Bacteria Belonging to Pseudomonas typographi sp. nov. from the Bark Beetle Ips typographus Have Genomic Potential to Aid in the Host Ecology
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation
2.2. DNA Extraction and Bacterial Identification
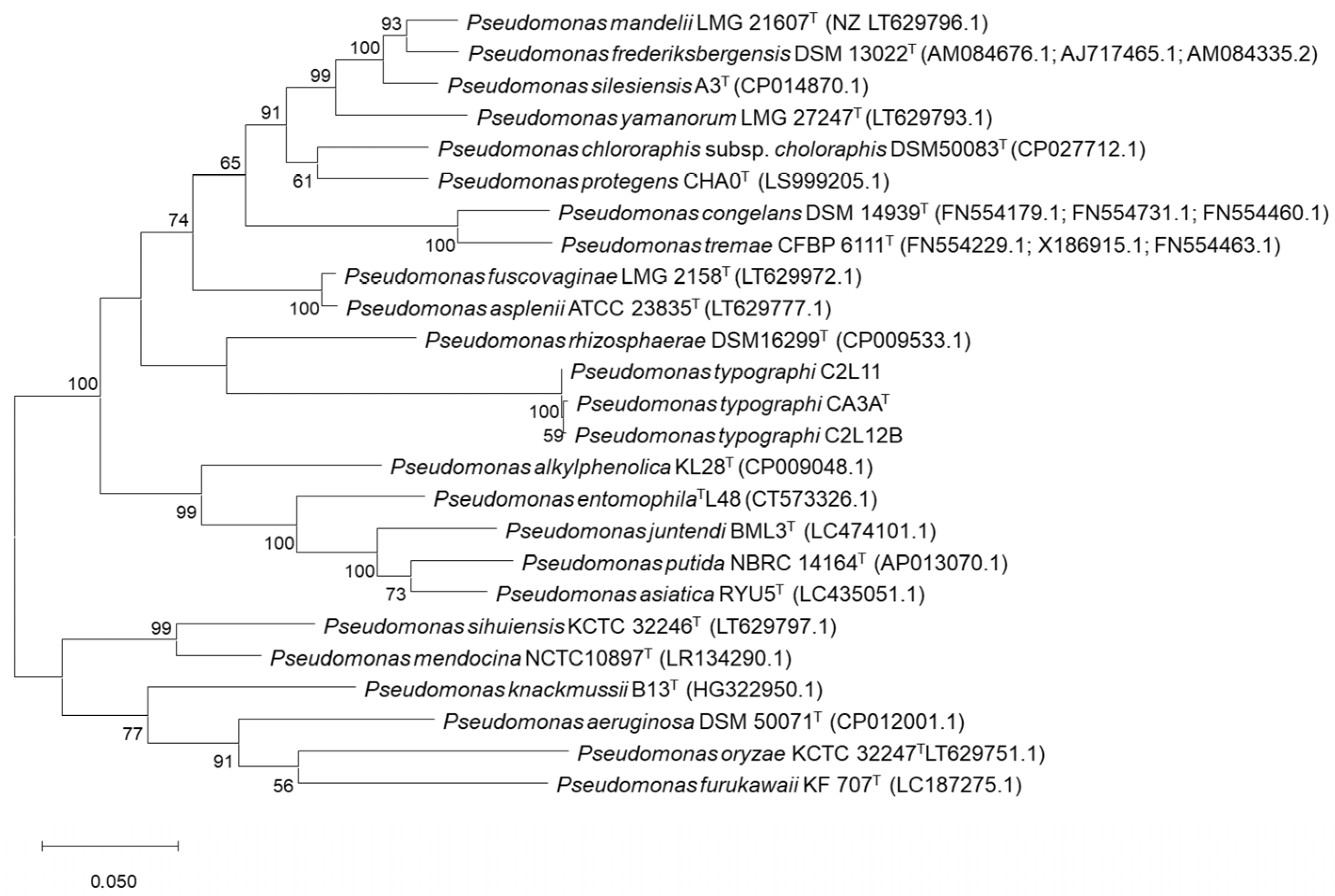
2.3. Phylogenetic Analyses
2.4. Genomic Analyses
2.5. Lignocellulose-Related Activity
2.6. Antimicrobial Potential
2.7. Bacterial Characterization
3. Results
3.1. Bacterial Identification
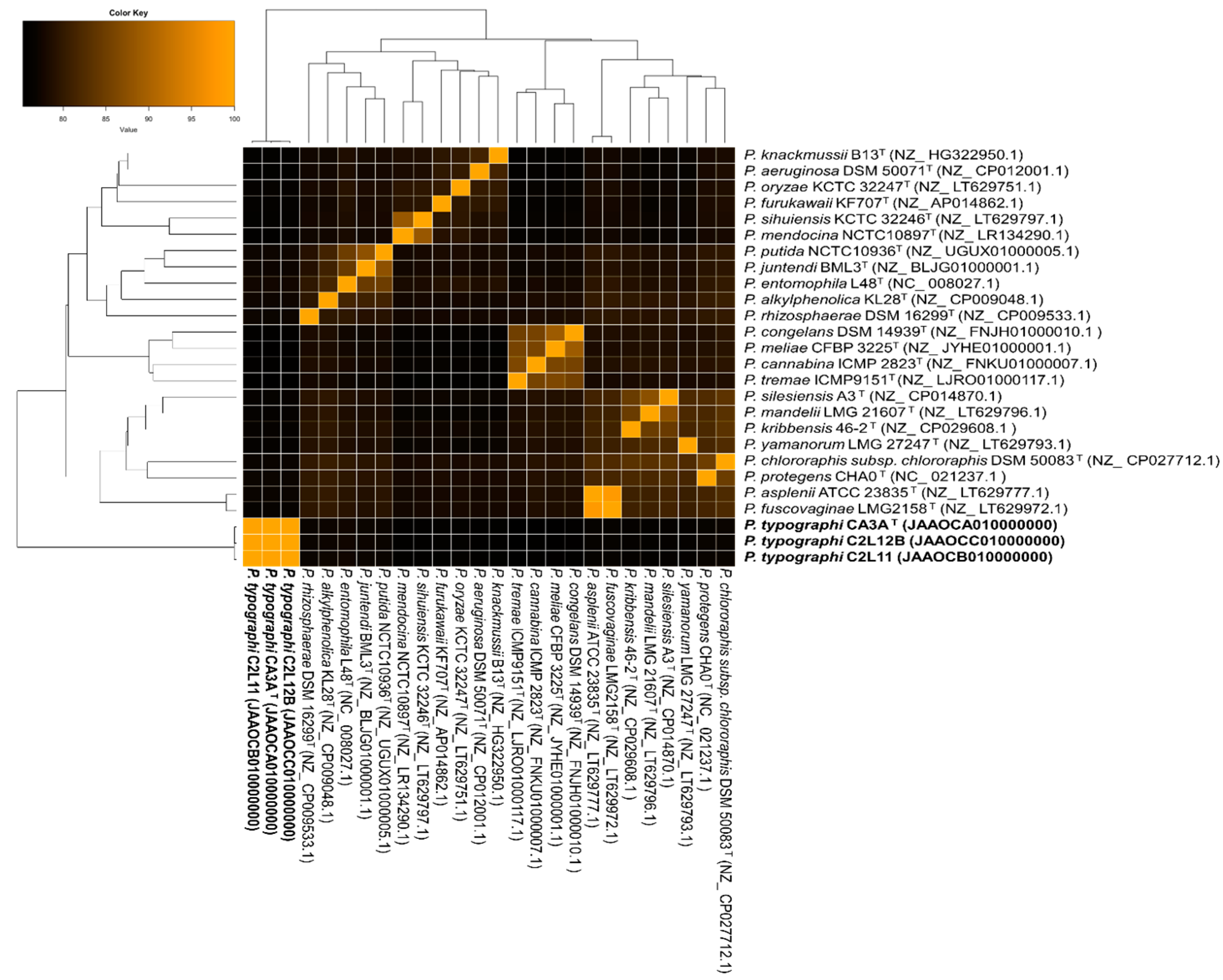
3.2. Genome Analysis
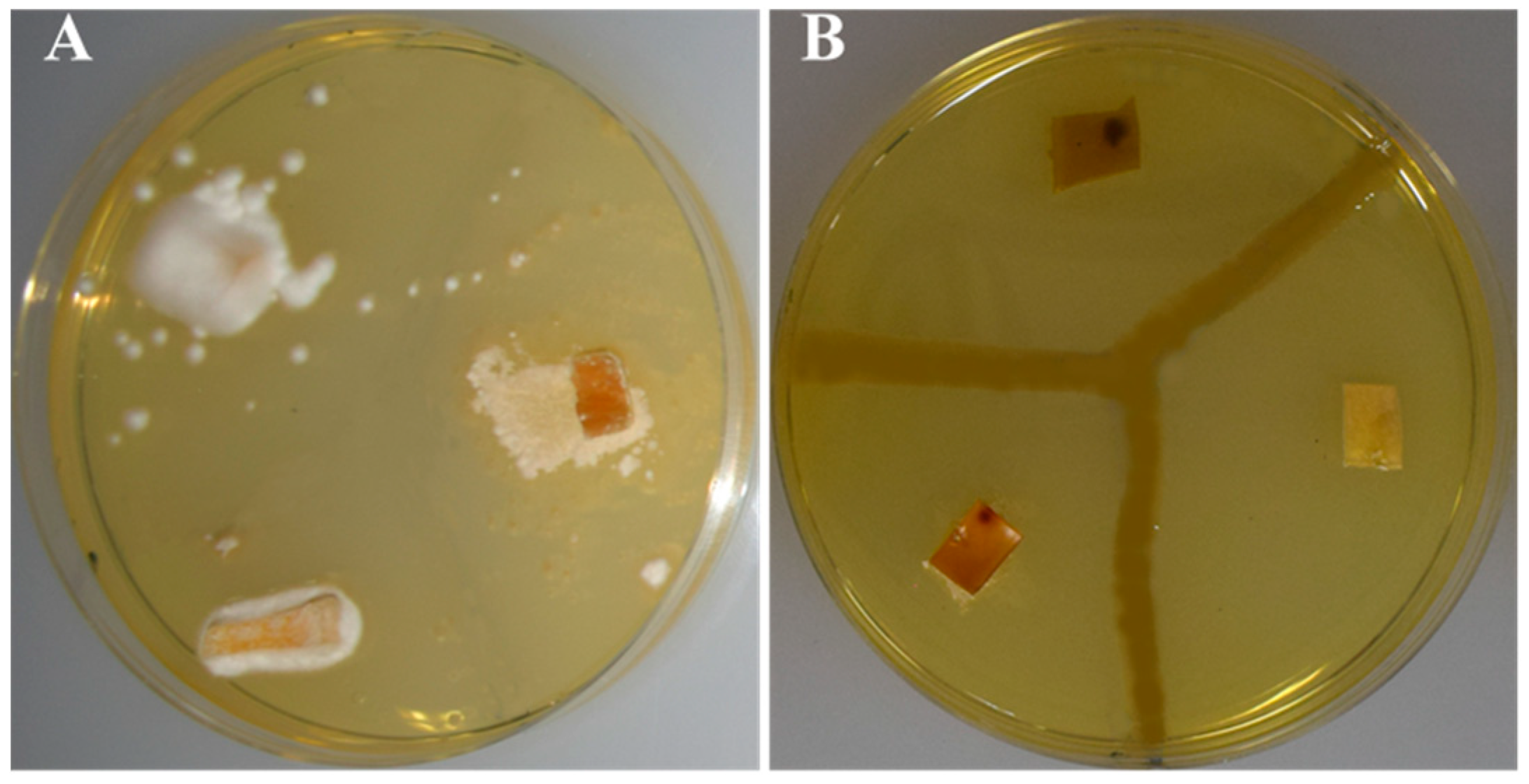
3.3. In Vitro Assays of Catalytic Reactions
3.4. Antibiosis Potential
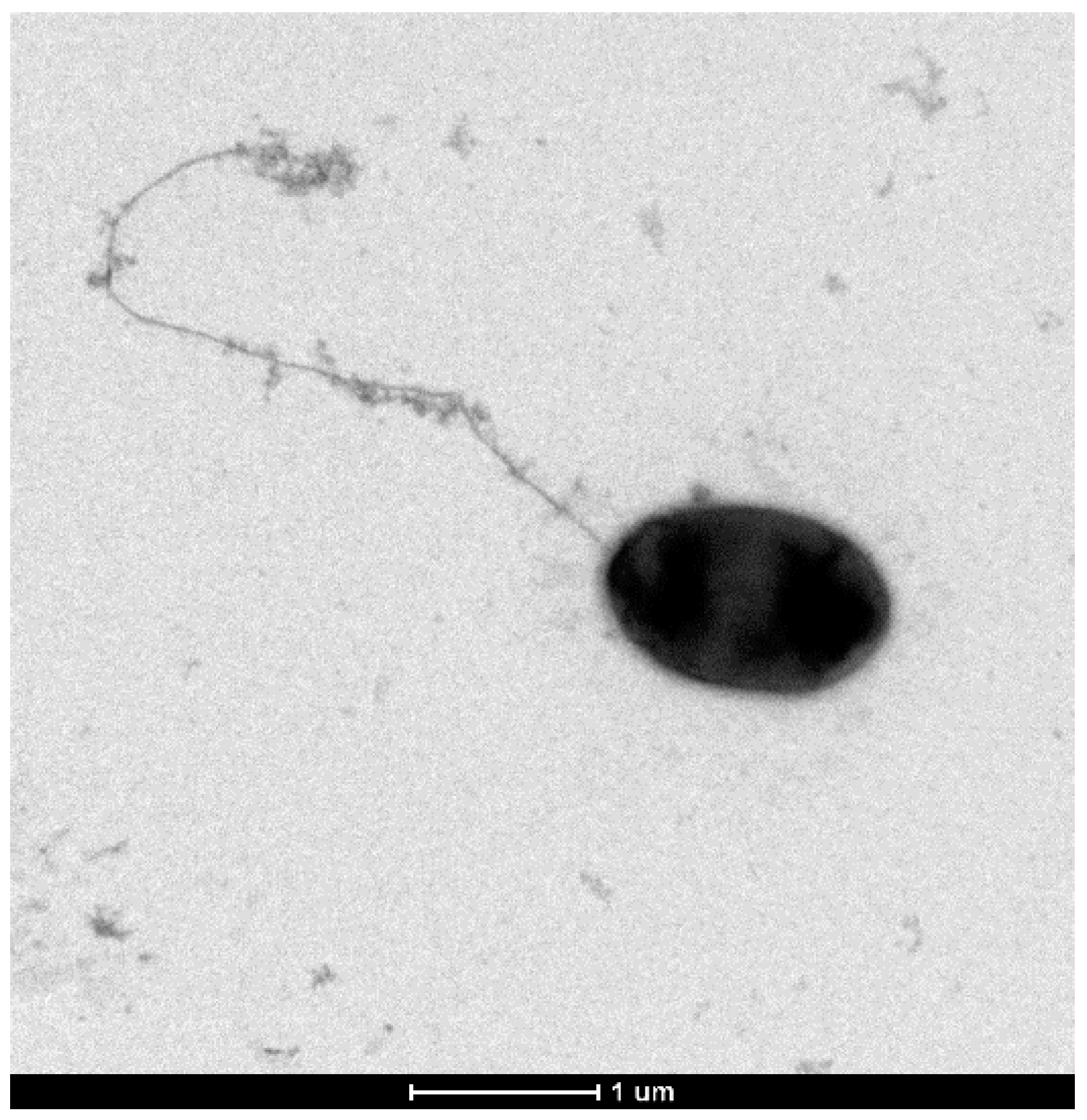
3.5. Other Phenotypic Traits of P. typographi
4. Discussion
5. Conclusions
Description of Pseudomonas typographi sp. nov.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- García-Fraile, P. Roles of bacteria in the bark beetle holobiont—How do they shape this forest pest? Ann. Appl. Biol. 2018, 172. [Google Scholar] [CrossRef]
- Six, D.L. The Bark Beetle Holobiont: Why Microbes Matter. J. Chem. Ecol. 2013, 39, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Species Profile Ips Typographus. Available online: http://www.iucngisd.org/gisd/species.php?sc=1441 (accessed on 3 July 2020).
- Fabryová, A.; Kostovčík, M.; Díez-Méndez, A.; Jiménez-Gómez, A.; Celador-Lera, L.; Saati-Santamaría, Z.; Sechovcová, H.; Menéndez, E.; Kolařik, M.; García-Fraile, P. On the bright side of a forest pest-the metabolic potential of bark beetles’ bacterial associates. Sci. Total Environ. 2018, 619–620, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wickham, J.D.; Chen, L.; Xu, D.; Lu, M.; Sun, J. Bacterial microbiota protect an invasive bark beetle from a pine defensive compound. Microbiome 2018, 6. [Google Scholar] [CrossRef]
