Population Genetic Structure and Demography of the Critically Endangered Chequered Blue Butterfly (Scolitantides orion) in a Highly Isolated Part of Its Distribution Range
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species
2.2. Study Sites
2.3. Adult Demography
2.4. Sampling for Genetic Studies and DNA Extraction
2.5. Microsatellite Analysis
2.6. PCR for Genes Amplification, Sequencing, and Genetic Diversity
2.7. Phylogenetic Analysis
3. Results
3.1. Adult Demography
3.2. Genetic Variation at Microsatellites Loci, Gene Flow and Effective Population Size
3.3. Sequence Polymorphism in Genes
3.4. Genetic Differentiation between S. orion Populations from Poland
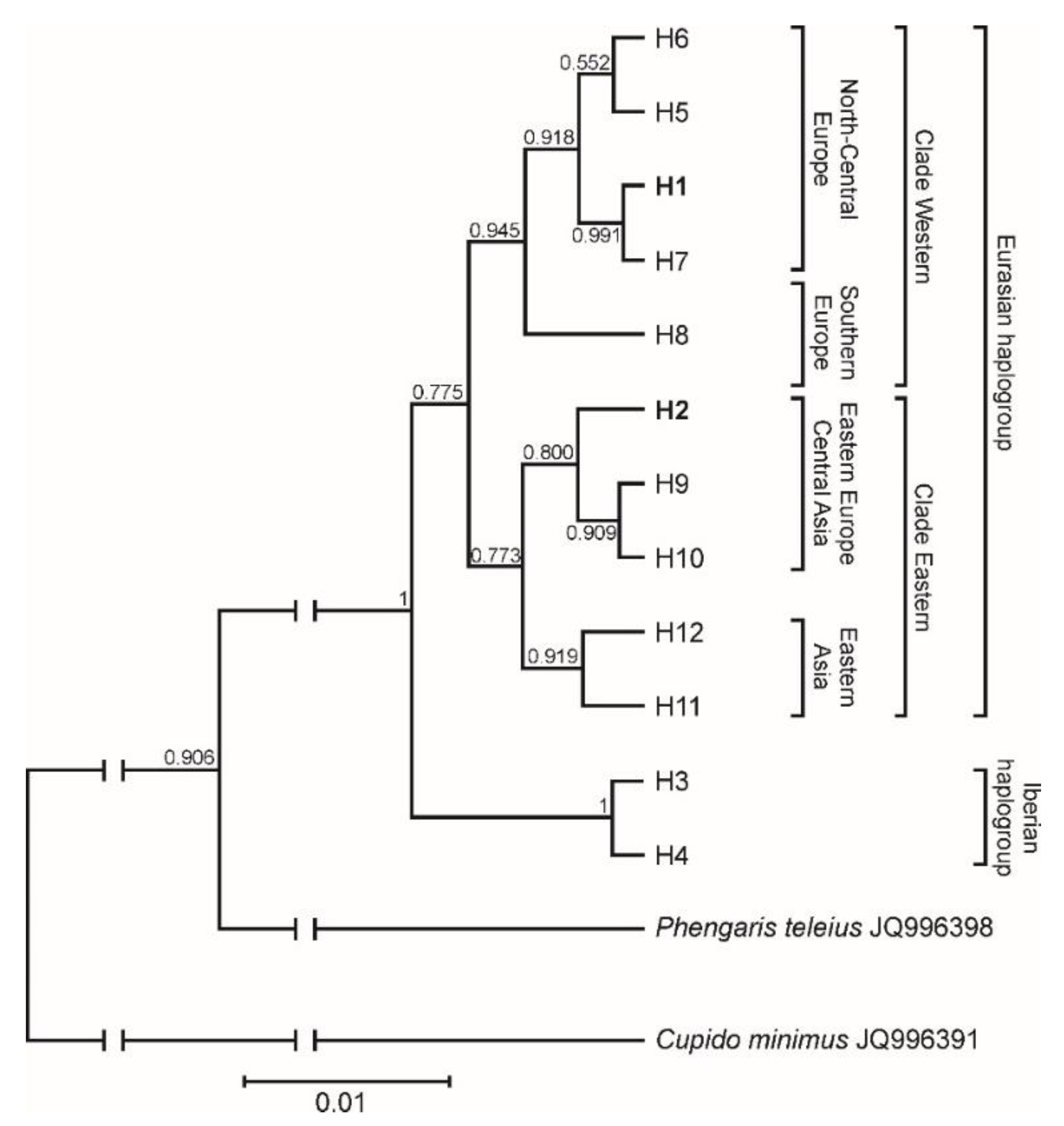
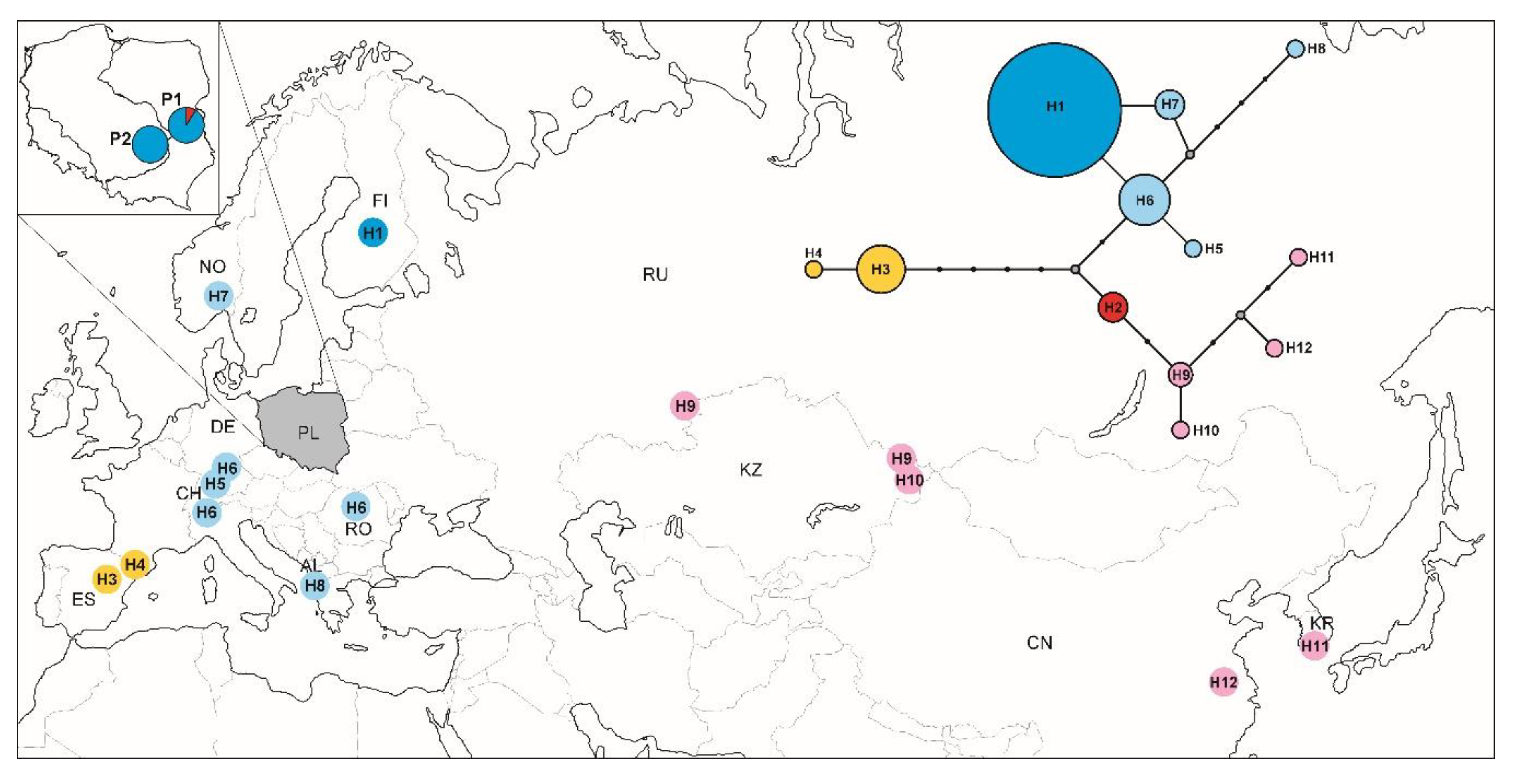
3.5. Phylogeographic Pattern Inferred from COI Sequences
4. Discussion
4.1. Adult Demography
4.2. Genetic Diversity and Differentiation between Populations
4.3. Phylogeographic Pattern
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thomas, J.A. Monitoring change in the abundance and distribution of insects using butterflies and other indicator groups. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.S.; Hill, J.K.; Thomas, J.A.; Asher, J.; Fox, R.; Huntley, B.; Roy, D.B.; Telfer, M.G.; Jeffcoate, S.; Harding, P.; et al. Rapid responses of British butterflies to opposing forces of climate and habitat change. Nature 2001, 414, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Besold, J.; Schmitt, T.; Tammaru, T.; Cassel-Lundhagen, A. Strong genetic impoverishment from the centre of distribution in southern Europe to peripheral Baltic and isolated Scandinavian populations of the pearly heath butterfly. J. Biogeogr. 2008, 35, 2090–2101. [Google Scholar] [CrossRef]
- Lande, R. Genetics and demography in biological conservation. Science 1988, 241, 1455–1460. [Google Scholar] [CrossRef] [PubMed]
- Saccheri, I.; Kuussaari, M.; Kankare, M.; Vikman, P.; Fortelius, W.; Hanski, I. Inbreeding and extinction in a butterfly metapopulation. Nature 1998, 392, 491–494. [Google Scholar] [CrossRef]
- Schmitt, T.; Hewitt, G.M. The genetic pattern of population threat and loss: A case study of butterflies. Mol. Ecol. 2004, 13, 21–31. [Google Scholar] [CrossRef]
