A Chemosensory Protein Detects Antifeedant in Locust (Locusta migratoria)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Reverse Transcription PCR (RT-PCR) and Quantitative Real-Time Reverse Transcription PCR (qRT-PCR)
2.3. The Expression and Purification of the Recombinant CSP
2.4. Western Blotting
2.5. Fluorescent Binding Assay
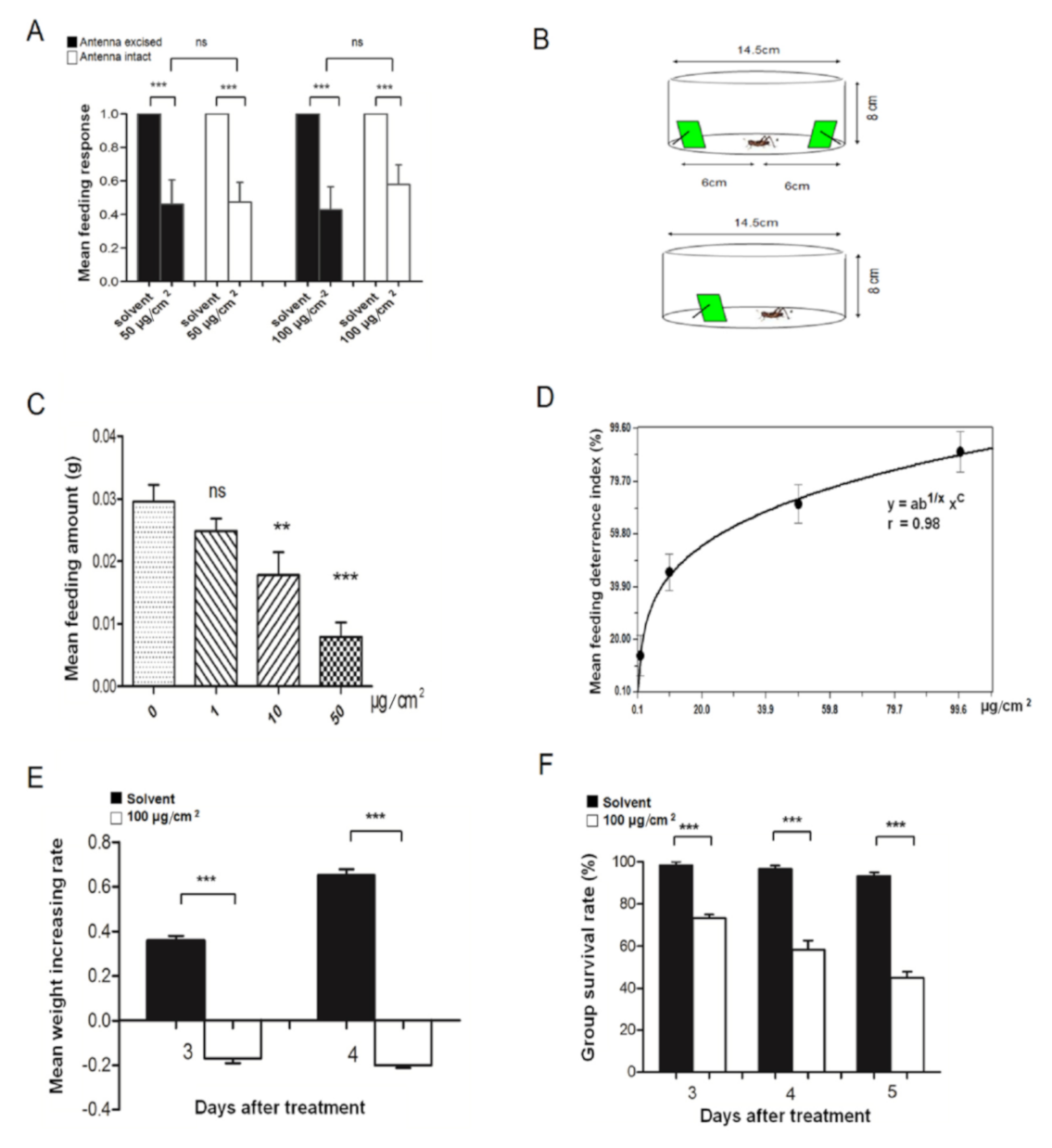
2.6. Testing the Role of α-Amylcinnamaldehyde in Locust Feeding
2.7. Detecting the Effects of Deficiency of LmigCSPIII by RNAi
2.8. Statistical Analyses
3. Results
3.1. LmigCSPIII Gene Is Expressed in Chemosensory Organs Throughout Development
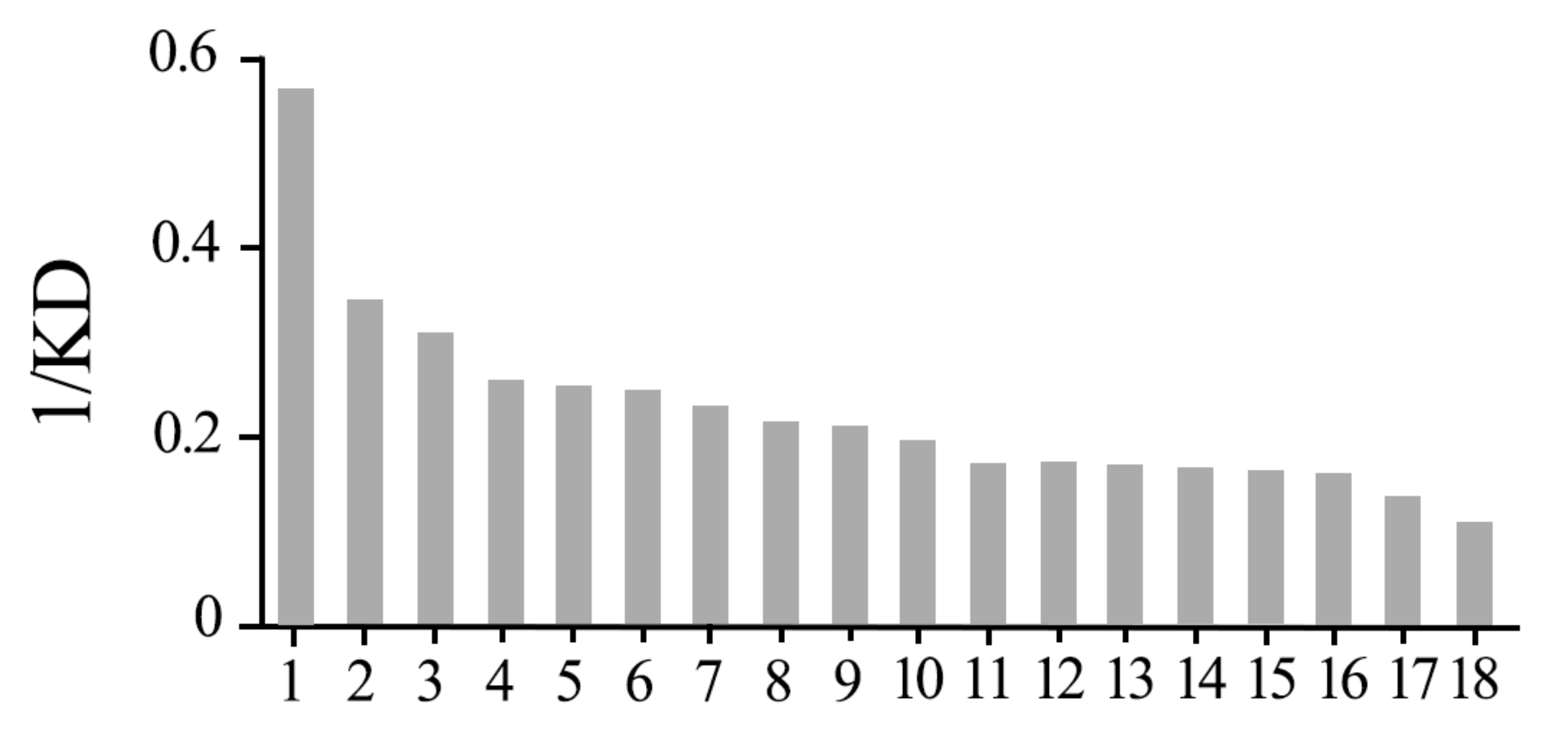
3.2. LmigCSPIII Has the Highest Binding Affinity to α-Amylcinnamaldehyde
3.3. A-Amylcinnamaldehyde Is a Locust Antifeedant
3.4. LmigCSPIII Deficient Mutants Have Decreased Sensitivity to α-Amylcinnamaldehyde
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Scott, K. Taste recognition: Food for thought. Neuron 2005, 48, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Tahvanainen, J.; Julkunen-Tiitto, R.; Kettunen, J. Phenolic glycosides govern the food selection pattern of willow feeding leaf beetles. Oecologia 1985, 67, 52–56. [Google Scholar] [CrossRef]
- Behmer, S.T.; Simpson, S.J.; Raubenheimer, D. Herbivore foraging in chemically heterogeneous environments: Nutrients and secondary metabolites. Ecology 2002, 83, 2489–2501. [Google Scholar] [CrossRef]
- Hallem, E.A.; Dahanukar, A.; Carlson, J.R. Insect odor and taste receptors. Annu. Rev. Entomol. 2006, 51, 113–135. [Google Scholar] [CrossRef]
- Bernays, E.A.; Chapman, R.F. Deterrent chemicals as a basis of oligophagy in Locusta migratoria(L.). Ecol. Entomol. 1977, 2, 1–18. [Google Scholar] [CrossRef]
- Bernays, E.A.; Chapman, R.F.; Singer, M.S. Sensitivity to chemically diverse phagostimulants in a single gustatory neuron of a polyphagous caterpillar. J. Comp. Physiol. A 2000, 186, 13–19. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Wanner, K.W.; Willis, L.G.; Theilmann, D.A.; Isman, M.B.; Feng, Q.; Plettner, E. Analysis of the insect os-d-like gene family. J. Chem. Ecol. 2004, 30, 889–911. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Vogt, R.G.; Miller, N.E.; Litvack, R.; Fandino, R.A.; Sparks, J.; Staples, J.; Friedman, R.; Dickens, J.C. The insect SNMP gene family. Insect Biochem. Mol. Biol. 2009, 39, 448–456. [Google Scholar] [CrossRef]