- Boone, C.K.; Keefover-Ring, K.; Mapes, A.C.; Adams, A.S.; Bohlmann, J.; Raffa, K.F. Bacteria Associated with a Tree-Killing Insect Reduce Concentrations of Plant Defense Compounds. J. Chem. Ecol. 2013, 39, 1003–1006. [Google Scholar] [CrossRef]
- Mattanovich, J.; Ehrenhofer, M.; Schafellner, C.; Tausz, M.; Fuhrer, E. The role of sulphur compounds for breeding success of Ips typographus L. (Col., Scolytidae) on Norway Spruce (Picea abies [L.] Karst.). J. Appl. Entomol. 2001, 125, 425–431. [Google Scholar] [CrossRef]
- Bozzano, L. Insect-Fungus Interactions, 1st ed.; Wilding, N., Collins, N.M., Hammond, P.M., Webber, J.F., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 1988. [Google Scholar]
- Morales-Jiménez, J.; de León, A.V.-P.; García-Domínguez, A.; Martínez-Romero, E.; Zúñiga, G.; Hernández-Rodríguez, C. Nitrogen-Fixing and Uricolytic Bacteria Associated with the Gut of Dendroctonus rhizophagus and Dendroctonus valens (Curculionidae: Scolytinae). Microb. Ecol. 2013, 66, 200–210. [Google Scholar] [CrossRef]
- Wegensteiner, R.; Weiser, J.; Führer, E. Observations on the occurrence of pathogens in the bark beetle Ips typographus L. (Col., Scolytidae). J. Appl. Entomol. 1996, 120, 199–204. [Google Scholar] [CrossRef]
- Wegensteiner, R.; Weiser, J. Annual variation of pathogen occurrence and pathogen prevalence in Ips typographus (Coleoptera, Scolytidae) from the BOKU University Forest Demonstration Centre. J. Pest Sci. 2004, 77, 221–228. [Google Scholar] [CrossRef]
- Takov, D.; Pilarska, D.; Wegensteiner, R. Entomopathogens in Ips typographus (Coleoptera: Scolytidae) from Several Spruce Stands in Bulgaria. Acta Zool. Bulg. 2006, 58, 409–420. [Google Scholar]
- Saati-Santamaría, Z.; López-Mondéjar, R.; Jiménez-Gómez, A.; Díez-Méndez, A.; Vetrovský, T.; Igual, J.M.; Velázquez, E.; Kolarik, M.; Rivas, R.; García-Fraile, P. Discovery of phloeophagus beetles as a source of pseudomonas strains that produce potentially new bioactive substances and description of pseudomonas bohemica sp. nov. Front. Microbiol. 2018, 9, 913. [Google Scholar] [CrossRef] [PubMed]