- Habel, J.C.; Rödder, D.; Lens, L.; Schmitt, T. The genetic signature of ecologically different grassland Lepidopterans. Biodivers. Conserv. 2013, 22, 2401–2411. [Google Scholar] [CrossRef]
- Maes, D.; Verovnik, R.; Wiemers, M.; Brosens, D.; Beshkov, S.; Bonelli, S.; Buszko, J.; Cantú-Salazar, L.; Cassar, L.F.; Collins, S.; et al. Integrating national Red Lists for prioritising conservation actions for European butterflies. J. Insect Conserv. 2019, 23, 301–330. [Google Scholar] [CrossRef]
- Tolman, T.; Lewington, R. Collins Butterfly Guide: The Most Complete Field Guide to the Butterflies of Britain and Europe; Harper Collins: London, UK, 2009. [Google Scholar]
- Kudrna, O.; Pennerstorfer, J.; Lux, K. Distribution atlas of European Butterflies and Skippers; PEKS: Schwanfeld, Germany, 2015. [Google Scholar]
- Van Swaay, C.A.M.; Cuttelod, A.; Collins, S.; Maes, D.; López Munguira, M.; Šašić, M.; Settele, J.; Verovnik, R.; Verstrael, T.; Warren, M.; et al. European Red List of European Butterflies; Publication Office of the European Union: Luxembourg, 2010. [Google Scholar]
- Van Swaay, C.A.M.; Warren, M.S. Red Data Book of European Butterflies (Rhopalocera); Nature and Environment Series No. 99; Council of Europe: Strasbourg, France, 1999. [Google Scholar]
- Marttila, O.; Saarinen, K.; Marttila, P. Six years from passing bell to recovery: Habitat restoration of the threatened chequered blue butterfly (Scolitantides orion) in SE Finland. Entomol. Fenn. 2000, 11, 113–117. [Google Scholar] [CrossRef]
- Coulondre, A. Systématique et répartition de Scolitantides orion (Pallas, 1771) (Lepidoptera: Lycaenidae). Linneana Belg. 1994, 14, 383–420. [Google Scholar]
- Komonen, A.; Tikkamäki, T.; Mattila, N.; Kotiaho, J.S. Patch size and connectivity influence the population turnover of the threatened chequered blue butterfly, Scolitantides orion (Lepidoptera: Lycaenidae). Eur. J. Entomol. 2008, 105, 131–136. [Google Scholar] [CrossRef]
- Tränkner, A.; Nuss, M. Risk spreading in the voltinism of Scolitantides orion orion (Pallas, 1771) (Lycaenidae). Nota Lepidopterol. 2005, 1, 55–64. [Google Scholar]
- Dawidowicz, Ł. Modraszek Orion Scolitanides orion (Pallas, 1771)—Biologia i Ekologia Gatunku Zagrożonego Wygięciem w Polsce. Ph.D. Thesis, Maria Curie-Sklodowska University, Lublin, Poland, 2018. [Google Scholar]