- Benton, R.; Vannice, K.S.; Gomez-Diaz, C.; Vosshall, L.B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 2009, 136, 149–162. [Google Scholar] [CrossRef] [Green Version]
- Clyne, P.J.; Warr, C.G.; Freeman, M.R.; Lessing, D.; Kim, J.; Carlson, J.R. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron 1999, 22, 327–338. [Google Scholar] [CrossRef] [Green Version]
- Vosshall, L.B.; Amrein, H.; Morozov, P.S.; Rzhetsky, A.; Axel, R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 1999, 96, 725–736. [Google Scholar] [CrossRef] [Green Version]
- Wetzel, C.H.; Behrendt, H.J.; Gisselmann, G.; Stortkuhl, K.F.; Hovemann, B.; Hatt, H. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. Proc. Natl. Acad. Sci. USA 2001, 98, 9377–9380. [Google Scholar] [CrossRef] [Green Version]
- Hansson, B.S.; Stensmyr, M.C. Evolution of insect olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [Green Version]
- Scott, K.; Brady, R., Jr.; Cravchik, A.; Morozov, P.; Rzhetsky, A.; Zuker, C.; Axel, R. A chemosensory gene family encoding candidate gustatory and olfactory receptor in Drosophila. Cell 2001, 104, 661–673. [Google Scholar] [CrossRef]
- Hill, C.A.; Fox, A.N.; Pitts, R.J.; Kent, L.B.; Tan, P.L.; Chrystal, M.A.; Cravchik, A.; Collins, F.H.; Robertson, H.M.; Zwiebel, L.J. G Protein-coupled receptors in Anopheles Gambiae. Science 2002, 298, 176–178. [Google Scholar] [CrossRef]
- Dahanukar, A.; Hallem, E.A.; Carlson, J.R. Insect chemoreception. Curr. Opin. Neurobiol. 2005, 15, 423–430. [Google Scholar] [CrossRef]
- Angeli, S.; Ceron, F.; Scaloni, A.; Monti, M.; Monteforti, G.; Minnocci, A.; Petacchi, R.; Pelosi, P. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 1999, 262, 745–754. [Google Scholar] [CrossRef]
- Picimbon, J.-F.; Dietrich, K.; Breer, H.; Krieger, J. Chemosensory proteins of Locusta migratoria (Orthoptera: Acrididae). Insect Biochem. Mol. Biol. 2000, 30, 233–241. [Google Scholar] [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Ab, E.; Wechselberger, R.; Angeli, S.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P.; et al. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 10606–10613. [Google Scholar] [CrossRef] [PubMed]
