Deep Sequencing of Small RNAs in the Whitefly Bemisia tabaci Reveals Novel MicroRNAs Potentially Associated with Begomovirus Acquisition and Transmission
Abstract
Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Whiteflies and Feeding Assays
2.2. RNA Isolation and Library Preparation
2.3. Initial Sequence Processing and Analysis of Reads for 18 Small RNA Libraries
2.4. Identification of Conserved and Novel miRNAs
2.5. Validation of Novel miRNAs by Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.6. Target Prediction and Gene Ontology Analysis
3. Results
3.1. Overview of miRNA Data
3.2. Identification and Validation of Novel B. tabaci-Specific miRNAs
3.3. Predicted Targets for Novel and Conserved B. tabaci miRNAs
3.4. Differentially Expressed miRNAs with a Potential Association with TYLCV Transmission
3.5. Evaluating Potential Inverse Relationships Between miRNA and mRNA Expression under the Same Treatment
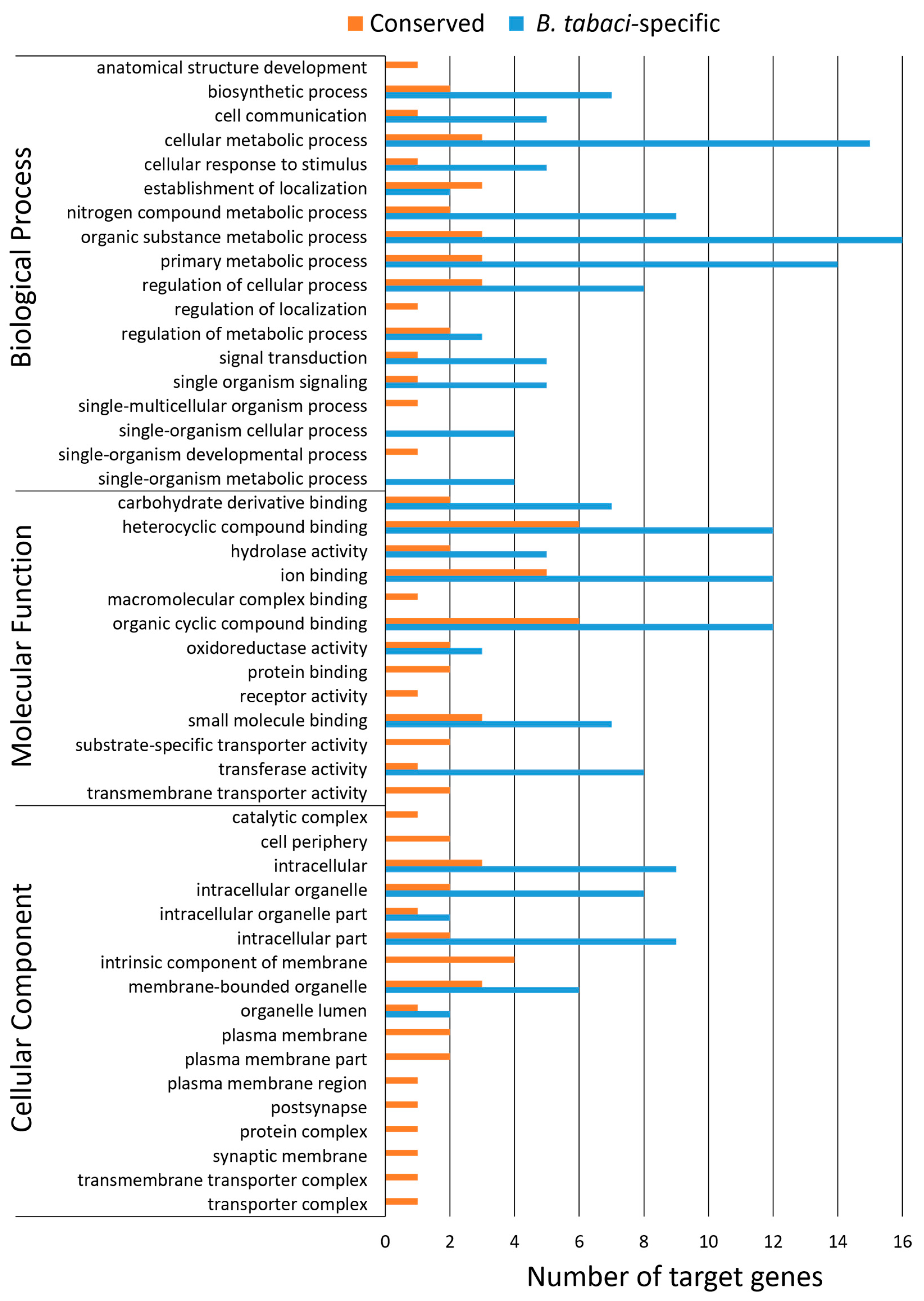
3.6. Gene Ontology Assignments for Predicted miRNA Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Stansly, P.A.; Naranjo, S.E. Bemisia: Bionomics and Management of a Global Pest; Springer Science + Business Media B.V.: Dordrecht, Netherland, 2010. [Google Scholar]
- Zang, L.-S.; Wei-Qiang, C.; Shu-Sheng, L. Comparison of performance on different host plants between the B biotype and a non-B biotype of Bemisia tabaci from Zhejiang, China. Entomol. Exp. Appl. 2006, 121, 221–227. [Google Scholar] [CrossRef]
- Houndété, T.A.; Kétoh, G.K.; Hema, O.S.; Brévault, T.; Glitho, I.A.; Martin, T. Insecticide resistance in field populations of Bemisia tabaci (Hemiptera: Aleyrodidae) in West Africa. Pest Manag. Sci. 2010, 66, 1181–1185. [Google Scholar] [CrossRef]
- Bedford, I.D.; Briddon, R.W.; Brown, J.K.; Rosell, R.C.; Markham, P.G. Geminivirus transmission and biological characterisation of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann. Appl. Biol. 1994, 125, 311–325. [Google Scholar] [CrossRef]
- Rosen, R.; Kanakala, S.; Kliot, A.; Cathrin Pakkianathan, B.; Farich, B.A.; Santana-Magal, N.; Elimelech, M.; Kontsedalov, S.; Lebedev, G.; Cilia, M.; et al. Persistent, circulative transmission of begomoviruses by whitefly vectors. Curr. Opin. Virol 2015, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bellows, T.S.; Perring, T.M.; Gill, R.J.; Headrick, D.H. Description of a species of Bemisia (Homoptera: Aleyrodidae). Ann. Entomol. Soc. Am. 1994, 87, 195–206. [Google Scholar] [CrossRef]
- Czosnek, H.; Hariton-Shalev, A.; Sobol, I.; Gorovits, R.; Ghanim, M. The incredible journey of begomoviruses in their whitefly vector. Viruses 2017, 9, 273. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olive, E.; Sanchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.M.; Varsani, A.; et al. The spread of tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog. 2010, 6, e1001164. [Google Scholar] [CrossRef]