- Buszko, J. A Distribution Atlas of Butterflies in Poland (Lepidoptera: Papilionoidea, Hesperioidea) 1986–1995; TURPRESS: Toruń, Poland, 1997; p. 59. [Google Scholar]
- Arnason, A.N.; Schwarz, C.J. Using POPAN-5 to analyse banding data. Bird Study 1999, 46, 157–168. [Google Scholar] [CrossRef]
- White, G.C.; Burnham, K.P. Program MARK: Survival estimation from populations of marked animals. Bird Study 1999, 46, 120–138. [Google Scholar] [CrossRef]
- Schtickzelle, N.; Le Boulenge, E.; Baguette, M. Metapopulation dynamics of the bog fritillary butterfly: Demographic processes in a patchy population. Oikos 2002, 97, 349–360. [Google Scholar] [CrossRef]
- Nowicki, P.; Vrabec, V. Evidence for positive density-dependent emigration in butterfly metapopulations. Oecologia 2011, 167, 657–665. [Google Scholar] [CrossRef]
- Osváth-Ferentcz, M.; Bonelli, S.; Nowicki, P.; Peregovits, L.; Rákosy, L.; Sielezniew, M.; Kostro-Ambroziak, A.; Dziekańska, I.; Kőrösi, Á. Population demography of the endangered large blue butterfly Maculinea arion in Europe. J. Insect Conserv. 2017, 21, 411–422. [Google Scholar] [CrossRef]
- Sielezniew, M.; Nowicki, P. Adult demography of an isolated population of the threatened butterfly Scarce Heath Coenonympha hero and its conservation implications. J. Insect Conserv. 2017, 21, 737–742. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Tsai, C. Regression and time series model selection in small samples. Biometrika 1989, 76, 297–307. [Google Scholar] [CrossRef]
- Nowicki, P.; Witek, M.; Skorka, P.; Settele, J.; Woyciechowski, M. Population ecology of the endangered butterflies Maculinea teleius and M. nausithous and the implications for conservation. Popul. Ecol. 2005, 47, 193–202. [Google Scholar] [CrossRef]
- Bubová, T.; Kulma, M.; Vrabec, V.; Nowicki, P. Adult longevity and its relationship with conservation status in European butterflies. J. Insect Conserv. 2016, 20, 1021–1032. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Zeisset, I.; Als, T.D.; Settele, J.; Boomsma, J.J. Microsatellite markers for the large blue butterflies Maculinea nausithous and Maculinea alcon (Lepidoptera: Lycaenidae) and their amplification in other Maculinea species. Mol. Ecol. Notes 2005, 5, 165–168. [Google Scholar] [CrossRef]