- Kostas, B.; Thomas, A.M. Insect Symbiosis; Kostas, B., Thomas, A.M., Eds.; CRC Press: Boca Raton, FL, USA, 2003; Volume 3. [Google Scholar]
- Morales-Jiménez, J.; Zúñiga, G.; Ramírez-Saad, H.C.; Hernández-Rodríguez, C. Gut-Associated Bacteria Throughout the Life Cycle of the Bark Beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and Their Cellulolytic Activities. Microb. Ecol. 2012, 64, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M. On the Multitrophic Interactions between Ips Typographus Their Tree Host, Associated Microorganisms, and a Predatory Medetera Fly; Institutionen för Växtskyddsbiologi, Sveriges Lantbruksuniversitet: Upsala, Sweden, 2019. [Google Scholar]
- Skrodenytee-Arbaciauskiene, V.; Radziute, S.; Stunzenas, V.; Buda, V. Erwinia typographi sp. nov., isolated from bark beetle (Ips typographus) gut. Int. J. Syst. Evol. Microbiol. 2012, 62, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 2009, 9, 1132–1147. [Google Scholar] [CrossRef] [PubMed]
- Marek-Kozaczuk, M.; Skorupska, A. Production of B-group vitamins by plant growth-promoting Pseudomonas fluorescens strain 267 and the importance of vitamins in the colonization and nodulation of red clover. Biol. Fertil. Soils 2001, 33, 146–151. [Google Scholar] [CrossRef]
- Rivas, R.; García-Fraile, P.; Mateos, P.F.; Martínez-Molina, E.; Velázquez, E. Characterization of xylanolytic bacteria present in the bract phyllosphere of the date palm Phoenix dactylifera. Lett. Appl. Microbiol. 2007, 44, 181–187. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and análisis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16s rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. gplots: Various R Programming Tools for Plotting Data. R Package Version 3.0.4. Available online: https://CRAN.R-project.org/package=gplots (accessed on 2 September 2020).