- Kitabayashi, A.N.; Arai, T.; Kubo, T.; Natori, S. Molecular cloning of cDNA for p10, a novel protein that increases in the regenerating legs of Periplaneta americana (American cockroach). Insect Biochem. Mol. Biol. 1998, 28, 785–790. [Google Scholar] [CrossRef]
- Maleszka, J.; Foret, S.; Saint, R.; Maleszka, R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 2007, 217, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, Y.; Sato, R.; Tsuchihara, K.; Ozaki, K.; Mita, K.; Asaoka, K.; Taniai, K. Ligand carrier protein genes expressed in larval chemosensory organs of Bombyx mori. Insect Biochem. Mol. Biol. 2011, 41, 545–562. [Google Scholar] [CrossRef]
- Ban, L.; Scaloni, A.; Brandazza, A.; Angeli, S.; Zhang, L.; Yan, Y.; Pelosi, P. Chemosensory proteins of Locusta migratoria. Insect Mol. Biol. 2003, 12, 125–134. [Google Scholar] [CrossRef]
- Zhou, J.J.; Kan, Y.; Antoniw, J.; Pickett, J.A.; Field, L.M. Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem. Senses 2006, 31, 453–465. [Google Scholar] [CrossRef]
- Jin, X.; Brandazza, A.; Navarrini, A.; Ban, L.; Zhang, S.; Steinbrecht, R.A.; Zhang, L.; Pelosi, P. Expression and immunolocalisation of odorant-binding and chemosensory proteins in locusts. Cell. Mol. Life Sci. 2005, 62, 1156–1166. [Google Scholar] [CrossRef]
- Zhou, X.H.; Ban, L.P.; Iovinella, I.; Zhao, L.J.; Gao, Q.; Felicioli, A.; Sagona, S.; Pieraccini, G.; Pelosi, P.; Zhang, L.; et al. Diversity, abundance, and sex-specific expression of chemosensory proteins in the reproductive organs of the locust Locusta migratoria manilensis. Biol. Chem. 2013, 394, 43–54. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, S.G.; Zhang, L. Expression of odorant-binding and chemosensory proteins and spatial map of chemosensilla on labial palps of Locusta migratoria (Orthoptera: Acrididae). Arthropod Struct. Dev. 2006, 35, 47–56. [Google Scholar] [CrossRef]
- Zhou, S.H.; Zhang, J.; Zhang, S.G.; Zhang, L. Expression of chemosensory proteins in hairs on wings of Locusta migratoria (Orthoptera: Acrididae). J. Appl. Entomol. 2008, 132, 439–450. [Google Scholar] [CrossRef]