- Ugelvig, L.V.N.; Andersen, A.; Boomsma, J.J.; Nash, D.R. Dispersal and gene flow in the rare, parasitic Large Blue butterfly Maculinea arion. Mol. Ecol. 2012, 21, 3224–3236. [Google Scholar] [CrossRef]
- Legrand, D.; Chaput-Bardy, A.; Turlure, C.; Dubois, Q.; Huet, M.; Schtickzelle, N.; Stevens, V.M.; Baguette, M. Isolation and characterization of 15 microsatellite loci in the specialist butterfly Boloria eunomia. Conserv. Genet. Resour. 2014, 6, 223–227. [Google Scholar] [CrossRef]
- Tartally, A.; Kelager, A.; Furst, M.A.; Nash, D.R. Host plant use drives genetic differentiation in syntopic populations of Maculinea alcon. PeerJ 2016, 4, e1865. [Google Scholar] [CrossRef]
- Harper, G.L.; Piyapattanakorn, S.; Goulson, D.; Maclean, N. The isolation of microsatellite markers from the Adonis blue butterfly (Lysandra bellargus). Mol. Ecol. 2000, 9, 1948–1949. [Google Scholar] [CrossRef]
- Ugelvig, L.V.N.; Nielsen, P.S.; Boomsma, J.J.; Nash, D.R. Reconstructing eight decades of genetic variation in an isolated Danish population of the large blue butterfly Maculinea alcon. BMC Evol. Biol. 2011, 11, 201. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A complete reimplementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (version 2.9.3): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Population. Vol 4. Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Piry, S.; Alapetite, A.; Cornuet, J.M.; Paetkau, D.; Baudouin, L.; Estoup, A. GENECLASS2: A software for genetic assignment and first-generation migrant detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S.; Do, C. LDNE: A program for estimating effective population size from data on linkage disequilibrium. Mol. Ecol. Resour. 2008, 8, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Brower, A.V.Z.; De Salle, R. Patterns of mitochondrial versus nuclear DNA sequence divergence among nymphalid butterflies: The utility of wingless as a source of characters for phylogenetic inference. Insect Mol. Biol. 1998, 7, 73–82. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H.J. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology; Krawetz, S., Misener, S., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 365–386. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequine suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. 2014. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 14 April 2018).
