, N.; Ho, S.L.; Nguyen, B.N.; Machida, R.J. GenBank is a reliable resource for 21st century biodiversity research. Proc. Natl. Acad. Sci. USA 2019, 116, 22651–22656. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Reeves, L.E.; Holderman, C.J.; Gillett-Kaufman, J.L.; Kawahara, A.Y.; Kaufman, P.E. Maintenance of host DNA integrity in field-preserved mosquito (Diptera: Culicidae) blood meals for identification by DNA barcoding. Parasit. Vectors 2016, 9, 503. [Google Scholar] [CrossRef]
| Technology: | Pros: | Cons: |
|---|---|---|
| Real-time and qPCR | - Gold standard of nucleic acid quantification - Sensitive, specific, rapid - Increasingly affordable | - Requires expensive instrumentation and reagents - Requires specific primer sets for anticipated animals |
| High Resolution Melting | - Compatible with real-time PCR equipment - Specific to DNA sequence, length, and GC content - Unique melt curves generated with universal primers - Useful for small amplicons and degraded DNA | - Requires expensive instrumentation - Must generate a unique library for comparison - Curves may overlap for unrelated species |
| Droplet Digital PCR | - Sensitive for low copy and rare DNA targets | - Requires specialized equipment - Accuracy and sensitivity are dependent on reaction conditions - Substantial and costly optimization |
| Next Generation Sequencing | - Target sequence agnostic - Can differentiate between mtDNA and NUMTs - Multiplexing capability - New, field-forward inexpensive portable instruments | - High equipment and sample processing costs - Data analysis requires specialized training - Partially digested DNA cannot be amplified/sequenced |
| Microarray and Microsphere | - Sensitive and high-throughput - Small probes can capture degraded DNA | - Requires specialized instrumentation - Unique probes must be designed for each target |
| Mass Spectrometry | - Targets proteins that are more stable during digestion than DNA - Can generate reference library of spectra from known protein sequences | - High equipment and sample processing costs - Relies on a comprehensive reference library - Project-specific reference libraries must be generated - Sample storage can influence profile quality |
| Stable Isotope Analysis | - Useful for samples with degraded DNA | - High equipment and sample processing costs - Specificity and species resolution remain a challenge - Project-specific reference libraries must be generated |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borland, E.M.; Kading, R.C. Modernizing the Toolkit for Arthropod Bloodmeal Identification. Insects 2021, 12, 37. https://doi.org/10.3390/insects12010037
Borland EM, Kading RC. Modernizing the Toolkit for Arthropod Bloodmeal Identification. Insects. 2021; 12(1):37. https://doi.org/10.3390/insects12010037
Chicago/Turabian StyleBorland, Erin M., and Rebekah C. Kading. 2021. "Modernizing the Toolkit for Arthropod Bloodmeal Identification" Insects 12, no. 1: 37. https://doi.org/10.3390/insects12010037
APA StyleBorland, E. M., & Kading, R. C. (2021). Modernizing the Toolkit for Arthropod Bloodmeal Identification. Insects, 12(1), 37. https://doi.org/10.3390/insects12010037