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Le Normand, M.; Edlund, U.; Holmbom, B.; Ek, M. Hot-water extraction and characterization of spruce bark non-cellulosic polysaccharides. Nord. Pulp Pap. Res. J. 2012, 27, 18–23. [Google Scholar] [CrossRef]
- BioFuel Region: Basic Chemical composition of the biomass components of pine, spruce and birch. Available online: http://biofuelregion.se/wp-content/uploads/2017/01/1_2_IS_2013-01-31_Basic_chemical_composition.pdf (accessed on 2 September 2020).
- Mateos, P.F.; Jimenez-Zurdo, J.I.; Chen, J.; Squartini, A.S.; Haack, S.K.; Martinez-Molina, E.; Hubbell, D.H.; Dazzo, F.B. Cell-associated pectinolytic and cellulolytic enzymes in Rhizobium leguminosarum biovar trifolii. Appl. Environ. Microbiol. 1992, 58, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- García-Fraile, P.; Rivas, R.; Willems, A.; Peix, A.; Martens, M.; Martínez-Molina, E.; Mateos, P.F.; Velázquez, E. Rhizobium cellulosilyticum sp. nov., isolated from sawdust of Populus alba. Int. J. Syst. Evol. Microbiol. 2007. [Google Scholar] [CrossRef]
- Kubátová, A.; Dvořák, L. Entomopathogenic fungi associated with insect hibernating in underground shelters The distribution of scorpion flies of the genus Panorpa in the West Palaearctic View project Libor Dvořák Municipal Museum Mariánské Lázně, Czech Republic. Czech Mycol. 2005, 57, 221–237. [Google Scholar] [CrossRef]
- Pažoutová, S.; Šrůtka, P.; Holuša, J.; Chudíčková, M.; Kolařík, M. Diversity of xylariaceous symbionts in Xiphydria woodwasps: Role of vector and a host tree. Fungal Ecol. 2010, 3, 392–401. [Google Scholar] [CrossRef]
- Wegensteiner, R.; Wermelinger, B.; Herrmann, M. Natural enemies of bark beetles: Predators, parasitoids, pathogens, and nematodes. In Bark Beetles: Biology and Ecology of Native and Invasive Species; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 247–304. ISBN 9780124171732. [Google Scholar]
- Jiménez-Gómez, A.; Saati-Santamaría, Z.; Igual, J.M.; Rivas, R.; Mateos, P.F.; García-Fraile, P. Genome insights into the novel species Microvirga brassicacearum, a rapeseed endophyte with biotechnological potential. Microorganisms 2019, 7, 354. [Google Scholar] [CrossRef]
- Doetsch, R.N. Determinative methods of light microscopy. In Manual of Methods for General Bacteriology; Gerdhardt, P., Murray, R.G.E., Costilow, R.N., Nester, E.W., Wood, W.A., Krieg, N.R., Phillips, G.B., Eds.; American Society for Microbiology: Washington, DC, USA, 1981; pp. 21–33. [Google Scholar]
- García-Fraile, P.; Chudíčková, M.; Benada, O.; Pikula, J.; Kolařík, M. Serratia myotis sp. nov. and Serratia vespertilionis sp. nov., isolated from bats hibernating in caves. Int. J. Syst. Evol. Microbiol. 2015, 65, 90–94. [Google Scholar] [CrossRef]
- Kovacs, N. Identification of Pseudomonas pyocyanea by the Oxidase Reaction. Nature 1956, 178, 703. [Google Scholar] [CrossRef]
- Ramírez-Bahena, M.-H.; Cuesta, M.J.; Flores-Félix, J.D.; Mulas, R.; Rivas, R.; Castro-Pinto, J.; Brañas, J.; Mulas, D.; González-Andrés, F.; Velázquez, E.; et al. Pseudomonas helmanticensis sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 2338–2345. [Google Scholar] [CrossRef]
- Ait Tayeb, L.; Ageron, E.; Grimont, F.; Grimont, P.A.D. Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Microbiol. Res. 2005, 156, 763–773. [Google Scholar] [CrossRef]
- Mulet, M.; Bennasar, A.; Lalucat, J.; García-Valdés, E. An rpoD-based PCR procedure for the identification of Pseudomonas species and for their detection in environmental samples. Mol. Cell. Probes 2009, 23, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Mulet, M.; Gomila, M.; Lemaitre, B.; Lalucat, J.; García-Valdés, E. Taxonomic characterisation of Pseudomonas strain L48 and formal proposal of Pseudomonas entomophila sp. nov. Syst. Appl. Microbiol. 2012, 35, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.; Ramírez-Bahena, M.-H.; Valverde, A.; Velázquez, E.; Zúñiga, D.; Velezmoro, C.; Peix, A. Pseudomonas punonensis sp. nov., isolated from straw. Int. J. Syst. Evol. Microbiol. 2013, 63, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.; Ramírez-Bahena, M.-H.; Cuesta, M.J.; Velázquez, E.; Peix, A. Pseudomonas guariconensis sp. nov., isolated from rhizospheric soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 4413–4420. [Google Scholar] [CrossRef]
- Menéndez, E.; Ramírez-Bahena, M.H.; Fabryová, A.; Igual, J.M.; Benada, O.; Mateos, P.F.; Peix, A.; Kolařík, M.; García-Fraile, P. Pseudomonas coleopterorum sp. nov., a cellulase-producing bacterium isolated from the bark beetle Hylesinus fraxini. Int. J. Syst. Evol. Microbiol. 2015, 65, 2852–2858. [Google Scholar] [CrossRef]
- Peix, A. Pseudomonas rhizosphaerae sp. nov., a novel species that actively solubilizes phosphate in vitro. Int. J. Syst. Evol. Microbiol. 2003, 53, 2067–2072. [Google Scholar] [CrossRef]
- Mulet, M.; Sánchez, D.; Lalucat, J.; Lee, K.; García-Valdés, E. Pseudomonas alkylphenolica sp. nov., a bacterial species able to form special aerial structures when grown on p-cresol. Int. J. Syst. Evol. Microbiol. 2015, 65, 4013–4018. [Google Scholar] [CrossRef]