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Saarinen, P. The ecology of Scolitantides orion (Lepidoptera: Lycaenidae) in a metapopulation in southern Finland 1991–92. Baptria 1995, 20, 195–198. [Google Scholar]
- Kan, P.; Kan-van Limburg Stirum, B. Scolitantides orion (Pallas, 1771), entre Sedum et fourmis (Lepidoptera: Lyceanidae). Lépidoptères 2011, 20, 88–96. [Google Scholar]
- Endrestøl, A.; Bengtson, R. Mapping of the Chequered Blue Butterfly Scolitantides orion in Norway 2015–2016. In NINA Rapport 1342; NINA: Trondheim, Norway, 2017. [Google Scholar]
- Čelechovský, A. Modrásek rozchodníkový a jeho výskyt na střední Moravě. Živa 2008, 3, 126–127. [Google Scholar]
- Crişan, A.; Sitar, C.; Craioveanu, M.C.; Vizauer, T.C.; Rakosy, L. Multiannual population size estimates and mobility of the endemic Pseudophilotes bavius hungarica (Lepidoptera: Lycaenidae) from Transylvania (Romania). North-West. J. Zool. 2014, 10, 115–124. [Google Scholar]
- Nowicki, P.; Deoniziak, K.; Dziekańska, I.; Kostro-Ambroziak, A.; Plazio, E.; Rutkowski, R.; Sielezniew, M. What keeps “living dead” alive: Demography of a small and isolated population of Maculinea (=Phengaris) alcon. J. Insect Conserv. 2019, 23, 291–300. [Google Scholar] [CrossRef]
- Bergman, K.-O.; Landin, J. Population structure and movements of a threatened butterfly (Lopinga achine) in a fragmented landscape in Sweden. Biol. Conserv. 2002, 108, 361–369. [Google Scholar] [CrossRef]
- Sielezniew, M.; Rutkowski, R.; Ponikwicka-Tyszko, D.; Ratkiewicz, M.; Dziekanska, I.; Švitra, G. Differences in genetic variability between two ecotypes of the endangered myrmecophilous butterfly Phengaris (=Maculinea) alcon- the setting of conservation priorities. Insect Conserv. Divers. 2012, 5, 223–236. [Google Scholar] [CrossRef]
- Ritter, S.; Michalski, S.G.; Settele, J.; Wiemers, M.; Fric, Z.F.; Sielezniew, M.; Šašić, M.; Rozier, Y.; Durka, W. Wolbachia infections mimic cryptic speciation in two parasitic butterfly species, Phengaris teleius and P. nausithous (Lepidoptera: Lycaenidae). PLoS ONE 2013, 8, e78107. [Google Scholar] [CrossRef]
- Andersen, A.; Simcox, D.J.; Thomas, J.A.; Nash, D.R. Assessing reintroduction schemes by comparing genetic diversity of reintroduced and source populations: A case study of the globally threatened large blue butterfly (Maculinea arion). Biol. Conserv. 2014, 175, 34–41. [Google Scholar] [CrossRef]
- Sielezniew, M.; Patricelli, D.; Rutkowski, R.; Witek, M.; Bonelli, S.; Buś, M.M. Population genetics of the endangered obligatorily myrmecophilous butterfly Phengaris (=Maculinea) arion in two areas of its European range. Insect Conserv. Diver. 2015, 8, 505–516. [Google Scholar] [CrossRef]
- Bereczki, J.; Póliska, S.; Váradi, A.; Tóth, J.P. Incipient sympatric speciation via host race formation in Phengaris arion (Lepidoptera: Lycaenidae). Org. Divers. Evol. 2020, 20, 63–76. [Google Scholar] [CrossRef]