- Kanakala, S.; Ghanim, M. Implication of the whitefly Bemisia tabaci cyclophilin B protein in the transmission of tomato yellow leaf curl virus. Front. Plant Sci. 2016, 7, 1702. [Google Scholar] [CrossRef]
- Hariton Shalev, A.; Sobol, I.; Ghanim, M.; Liu, S.-S.; Czosnek, H. The whitefly Bemisia tabaci knottin-1 gene is implicated in regulating the quantity of tomato yellow leaf curl virus ingested and transmitted by the insect. Viruses 2016, 8, 205. [Google Scholar] [CrossRef]
- Morin, S.; Ghanim, M.; Sobol, I.; Czosnek, H. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast two-hybrid system. Virology 2000, 276, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, D.K.; Chen, W.; Zheng, Y.; Kaur, N.; Wintermantel, W.M.; Simmons, A.M.; Fei, Z.; Ling, K.-S. Comparative transcriptome analysis reveals networks of genes activated in the whitefly, Bemisia tabaci when fed on tomato plants infected with tomato yellow leaf curl virus. Virology 2018, 513, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.; Qian, L.-X.; Shao, R.-X.; Liu, Y.-Q.; Liu, S.-S.; Wang, X.-W. Transcriptome profiling of whitefly guts in response to tomato yellow leaf curl virus infection. Virol. J. 2018, 15, 14. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, J.; Su, Y.-L. Transcriptome analysis of gene expression profiles of tomato yellow leaf curl virus-infected whiteflies over different viral acquisition access periods. Insects 2020, 11, 297. [Google Scholar] [CrossRef]
- Luan, J.B.; Li, J.M.; Varela, N.; Wang, Y.L.; Li, F.F.; Bao, Y.Y.; Zhang, C.X.; Liu, S.S.; Wang, X.W. Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 2011, 85, 3330–3340. [Google Scholar] [CrossRef]
- Guo, Q.; Tao, Y.L.; Chu, D. Characterization and comparative profiling of miRNAs in invasive Bemisia tabaci (Gennadius) B and Q. PLoS ONE 2013, 8, e59884. [Google Scholar] [CrossRef]
- Wang, B.; Wang, L.; Chen, F.; Yang, X.; Ding, M.; Zhang, Z.; Liu, S.S.; Wang, X.W.; Zhou, X. MicroRNA profiling of the whitefly Bemisia tabaci Middle East-Aisa Minor I following the acquisition of tomato yellow leaf curl China virus. Virol. J. 2016, 13, 20. [Google Scholar] [CrossRef]
- Li, H.; Wei, X.; Ding, T.; Chu, D. Genome-wide profiling of cardinium-responsive microRNAs in the exotic whitefly, Bemisia tabaci (Gennadius) Biotype, Q. Front. Physiol. 2018, 9, 1580. [Google Scholar] [CrossRef]
- Wong, R.R.; Abd-Aziz, N.; Affendi, S.; Poh, C.L. Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci. 2020, 27, 4. [Google Scholar] [CrossRef]