- Behrendt, U.; Ulrich, A.; Schumann, P. Fluorescent pseudomonads associated with the phyllosphere of grasses; Pseudomonas trivialis sp. nov., Pseudomonas poae sp. nov. and Pseudomonas congelans sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1461–1469. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Menendez, E.; Garcia-Fraile, P.; Rivas, R. Biotechnological applications of bacterial cellulases. AIMS Bioeng. 2015, 2, 163–182. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Guo, P.; Wang, T.; Gao, L.; Yin, H.; Cai, C.; Gu, J.; Lü, X. Metagenomic mining pectinolytic microbes and enzymes from an apple pomace-adapted compost microbial community. Biotechnol. Biofuels 2017, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bertoldo, C.; Antranikian, G. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 2002, 6, 151–160. [Google Scholar] [CrossRef]
- Boller, T. Antimicrobial functions of the plant hydrolases, chitinase and ß-1,3-glucanase. Developments in Plant Pathology. In Mechanisms of Plant Defense Responses; Fritig, B., Legrand, M., Eds.; Springer: Dordrecht, The Netherlands, 1993; pp. 391–400. ISBN 978-94-011-1737-1. [Google Scholar]
- Arora, N.K.; Khare, E.; Oh, J.H.; Kang, S.C.; Maheshwari, D.K. Diverse mechanisms adopted by fluorescent Pseudomonas PGC2 during the inhibition of Rhizoctonia solani and Phytophthora capsici. World J. Microbiol. Biotechnol. 2008, 24, 581–585. [Google Scholar] [CrossRef]
- Rojas Murcia, N.; Lee, X.; Waridel, P.; Maspoli, A.; Imker, H.J.; Chai, T.; Walsh, C.T.; Reimmann, C. The Pseudomonas aeruginosa antimetabolite L -2-amino-4-methoxy-trans-3-butenoic acid (AMB) is made from glutamate and two alanine residues via a thiotemplate-linked tripeptide precursor. Front. Microbiol. 2015, 6, 170. [Google Scholar] [CrossRef]
- Baars, O.; Zhang, X.; Gibson, M.I.; Stone, A.T.; Morel, F.M.M.; Seyedsayamdost, M.R. Crochelins: Siderophores with an Unprecedented Iron-Chelating Moiety from the Nitrogen-Fixing Bacterium Azotobacter chroococcum. Angew. Chem. Int. Ed. Engl. 2018, 57, 536–541. [Google Scholar] [CrossRef]
- Lee, X.; Reimmann, C.; Greub, G.; Sufrin, J.; Croxatto, A. The Pseudomonas aeruginosa toxin L-2-amino-4-methoxy-trans-3-butenoic acid inhibits growth and induces encystment in Acanthamoeba castellanii. Microbes Infect. 2012, 14, 268–272. [Google Scholar] [CrossRef]
- Sulochana, M.B.; Jayachandra, S.Y.; Kumar, S.K.A.; Dayanand, A. Antifungal attributes of siderophore produced by the Pseudomonas aeruginosa JAS-25. J. Basic Microbiol. 2013, 54, 418–424. [Google Scholar] [CrossRef]
- Cvikrová, M.; Malá, J.; Hrubcová, M.; Eder, J. Soluble and cell wall-bound phenolics and lignin in Ascocalyx abietina infected Norway spruces. Plant Sci. 2006, 170, 563–570. [Google Scholar] [CrossRef]
- Adams, A.S.; Aylward, F.O.; Adams, S.M.; Erbilgin, N.; Aukema, B.H.; Currie, C.R.; Suen, G.; Raffa, K.F. Mountain pine beetles colonizing historical and naïve host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl. Environ. Microbiol. 2013, 79, 3468–3475. [Google Scholar] [CrossRef] [PubMed]
- Sagot, B.; Gaysinski, M.; Mehiri, M.; Guigonis, J.-M.; Le Rudulier, D.; Alloing, G. Osmotically induced synthesis of the dipeptide N-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 12652–12657. [Google Scholar] [CrossRef] [PubMed]
- Sedkova, N.; Tao, L.; Rouvière, P.E.; Cheng, Q. Diversity of Carotenoid Synthesis Gene Clusters from Environmental Enterobacteriaceae Strains. Appl. Environ. Microbiol. 2005, 71, 8141–8146. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Moran, N.A. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 2012, 8, 986–989. [Google Scholar] [CrossRef]

| pNP Substrate | Enzyme Tested |
|---|---|
| PNP-α-d-glucopyranoside | α-glucosidase |
| PNP-β-d-glucopyranoside | β-glucosidase |
| PNP-β-d-cellobioside | cellobiohydrolase |
| PNP-α-d-xylopyranoside | α-xylosidase |
| PNP-β-l-xylopyranoside | β-xylosidase |
| Genomes | P. typographi CA3AT (JAAOCA000000000) | P. typographi C2L11 (JAAOCB000000000) | P. typographi C2L12B (JAAOCC000000000) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ANIb | dDDH | G + C Diff. | ANIb | dDDH | G + C Diff. | ANIb | dDDH | G + C Diff. | |
| P. typographi CA3AT (JAAOCA000000000) | - | - | - | 99.6 | 97.7 | 0.12 | 99.7 | 97.7 | 0.16 |
| P. typographi C2L11 (JAAOCB000000000 | - | - | - | - | - | - | 99.7 | 97.7 | 0.16 |
| P. alkylphenolica KL28T (NZ_P009048.1) | 77.2 | 24.1 | 1.98 | 77.3 | 24.2 | 1.86 | 77.3 | 24.1 | 2.02 |
| P. congelans DSM 14939T (NZ_FNJH01000010.1) | 75.7 | 23.0 | 3.28 | 75.7 | 23.1 | 3.16 | 75.7 | 23.0 | 3.32 |
| P. entomophila L48T (NC_008027.1) | 77.6 | 24.6 | 1.56 | 77.5 | 24.6 | 1.67 | 77.6 | 24.5 | 1.51 |
| P. kribbensis 46-2T (NZ_CP029608.1) | 76.5 | 23.5 | 2.06 | 76.5 | 23.5 | 1.94 | 76.5 | 23.5 | 2.10 |
| P. mandelii LMG 21607T (NZ_LT629796.1) | 76.3 | 23.1 | 3.40 | 76.3 | 23.2 | 3.29 | 76.4 | 23.2 | 3.45 |
| P. meliae CFBP 3225T (NZ_JYHE01000001.1) | 75.5 | 22.9 | 4.20 | 75.4 | 23.0 | 4.09 | 75.4 | 23.0 | 4.24 |