- Nève, G. Population genetics of butterflies. In Ecology of Butterflies in Europe; Settele, J., Konvička, M., Shreeve, T.G., Van Dyck, H., Eds.; Cambridge University Press: Cambridge, UK, 2009; pp. 107–129. [Google Scholar]
- Vanden Broeck, A.; Maes, D.; Kelager, A.; Wynhoff, I.; WallisDeVries, M.F.; Nash, D.R.; Oostermeijer, J.G.B.; Van Dyck, H.; Mergeay, J. Gene flow and effective population sizes of the butterfly Maculinea alcon in a highly fragmented anthropogenic landscape. Biol. Conserv. 2017, 209, 89–97. [Google Scholar] [CrossRef]
- Sielezniew, M.; Rutkowski, R. Population isolation rather than ecological variation explains the genetic structure of endangered myrmecophilous butterfly Phengaris (=Maculinea) arion. J. Insect Conserv. 2012, 16, 39–50. [Google Scholar] [CrossRef]
- Sucháčková Bartoňová, A.; Konvička, M.; Marešová, J.; Kolev, Z.; Wahlberg, N.; Fric, Z.F. Recently lost connectivity in the Western Palaearctic steppes: The case of a scarce specialist butterfly. Conserv. Genet. 2020, 21, 561–575. [Google Scholar] [CrossRef]
- Orsini, L.; Jorander, J.; Alasentie, A.; Hanski, I. Genetic spatial structure in a butterfly metapopulation correlates better with past than present demographic structure. Mol. Ecol. 2008, 17, 2629–2642. [Google Scholar] [CrossRef]
- Mace, G.M.; Collar, N.J.; Gaston, K.J.; Hilton-Taylor, C.; Akc¸akaya, H.R.; Leader-Williams, N.; Milner-Gulland, E.J.; Stuart, S.N. Quantification of extinction risk: IUCN’s system for classifying threatened species. Conserv. Biol. 2008, 6, 1424–1442. [Google Scholar] [CrossRef]
- Dennis, R.L.H.; Shreeve, T.G. Butterfly wing morphology variation in the British Isles: The influence of climate, behavioural posture and the hostplant-habitat. Biol. J. Linn. Soc. 1989, 38, 323–348. [Google Scholar] [CrossRef]
- Sielezniew, M.; Dziekańska, I. Geographical variation in wing pattern in Phengaris (=Maculinea) arion (L.) (Lepidoptera: Lycaenidae): Subspecific differentiation or clinal adaptation? Ann. Zool. 2011, 61, 739–750. [Google Scholar] [CrossRef]
- Kajtoch, Ł.; Cieślak, E.; Varga, Z.; Paul, W.; Mazur, M.A.; Sramkó, G.; Kubisz, D. Phylogeographic patterns of steppe species in Eastern Central Europe: A review and the implications for conservation. Biodivers. Conserv. 2016, 25, 2309–2339. [Google Scholar] [CrossRef]
- Wahlberg, N.; Saccheri, I. The effects of Pleistocene glaciations on the phylogeography of Melitaea cinxia (Lepidoptera: Nymphalidae). Eur. J. Entomol. 2007, 104, 675–684. [Google Scholar] [CrossRef]
- Sielezniew, M.; Ponikwicka, D.; Ratkiewicz, M.; Rutkowski, R.; Dziekańska, I.; Kostro-Ambroziak, A. Diverging patterns of mitochondrial and nuclear diversity in the specialized butterfly Plebejus argus (Lepidoptera: Lycaenidae). Eur. J. Entomol. 2011, 108, 537–545. [Google Scholar] [CrossRef]
- Marešová, J.; Habel, J.C.; Neve, G.; Sielezniew, M.; Bartoňová, A.; Kostro-Ambroziak, A.; Fric, Z.F. Cross-continental phylogeography of two Holarctic Nymphalid butterflies, Boloria eunomia and Boloria selene. PLoS ONE 2019, 14, e0214483. [Google Scholar] [CrossRef]