- Zhou, S.H.; Zhang, S.G.; Zhang, L. The chemosensilla on tarsi of Locusta migratoria (Orthoptera: Acrididae): Distribution, ultrastructure, expression of chemosensory proteins. J. Morphol. 2009, 270, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Ban, L.; Zhang, L.; Yan, Y.; Pelosi, P. Binding properties of a locust’s chemosensory protein. Biochem. Biophys. Res. Commun. 2002, 293, 50–54. [Google Scholar] [CrossRef]
- Sun, Y.F.; De Biasio, F.; Qiao, H.L.; Iovinella, I.; Yang, S.X.; Ling, Y.; Riviello, L.; Battaglia, D.; Falabella, P.; Yang, X.L.; et al. Two Odorant-Binding Proteins Mediate the Behavioural Response of Aphids to the Alarm Pheromone (E)-ß-farnesene and Structural Analogues. PLoS ONE 2012, 7, e32759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, D.A.; Isman, M.B. Antifeedant and toxic activity of Trichilia americana extract against the larvae of Spodoptera litura. Entomol. Exp. Appl. 2001, 98, 9–16. [Google Scholar] [CrossRef]
- Akhtar, Y.; Yeoung, Y.R.; Isman, M.B. Comparative bioactivity of selected extracts from Meliaceae and some commercial botanical insecticides against two noctuid caterpillars, Trichoplusia ni and Pseudaletia unipuncta. Phytochem. Rev. 2008, 7, 77–88. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Zhang, J.; Li, D.; Sun, Y.; Guo, Y.; Ma, E.; Zhu, K.Y. Silencing of two alternative splicing-derived mRNA variants of chitin synthase 1 gene by RNAi is lethal to the oriental migratory locust, Locusta migratoria manilensis (Meyen). Insect Biochem. Mol. Biol. 2010, 40, 824–833. [Google Scholar] [CrossRef]
- Marchal, E.; Verlinden, H.; Badisco, L.; Van Wielendaele, P.; Vanden Broeck, J. RNAi-mediated knockdown of Shade negatively affects ecdysone-20-hydroxylation in the desert locust, Schistocerca gregaria. J. Insect Physiol. 2012, 58, 890–896. [Google Scholar] [CrossRef]
- Tobback, J.; Vuerinckx, K.; Boerjan, B.; Huybrechts, R. RNA interference mortality points to noncircadian functions for the clock gene in the desert locust Schistocerca gregaria. Insect Mol. Biol. 2012, 21, 369–381. [Google Scholar] [CrossRef]
- US, E.P.A. Initial Risk-Based Prioritization of High Production Volume (HPV) Chemicals, Cinnamyl Derivatives Category, Alpha-Amylcinnamaldehyde (CASRN 122-40-7). 2009. Available online: http://www.epa.gov/hpvis/rbp/Category_Cinnamyl%20Derivatives_Web_April%202009.pdf (accessed on 18 December 2020).