| P. rhizosphaerae DSM 16299T (NZ_CP009533.1) | 77.2 | 24.8 | 0.61 | 77.2 | 24.9 | 0.49 | 77.2 | 24.8 | 0.65 |
| P. silesiensis A3T (NZ_CP014870.1) | 76.6 | 23.7 | 3.02 | 76.5 | 23.6 | 2.91 | 76.5 | 23.6 | 3.06 |
| P. tremae ICMP9151T (NZ_LJRO01000117.1) | 75.3 | 22.5 | 4.81 | 75.4 | 22.9 | 4.69 | 75.3 | 22.7 | 4.85 |
| Atribute | Strain (Value) | ||
|---|---|---|---|
| CA3AT | C2L11 | C2L12B | |
| Genome size (bp) | 5,982,847 | 6,069,978 | 5,860,340 |
| G + C content (%) | 62.1 | 62.0 | 62.2 |
| N50 value | 141,379 | 127,151 | 133,313 |
| L50 value | 15 | 16 | 11 |
| Number of contigs (with protein encoding genes) | 229 | 158 | 174 |
| Number of subsystems | 364 | 362 | 364 |
| Number of coding sequences | 5965 | 5954 | 5775 |
| Number of RNAs | 63 | 60 | 64 |
| Number of genes related to: | |||
| Cofactors, vitamins, prosthetic groups, pigments | 182 | 182 | 179 |
| Cell wall and capsule | 59 | 57 | 49 |
| Virulence, disease, and defense | 49 | 49 | 49 |
| Potassium metabolism | 7 | 7 | 7 |
| Miscellaneous | 44 | 44 | 44 |
| Phages, prophages, transposable elements, plasmids | 49 | 18 | 48 |
| Membrane transport | 122 | 107 | 103 |
| Iron acquisition and metabolism | 11 | 11 | 11 |
| RNA metabolism | 53 | 54 | 53 |
| Nucleosides and nucleotides | 102 | 101 | 102 |
| Protein metabolism | 201 | 198 | 200 |
| Motility and chemotaxis | 73 | 73 | 73 |
| Regulation and cell signaling | 36 | 37 | 36 |
| Secondary metabolism | 5 | 4 | 4 |
| DNA metabolism | 92 | 88 | 84 |
| Fatty acids, lipids, and isoprenoids | 126 | 119 | 120 |
| Nitrogen metabolism | 18 | 18 | 18 |
| Respiration | 123 | 124 | 121 |
| Stress response | 102 | 98 | 96 |
| Metabolism of aromatic compounds | 58 | 57 | 62 |
| Amino acids and derivatives | 463 | 458 | 455 |
| Sulfur metabolism | 87 | 85 | 79 |
| Phosphorus metabolism | 25 | 25 | 25 |
| Carbohydrates | 305 | 298 | 300 |
| EF * | R * | T * | CARBOHYDRATE-ACTIVE ENZYMES (CAZymes) |
|---|---|---|---|
| AA | |||
| 3 | 4 | 2 | cellobiose dehydrogenase (EC 1.1.99.18); glucose 1-oxidase (EC 1.1.3.4); aryl alcohol oxidase (EC 1.1.3.7); alcohol oxidase (EC 1.1.3.13); pyranose oxidase (EC 1.1.3.10) |
| 6 | 2 | 2 | 1,4-benzoquinone reductase (EC. 1.6.5.6) |
| 12 | 1 | 2 | pyrroloquinoline quinone-dependent oxidoreductase |
| CBM | |||
| 50 | 1 | 2 | 50 residues found attached to various enzymes from families GH18, GH19, GH23, GH24, GH25, and GH73 |
| CE | |||
| 4 | 3 | 3 | acetyl xylan esterase (EC 3.1.1.72); chitin deacetylase (EC 3.5.1.41); chitooligosaccharide deacetylase (EC 3.5.1.-); peptidoglycan GlcNAc deacetylase (EC 3.5.1.-); peptidoglycan N-acetylmuramic acid deacetylase (EC 3.5.1.-) |
| 11 | 1 | 3 | UDP-3-0-acyl N-acetylglucosamine deacetylase (EC 3.5.1.108). |
| GH | |||
| 0 | 1 | 2 | Glycoside hydrolases not yet assigned to a family. |
| 3 | 2 | 3 | β-glucosidase (EC 3.2.1.21); xylan 1,4-β-xylosidase (EC 3.2.1.37); β-glucosylceramidase (EC 3.2.1.45); β-N-acetylhexosaminidase (EC 3.2.1.52); α-l-arabinofuranosidase (EC 3.2.1.55); glucan 1,3-β-glucosidase (EC 3.2.1.58); glucan 1,4-β-glucosidase (EC 3.2.1.74); coniferin β-glucosidase (EC 3.2.1.126); exo-1,3-1,4-glucanase (EC 3.2.1.-); β-N-acetylglucosaminide phosphorylases (EC 2.4.1.-); β-1,2-glucosidase (EC 3.2.1.-); β-1,3-glucosidase (EC 3.2.1.-); xyloglucan-specific exo-β-1,4-glucanase/exo-xyloglucanase (EC 3.2.1.155) and other. |
| 10 | 1 | 3 | α-amylase (EC 3.2.1.1); pullulanase (EC 3.2.1.41); cyclomaltodextrin glucanotransferase (EC 2.4.1.19); neopullulanase (EC 3.2.1.135); α-glucosidase (EC 3.2.1.20); maltotetraose-forming α-amylase (EC 3.2.1.60); isoamylase (EC 3.2.1.68); amylosucrase (EC 2.4.1.4); amylo-α-1,6-glucosidase (EC 3.2.1.33); α-1,4-glucan: phosphate α-maltosyltransferase (EC 2.4.99.16) and other. |
| 13_11 + CBM48 | 2 | 3 | α-amylase (EC 3.2.1.1); pullulanase (EC 3.2.1.41); cyclomaltodextrin glucanotransferase (EC 2.4.1.19); cyclomaltodextrinase (EC 3.2.1.54); trehalose-6-phosphate hydrolase (EC 3.2.1.93); oligo-α-glucosidase (EC 3.2.1.10); maltogenic amylase (EC 3.2.1.133); neopullulanase (EC 3.2.1.135); α-glucosidase (EC 3.2.1.20); isoamylase (EC 3.2.1.68); maltotriose-forming α-amylase (EC 3.2.1.116); branching enzyme (EC 2.4.1.18); trehalose synthase (EC 5.4.99.16); 4-α-glucanotransferase (EC 2.4.1.25); maltopentaose-forming α-amylase (EC 3.2.1.-); amylosucrase (EC 2.4.1.4); amylo-α-1,6-glucosidase (EC 3.2.1.33) and other. |
| 13_16 | 1 | 3 | α-amylase (EC 3.2.1.1); pullulanase (EC 3.2.1.41); cyclomaltodextrin glucanotransferase (EC 2.4.1.19); cyclomaltodextrinase (EC 3.2.1.54); trehalose-6-phosphate hydrolase (EC 3.2.1.93); oligo-α-glucosidase (EC 3.2.1.10); maltogenic amylase (EC 3.2.1.133); neopullulanase (EC 3.2.1.135); α-glucosidase (EC 3.2.1.20); maltotetraose-forming α-amylase (EC 3.2.1.60); isoamylase (EC 3.2.1.68); glucodextranase (EC 3.2.1.70); maltohexaose-forming α-amylase (EC 3.2.1.98); maltotriose-forming α-amylase (EC 3.2.1.116); branching enzyme (EC 2.4.1.18); trehalose synthase (EC 5.4.99.16); 4-α-glucanotransferase (EC 2.4.1.25); maltopentaose-forming α-amylase (EC 3.2.1); amylo-α-1,6-glucosidase (EC 3.2.1.33) and other |