- Kajtoch, Ł.; Kubisz, D.; Gutowski, J.M.; Babik, W. Evolutionary units of Coraebus elatus (Coleoptera: Buprestidae) in central and eastern Europe—Implications for origin and conservation. Insect Conserv. Divers. 2014, 7, 41–54. [Google Scholar] [CrossRef]
- Banaszek, A.; Jadwiszczak, K.A.; Ratkiewicz, M.; Ziomek, J. Low genetic diversity and significant structuring of the common hamster populations Cricetus cricetus in Poland revealed by the mtDNA control region sequence variation. Acta Theriol. 2009, 54, 289–295. [Google Scholar] [CrossRef]
- Patricelli, D.; Sielezniew, M.; Ponikwicka-Tyszko, D.; Ratkiewicz, M.; Bonelli, S.; Barbero, F.; Witek, M.; Buś, M.M.; Rutkowski, R.; Balletto, E. Contrasting genetic structure of rear edge and continuous range populations of a parasitic butterfly infected by Wolbachia. BMC Evol. Biol. 2013, 13, 14. [Google Scholar] [CrossRef]
| Locus | References | Dye | Mix | No. of Cycles | µM | Size Range | Fnull # | |
|---|---|---|---|---|---|---|---|---|
| Macu15 * | [30] | 6-FAM | SorI (1) | 30 | 0.3 | 146 | ND | ND |
| Macu44 | [31] | VIC | SorI (1) | 0.3 | 180–186 | 0.013 | 0.013 | |
| boleun01 | [32] | NED | SorI (1) | 1.8 | 144–188 | −0.191 | −0.311 | |
| Macu5 | [30] | 6-FAM | SorII (2) | 40 | 0.3 | 174–240 | −0.057 | −0.063 |
| Macu40* | [33] | NED | SorII (2) | 0.3 | 85–167 | 0.213 | 0.169 | |
| Macu16 | [30] | VIC | SorIII (3) | 45 | 0.3 | 217–303 | −0.018 | −0.054 |
| Lb4/18 * | [34] | PET | SorIII (3) | 0.3 | 201–287 | 0.119 | 0.091 | |
| Malc169 | [30] | 6-FAM | SorIII (3) | 0.3 | 248–332 | 0.030 | 0.029 | |
| Macu9 * | [30] | NED | SorIII (3) | 0.3 | 181 | ND | ND | |
| Macari16 * | [35] | VIC | SorIV (4) | 45 | 0.3 | 80–228 | 0.082 | 0.073 |
| Macari18 | [35] | PET | SorIV (4) | 0.3 | 75–157 | −0.122 | −0.150 | |
| Macu29 * | [33] | VIC | 45 | 0.3 | 91–217 | 0.190 | 0.130 | |
| Population | Year | Number of Marked Butterflies | Estimated Population Size N ± SE (95% CI) | |||
|---|---|---|---|---|---|---|
| M | F | M | F | All | ||
| P1 | 2015 | 6 | 13 | 8 ± 3 (6–22) | 15 ± 3 (13–29) | 23 ± 4 (20–39) |
| 2016 | 31 | 20 | 48 ± 6 (40–63) | 37 ± 6 (29–52) | 85 ± 8 (66–90) | |
| P2 | 2015 | 23 | 13 | 40 ± 6 (32–55) | 30 ± 6 (22–45) | 70 ± 8 (57–90) |
| 2016 | 47 | 27 | 92 ± 10 (76–116) | 72 ± 10 (56–96) | 164 ± 14 (138–200) | |
| Population | Year | Sex | Survival Rate φ ± SE (95% CI) | Mean Lifespan (Days) e ± SE (95% CI) |
|---|---|---|---|---|
| P1 | 2015 | All | 0.84 ± 0.04 (0.75–0.91) | 5.93 ± 0.11 (3.49–10.35) |
| 2016 | M | 0.91 ± 0.04 (0.81–0.96) | 10.68 ± 0.04 (4.82–24.49) | |
| 2016 | F | 0.87 ± 0.06 (0.71–0.95) | 7.39 ± 0.06 (2.94–19.99) | |
| P2 | 2015 | M | 0.87 ± 0.04 (0.76–0.93) | 6.93 ± 0.04 (3.72–13.34) |
| 2015 | F | 0.73 ± 0.12 (0.45–0.90) | 3.17 ± 0.12 (1.33–9.04) | |
| 2016 | All | 0.85 ± 0.03 (0.78–0.90) | 6.19 ± 0.03 (4.00–9.74) |