- Pass, G.J.; Foley, W.J. Plant secondary metabolites as mammalian feeding deterrents: Separating the effects of the taste of salicin from its post-ingestive consequences in the common brushtail possum (Trichosurus vulpecula). J. Comp. Physiol. B 2000, 170, 185–192. [Google Scholar] [CrossRef]
- Chapman, R.F.; Joern, A. Biology of Grasshoppers; John Wiley & Sons: Hoboken, NJ, USA, 1990. [Google Scholar]
- Bernays, E.A.; Chapman, R.F. Plant Chemistry and Acridoid Feeding Behaviour; Academic Press: London, UK, 1978. [Google Scholar]
- Jacquin-Joly, E.; Vogt, R.G.; Francois, M.C.; Nagnan-Le Meillour, P. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Senses 2001, 26, 833–844. [Google Scholar] [CrossRef]
- Celorio-Mancera, M.d.l.P.; Sundmalm, S.M.; Vogel, H.; Rutishauser, D.; Ytterberg, A.J.; Zubarev, R.A.; Janz, N. Chemosensory proteins, major salivary factors in caterpillar mandibular glands. Insect Biochem. Mol. Biol. 2012, 42, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Wu, C.; Zhang, L. Electrophysiological response patterns of 16 olfactory neurons from the trichoid sensilla to odorant from fecal volatiles in the locust, Locusta migratoria manilensis. Arch. Insect Biochem. Physiol. 2011, 77, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, X.; Ma, Z.; Xue, L.; Han, J.; Yu, D.; Kang, L. CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet. 2011, 7, e1001291. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Calvello, M.; Ban, L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses 2005, 30, i291–i292. [Google Scholar] [CrossRef] [Green Version]
- Kulmuni, J.; Wurm, Y.; Pamilo, P. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 2013, 110, 538–547. [Google Scholar] [CrossRef] [Green Version]
- Sinoir, Y. The role of the feelers and labrum in the eating behavior of the migratory cricket iarva. Ann. de la Nutr. et de l’Aliment. 1969, 23, 167–194. [Google Scholar]
- Linton, Y.M.; Nisbet, A.J.; Mordue (L), A.J. The effects of azadirachtin on the testes of the desert locust, Schistocerca gregaria (Forskal). J. Insect Physiol. 1997, 43, 1077–1084. [Google Scholar] [CrossRef]
- Woodhead, S. Surface chemistry of Sorghum bicolor and its importance in feeding by Locusta migratoria. Physiol. Entomol. 1983, 8, 345–352. [Google Scholar] [CrossRef]
- Chapman, R. The chemical inhibition of feeding by phytophagous insects a review. Bull. Entomol. Res. 1974, 643, 339–363. [Google Scholar] [CrossRef]
- Vogt, R.G. Molecular Basis of Pheromone Detection in Insects; Elsevier: Amsterdam, The Netherlands, 2005; Volume 3, pp. 753–804. [Google Scholar] [CrossRef]
- Jiao, Y.; Moon, S.J.; Montell, C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc. Natl. Acad. Sci. USA 2007, 104, 14110–14115. [Google Scholar] [CrossRef] [Green Version]
- Montell, C. A taste of the Drosophila gustatory receptors. Curr. Opin. Neurobiol. 2009, 19, 345–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cande, J.; Prud’homme, B.; Gompel, N. Smells like evolution: The role of chemoreceptor evolution in behavioral change. Curr. Opin. Neurobiol. 2013, 23, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Melcher, C.; Pankratz, M.J. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 2005, 3, e305. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Primer Names | Sequences (5’-3’) | |