| 13_26 | 1 | 3 | α-amylase (EC 3.2.1.1); pullulanase (EC 3.2.1.41); cyclomaltodextrin glucanotransferase (EC 2.4.1.19); cyclomaltodextrinase (EC 3.2.1.54); trehalose-6-phosphate hydrolase (EC 3.2.1.93); oligo-α-glucosidase (EC 3.2.1.10); maltogenic amylase (EC 3.2.1.133); neopullulanase (EC 3.2.1.135); α-glucosidase (EC 3.2.1.20); maltotetraose-forming α-amylase (EC 3.2.1.60); isoamylase (EC 3.2.1.68); maltohexaose-forming α-amylase (EC 3.2.1.98); maltotriose-forming α-amylase (EC 3.2.1.116); branching enzyme (EC 2.4.1.18); amylosucrase (EC 2.4.1.4); amylo-α-1,6-glucosidase (EC 3.2.1.33); and other. |
| 13_3 | 1 | 3 | α-amylase (EC 3.2.1.1); pullulanase (EC 3.2.1.41); cyclomaltodextrin glucanotransferase (EC 2.4.1.19); cyclomaltodextrinase (EC 3.2.1.54); trehalose-6-phosphate hydrolase (EC 3.2.1.93); oligo-α-glucosidase (EC 3.2.1.10); maltogenic amylase (EC 3.2.1.133); neopullulanase (EC 3.2.1.135); α-glucosidase (EC 3.2.1.20); maltotetraose-forming α-amylase (EC 3.2.1.60); isoamylase (EC 3.2.1.68); glucodextranase (EC 3.2.1.70); maltohexaose-forming α-amylase (EC 3.2.1.98); maltotriose-forming α-amylase (EC 3.2.1.116); maltopentaose-forming α-amylase (EC 3.2.1.-); amylosucrase (EC 2.4.1.4); malto-oligosyltrehalose synthase (EC 5.4.99.15); amylo-α-1,6-glucosidase (EC 3.2.1.33) and other |
| 13_33 | 1 | 2 | α-amylase (EC 3.2.1.1); pullulanase (EC 3.2.1.41); cyclomaltodextrin glucanotransferase (EC 2.4.1.19); cyclomaltodextrinase (EC 3.2.1.54); trehalose-6-phosphate hydrolase (EC 3.2.1.93); oligo-α-glucosidase (EC 3.2.1.10); maltogenic amylase (EC 3.2.1.133); neopullulanase (EC 3.2.1.135); α-glucosidase (EC 3.2.1.20); maltotetraose-forming α-amylase (EC 3.2.1.60); isoamylase (EC 3.2.1.68); glucodextranase (EC 3.2.1.70); maltohexaose-forming α-amylase (EC 3.2.1.98); maltotriose-forming α-amylase (EC 3.2.1.116); branching enzyme (EC 2.4.1.18); trehalose synthase (EC 5.4.99.16); maltopentaose-forming α-amylase (EC 3.2.1.-); amylosucrase (EC 2.4.1.4); amylo-α-1,6-glucosidase (EC 3.2.1.33) and other. |
| 15 | 2 | 3 | glucoamylase (EC 3.2.1.3); glucodextranase (EC 3.2.1.70); α,α-trehalase (EC 3.2.1.28); dextran dextrinase (EC 2.4.1.2) |
| 19 | 2 | 2 | chitinase (EC 3.2.1.14); lysozyme (EC 3.2.1.17) |
| 23 | 2 | lysozyme type G (EC 3.2.1.17); peptidoglycan lyase (EC 4.2.2.n1); chitinase (EC 3.2.1.14) | |
| 23 + CBM50 | 4 | 3 | lysozyme type G (EC 3.2.1.17); peptidoglycan lyase (EC 4.2.2.n1); chitinase (EC 3.2.1.14) |
| 24 | CA3AT | 2 | lysozyme (EC 3.2.1.17) |
| 39 | 1 | 3 | α-l-iduronidase (EC 3.2.1.76); β-xylosidase (EC 3.2.1.37); α-l-arabinofuranosidase (EC 3.2.1.55); β-glucosidase (EC 3.2.1.21); β-galactosidase (EC 3.2.1.23) |
| 65 | 1 | 3 | α,α-trehalase (EC 3.2.1.28); maltose phosphorylase (EC 2.4.1.8); trehalose phosphorylase (EC 2.4.1.64); kojibiose phosphorylase (EC 2.4.1.230); trehalose-6-phosphate phosphorylase (EC 2.4.1.216); nigerose phosphorylase (EC 2.4.1.279); 3-O-α-glucopyranosyl-l-rhamnose phosphorylase (EC 2.4.1.282); 2-O-α-glucopyranosylglycerol: phosphate β-glucosyltransferase (EC 2.4.1.-); α-glucosyl-1,2-β-galactosyl-L-hydroxylysine α-glucosidase (EC 3.2.1.107) |
| 73 | 1 | 3 | lysozyme (EC 3.2.1.17); mannosyl-glycoprotein endo-β-N-acetylglucosaminidase (EC 3.2.1.96); peptidoglycan hydrolase with endo-β-N-acetylglucosaminidase specificity (EC 3.2.1.-) |
| 77 | 1 | 3 | amylomaltase or 4-α-glucanotransferase (EC 2.4.1.25) |
| 94 + GT84 | 1 | 3 | cellobiose phosphorylase (EC 2.4.1.20); laminaribiose phosphorylase (EC 2.4.1.31); cellodextrin phosphorylase (EC 2.4.1.49); chitobiose phosphorylase (EC 2.4.1.-); cyclic β-1,2-glucan synthase (EC 2.4.1.-); cellobionic acid phosphorylase (EC 2.4.1.321); β-1,2-oligoglucan phosphorylase (EC 2.4.1.-) |
| 102 | 1 | 3 | peptidoglycan lytic transglycosylase (EC 3.2.1.-) |
| 103 | 2 | 3 | peptidoglycan lytic transglycosylase (EC 3.2.1.-) |
| 129 | 1 | 3 | α-N-acetylgalactosaminidase (EC 3.2.1.49); |
| GT | |||
| 0 | 1 | 2 | Glycosyltransferases not yet assigned to a family |
| 1 | 1 | 3 | UDP-glucuronosyltransferase (EC 2.4.1.17); zeatin O-β-xylosyltransferase (EC 2.4.2.40); 2-hydroxyacylsphingosine 1-β-galactosyltransferase (EC 2.4.1.45); N-acylsphingosine galactosyltransferase (EC 2.4.1.47); flavonol 3-O-glucosyltransferase (EC 2.4.1.91); anthocyanidin 3-O-glucosyltransferase (EC 2.4.1.115); sinapate 1-glucosyltransferase (EC 2.4.1.120); indole-3-acetate β-glucosyltransferase (EC 2.4.1.121); flavonol l-rhamnosyltransferase (EC 2.4.1.159) and other |
| 2 | 5 | 3 | cellulose synthase (EC 2.4.1.12); chitin synthase (EC 2.4.1.16); dolichyl-phosphate β-d-mannosyltransferase (EC 2.4.1.83); dolichyl-phosphate β-glucosyltransferase (EC 2.4.1.117); N-acetylglucosaminyltransferase (EC 2.4.1.-); N-acetylgalactosaminyltransferase (EC 2.4.1.-); hyaluronan synthase (EC 2.4.1.212); chitin oligosaccharide synthase (EC 2.4.1.-); β-1,3-glucan synthase (EC 2.4.1.34) and other. |