| Population | All (N = 65) | P1 (N = 34) | P2 (N = 31) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | A | HE | HO | PIC | A | HE | HO | PIC | PHW | A | HE | HO | PIC | PHW |
| Macu44 | 4 | 0.54 | 0.52 | 0.44 | 2 | 0.51 | 0.53 | 0.37 | 1.000 | 4 | 0.56 | 0.52 | 0.49 | 0.668 |
| boleun01 | 6 | 0.53 | 0.74 | 0.46 | 4 | 0.54 | 0.79 | 0.44 | 0.003 | 5 | 0.52 | 0.68 | 0.47 | 0.347 |
| Macu5 | 12 | 0.55 | 0.57 | 0.52 | 7 | 0.58 | 0.71 | 0.52 | 0.271 | 9 | 0.51 | 0.42 | 0.49 | 0.198 |
| Macu16 | 6 | 0.11 | 0.11 | 0.10 | 5 | 0.14 | 0.15 | 0.14 | 1.000 | 3 | 0.06 | 0.07 | 0.06 | 1.000 |
| Malc169 | 12 | 0.67 | 0.59 | 0.65 | 10 | 0.68 | 0.58 | 0.64 | 0.015 | 8 | 0.64 | 0.61 | 0.60 | 0.107 |
| Macari18 | 7 | 0.61 | 0.74 | 0.54 | 7 | 0.66 | 0.77 | 0.60 | 0.132 | 5 | 0.56 | 0.71 | 0.47 | 0.316 |
| Overall | 7.83 | 0.50 | 0.55 | 0.45 | 5.83 | 0.52 | 0.59 | 0.45 | 5.67 | 0.48 | 0.50 | 0.43 | ||
| Nh | h (SE) | π (SE) | S | |||||
|---|---|---|---|---|---|---|---|---|
| Population | P1 | P2 | P1 | P2 | P1 | P2 | P1 | P2 |
| COI 632 bp | 2 | 1 | 0.166 (0.080) | 0 | 0.001 (0.001) | 0 | 4 | 0 |
| ND5 570 bp | 2 | 2 | 0.121 (0.075) | 0.443 (0.069) | 0.0002 (0.0004) | 0.0008 (0.0008) | 1 | 1 |
| COI_ND5 1203 bp | 3 | 2 | 0.234 (0.095) | 0.443 (0.069) | 0.0005 (0.0005) | 0.0004 (0.0004) | 5 | 1 |
| EF-1α 573 bp | 4 | 2 | 0.118 (0.054) | 0.259 (0.065) | 0.0004 (0.0005) | 0.0005 (0.0006) | 4 | 1 |
| Wgl 338 bp | 3 | 2 | 0.536 (0.024) | 0.474 (0.033) | 0.002 (0.002) | 0.001 (0.001) | 2 | 1 |
| EF-1α_Wgl 965 bp | 5 | 4 | 0.568 (0.029) | 0.624 (0.034) | 0.0008 (0.0007) | 0.0008 (0.0006) | 6 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czajkowska, M.; Dawidowicz, Ł.; Borkowska, A.; Dziekańska, I.; Sielezniew, M. Population Genetic Structure and Demography of the Critically Endangered Chequered Blue Butterfly (Scolitantides orion) in a Highly Isolated Part of Its Distribution Range. Insects 2020, 11, 608. https://doi.org/10.3390/insects11090608
Czajkowska M, Dawidowicz Ł, Borkowska A, Dziekańska I, Sielezniew M. Population Genetic Structure and Demography of the Critically Endangered Chequered Blue Butterfly (Scolitantides orion) in a Highly Isolated Part of Its Distribution Range. Insects. 2020; 11(9):608. https://doi.org/10.3390/insects11090608
Chicago/Turabian StyleCzajkowska, Magdalena, Łukasz Dawidowicz, Anetta Borkowska, Izabela Dziekańska, and Marcin Sielezniew. 2020. "Population Genetic Structure and Demography of the Critically Endangered Chequered Blue Butterfly (Scolitantides orion) in a Highly Isolated Part of Its Distribution Range" Insects 11, no. 9: 608. https://doi.org/10.3390/insects11090608
APA StyleCzajkowska, M., Dawidowicz, Ł., Borkowska, A., Dziekańska, I., & Sielezniew, M. (2020). Population Genetic Structure and Demography of the Critically Endangered Chequered Blue Butterfly (Scolitantides orion) in a Highly Isolated Part of Its Distribution Range. Insects, 11(9), 608. https://doi.org/10.3390/insects11090608