|---|---|---|
| RT-PCR | LmigCSPIII-s LmigCSPIII-as | ACGCCTCCTCAAGTCCTACA AGCTGTTTCACTTATTCACAGAGT |
| qPCR | LmigCSPIII-s LmigCSPIII-as | AAGGGGTGGGAGACGGCCTG CAGCTCCTCCCCAACGACAGC |
| dsLmigCSPIII | LmigCSPIII-s | TAATACGACTCACTATAGG AAGGGGTGGGAGACGGCCTG |
| LmigCSPIII-as | TAATACGACTCACTATAGG CAGCTCCTCCCCAACGACAGC | |
| Clone | LmigCSPIII-s | CATATAGGGCCACTCAGGACCCGCTG Nde1 |
| LmigCSPIII-as | GAATTCTCAGAAGTTGATGCCGCGGTG EcoR1 | |
| Control | LmigActin-s LmigActin-as | GCAAAGCTGGCTTCGCCG ATGTTCCTCGGGCGCCAC |
| Ligands | |
|---|---|
| Aliphatic acid | Aliphatic ester derivatives |
| Butanoic acid | Methyl isovalate |
| Pentadecanoic acid | Ethyl cis-3-hexenoate |
| Palmitic acid | Ethyl caprylate |
| Aliphatic alcohols | Ethyl nonanoate |
| 3-Methy-1-butanol | Ethyl caprate |
| 2-Hexanol | Ethyl undecanoate |
| 3-Hexanol | Ethyl laurate |
| 3- Octanol | Ethyl tridecanoate |
| 5-Nonanol | Ethyl myristate |
| Linalool | Dodecyl 2-methylacrylate |
| 3,7-Dimethyl octanol | Ethyl palmitate |
| cis-3,7-Dimethyl-2,6-octadien-1-ol | Oleamide |
| 10-Undecylenylalcohol | Ethyl sterate |
| Undecanol | Geranyl acetate |
| 2,2,4-Trimethyl-3-nonanol | Orbicular compounds |
| Tridecanol | Cyclohexanol |
| Dodecanol | 2,5-Dimethyl cyclohexanone |
| Tetradecanol | α-Pinene |
| Pentadecanol | 2,6-Dimethyl cyclohexanone |
| Hexadecanol | 2,6,6-Trimethyl-2-cyclohexene-1,4-dione |
| Heptadecanol | Heterocyclic compounds |
| 1-Hydroxyoctadecane | 2,5-Dimetylpyrazine |
| Guaicacol | β-Cyclocitral |
| Aliphatic ketones | 1-Aminoanthracen |
| 2-Heptaone | Aromatic compounds |
| 6-Dimethyl-4-heptanone | Phenol |
| 2-Undecanone | Benzaldehyde |
| 6-Undecanone | Benzyl alcohol |
| 2-Dodecanone | DL-sec-phenethyl alcohol |
| 2-Tridocanone | Benzeneacetonitrile |
| 2-Pentadecanone | 4-Tert-butylphenol |
| 2-Hexadecanone | Dimethyl phthalate |
| 2-Heptadecanone | Diethyl phthalate |
| 2-Octadecanone | α-Amylcinnamaldehyde |
| 2-Nonadecanone | Benzyl benzoate |
| 6-Methyl-5-Hepten-2-one | Dibutyl phthalate |
| Aliphatic aldehydes | Di (.alpha.-ethylhexyl) phthalate |
| trans-2-Hexenal | Aliphatic alkane |
| Hexanal | 1-Undecene |
| Octanal | 1-Hexadecene |
| Nonanal | 1-Nonadecene |
| Decanal | |
| Undecylic aldehyde | |
| Dodecanal | |
| Tridecanal | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, X.; Xu, H.; Zheng, N.; Yin, X.; Zhang, L. A Chemosensory Protein Detects Antifeedant in Locust (Locusta migratoria). Insects 2021, 12, 1. https://doi.org/10.3390/insects12010001
Jiang X, Xu H, Zheng N, Yin X, Zhang L. A Chemosensory Protein Detects Antifeedant in Locust (Locusta migratoria). Insects. 2021; 12(1):1. https://doi.org/10.3390/insects12010001
Chicago/Turabian StyleJiang, Xingcong, Haozhi Xu, Nan Zheng, Xuewei Yin, and Long Zhang. 2021. "A Chemosensory Protein Detects Antifeedant in Locust (Locusta migratoria)" Insects 12, no. 1: 1. https://doi.org/10.3390/insects12010001
APA StyleJiang, X., Xu, H., Zheng, N., Yin, X., & Zhang, L. (2021). A Chemosensory Protein Detects Antifeedant in Locust (Locusta migratoria). Insects, 12(1), 1. https://doi.org/10.3390/insects12010001