| 4 | 6 | 3 | sucrose synthase (EC 2.4.1.13); sucrose-phosphate synthase (EC 2.4.1.14); α-glucosyltransferase (EC 2.4.1.52); lipopolysaccharide N-acetylglucosaminyltransferase (EC 2.4.1.56); phosphatidylinositol α-mannosyltransferase (EC 2.4.1.57); GDP-Man: Man1GlcNAc2-PP-dolichol α-1,3-mannosyltransferase (EC 2.4.1.132) and other. |
| 5 | 1 | 3 | UDP-Glc: glycogen glucosyltransferase (EC 2.4.1.11); ADP-Glc: starch glucosyltransferase (EC 2.4.1.21); NDP-Glc: starch glucosyltransferase (EC 2.4.1.242); UDP-Glc: α-1,3-glucan synthase (EC 2.4.1.183) UDP-Glc: α-1,4-glucan synthase (EC 2.4.1.-) |
| 9 | 2 | 3 | lipopolysaccharide N-acetylglucosaminyltransferase (EC 2.4.1.56); heptosyltransferase (EC 2.4.-.-). |
| 19 | 1 | 3 | lipid-A-disaccharide synthase (EC 2.4.1.182). |
| 28 | 1 | 3 | 1,2-diacylglycerol 3-β-galactosyltransferase (EC 2.4.1.46); 1,2-diacylglycerol 3-β-glucosyltransferase (EC 2.4.1.157); UDP-GlcNAc: Und-PP-MurAc-pentapeptide β-N-acetylglucosaminyltransferase (EC 2.4.1.227); digalactosyldiacylglycerol synthase (EC 2.4.1.241) |
| 30 | 1 | 3 | CMP-β-KDO: α-3-deoxy-d-manno-octulosonic-acid (KDO) transferase (EC 2.4.99.-). |
| 35 | 1 | 3 | glycogen or starch phosphorylase (EC 2.4.1.1). |
| 51 | 3 | 3 | murein polymerase (EC 2.4.1.129). |
| 104 | 1 | 2 | dTDP-β-l-Rhap: arginine α-l-rhamnosyltransferase (EC 2.4.1.-) |
| PL | |||
| 4 | 1 | 2 | rhamnogalacturonan endolyase (EC 4.2.2.23). |
| 5_1 | 1 | 3 | alginate lyase (EC 4.2.2.3); endo-β-1,4-glucuronan lyase (EC 4.2.2.14) |
| 7_1 | 4 | 2 | poly(β-mannuronate) lyase/M-specific alginate lyase (EC 4.2.2.3); α-L-guluronate lyase/G-specific alginate lyase (EC 4.2.2.11); poly-(MG)-lyase/MG-specific alginate lyase (EC 4.2.2.-); endo-β-1,4-glucuronan lyase (EC 4.2.2.14); oligoalginate lyase/exo-alginate lyase (EC 4.2.2.26) |
| 26 | 1 | 2 | rhamnogalacturonan exolyase (EC 4.2.2.24). |
| Strain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| CA3AT | T | T | T | PI | PI | PI | T | T |
| C2L11 | T | T | T | T | T | T | T | T |
| C2L12B | PI | PI | T | T | T | T | PI | PI |
| TEST | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Oxidase | + | + | + | + | + | - | |
| Growth: | |||||||
| >37 °C | + | + | + | + | ND | + | ND |
| >7% NaCl | + | + | + | ND | + | - | ND |
| API 20NE | |||||||
| Reduction of nitrates | W | W | + | + | + | - | - |
| Glucose fermentation | W | W | W | - | ND | - | - |
| Arginine dihydrolase | + | + | + | - | ND | + | - |
| Urease | + | + | + | - | ND | - | - |
| Esculin hydrolysis | - | - | - | - | ND | - | + |
| Gelatin hydrolysis | - | - | - | - | + | + | + |
| Assimilation of: | |||||||
| l-Arabinose | + | + | + | + | + | - | + |
| d-Mannose | W | W | W | + | + | + | + |
| N-Acetyl-glucosamine | - | - | - | - | + | + | - |
| Adipate | W | W | W | - | ND | - | ND |
| Malate | W | W | W | + | ND | + | + |
| Trisodium citrate | + | W | W | + | ND | + | ND |
| Phenylacetate | - | - | - | - | + | + | ND |
| API 50CH | |||||||
| Glycerol | W | + | - | + | ND | ND | + |
| Erythritol | + | - | - | + | ND | - | + |
| d-Arabinose | - | - | - | + | ND | ND | ND |
| d-Adonitol | - | - | - | + | ND | - | - |
| d-Galactose | + | + | + | + | ND | - | + |
| l-Sorbose | - | - | - | + | ND | ND | ND |
| l-Rhamnose | + | W | W | + | ND | - | - |
| Dulcitol | - | - | - | + | ND | ND | ND |
| Inositol | W | - | - | + | ND | ND | + |
| d-Sorbitol | + | - | - | + | ND | - | + |
| d-Melibiose | - | W | W | ND | - | - | - |
| d-Saccharose (sucrose) | - | - | - | ND | ND | - | + |
| d-Trehalose | - | - | - | ND | ND | - | + |
| Glycogen | - | - | - | ND | W | W | - |
| Gentiobiose | W | + | + | + | ND | - | - |
| d-Turanose | + | - | - | + | ND | - | - |
| d-Tagatose | - | - | - | + | ND | ND | ND |
| l-Fucose | - | - | - | + | ND | W | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peral-Aranega, E.; Saati-Santamaría, Z.; Kolařik, M.; Rivas, R.; García-Fraile, P. Bacteria Belonging to Pseudomonas typographi sp. nov. from the Bark Beetle Ips typographus Have Genomic Potential to Aid in the Host Ecology. Insects 2020, 11, 593. https://doi.org/10.3390/insects11090593
Peral-Aranega E, Saati-Santamaría Z, Kolařik M, Rivas R, García-Fraile P. Bacteria Belonging to Pseudomonas typographi sp. nov. from the Bark Beetle Ips typographus Have Genomic Potential to Aid in the Host Ecology. Insects. 2020; 11(9):593. https://doi.org/10.3390/insects11090593
Chicago/Turabian StylePeral-Aranega, Ezequiel, Zaki Saati-Santamaría, Miroslav Kolařik, Raúl Rivas, and Paula García-Fraile. 2020. "Bacteria Belonging to Pseudomonas typographi sp. nov. from the Bark Beetle Ips typographus Have Genomic Potential to Aid in the Host Ecology" Insects 11, no. 9: 593. https://doi.org/10.3390/insects11090593
APA StylePeral-Aranega, E., Saati-Santamaría, Z., Kolařik, M., Rivas, R., & García-Fraile, P. (2020). Bacteria Belonging to Pseudomonas typographi sp. nov. from the Bark Beetle Ips typographus Have Genomic Potential to Aid in the Host Ecology. Insects, 11(9), 593. https://doi.org/10.3390/insects11090593