- Blair, C.D.; Olson, K.E. The role of RNA interference (RNAi) in arbovirus-vector interactions. Viruses 2015, 7, 820–843. [Google Scholar] [CrossRef]
- Lucas, K.; Raikhel, A.S. Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 2013, 43, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Mongelli, V.; Saleh, M.C. Bugs are not to be silenced: Small RNA pathways and antiviral responses in insects. Annu. Rev. Virol. 2016, 3, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Shatters, R.G.; Powell, C.A.; Boykin, L.M.; Liansheng, H.; McKenzie, C.L. Improved DNA barcoding method for Bemisia tabaci and related Aleyrodidae: Development of universal and Bemisia tabaci biotype-specific mitochondrial cytochrome c oxidase I polymerase chain reaction primers. J. Econ. Entomol. 2009, 102, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-R.; Zheng, Y.; Liu, B.; Zhong, S.; Giovannoni, J.; Fei, Z. A cost-effective method for Illumina small RNA-Seq library preparation using T4 RNA ligase 1 adenylated adapters. Plant Methods 2012, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Gao, S.; Padmanabhan, C.; Li, R.; Galvez, M.; Gutierrez, D.; Fuentes, S.; Ling, K.-S.; Kreuze, J.; Fei, Z. VirusDetect: An automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 2017, 500, 130–138. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Chen, W.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.; Stensmyr, M.C.; Zheng, Y.; Liu, W.; Sun, H.; et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016, 14, 110. [Google Scholar] [CrossRef]
- Shi, R.; Sun, Y.-H.; Zhang, X.-H.; Chiang, V.L. Poly(T) adaptor RT-PCR. Methods Mol. Biol. 2012, 822, 53–66. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Shamimuzzaman, M.; Hasegawa, D.K.; Chen, W.; Simmons, A.M.; Fei, Z.; Ling, K.-S. Genome-wide profiling of piRNAs in the whitefly Bemisia tabaci reveals cluster distribution and association with begomovirus transmission. PLoS ONE 2019, 14, e0213149. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Chen, C.; Yang, Z.; Guo, L.; Yang, X.; Wang, D.; Chen, M.; Huang, J.; Wen, Y.; Zeng, Y.; et al. Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. Gigascience 2017, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Kollenberg, M.; Winter, S.; Gotz, M. Quantification and localization of watermelon chlorotic stunt virus and tomato yellow leaf curl virus (Geminiviridae) in populations of Bemisia tabaci (Hemiptera, Aleyrodidae) with differential virus transmission characteristics. PLoS ONE 2014, 9, e111968. [Google Scholar] [CrossRef]
- Czosnek, H.; Ghanim, M. Back to Basics: Are begomoviruses whitefly pathogens? J. Integr. Agric. 2012, 11, 225–234. [Google Scholar] [CrossRef]
- Hunter, W.B.; Hiebert, E.; Webb, S.E.; Tsai, J.H.; Polston, J.E. Location of geminiviruses in the whitefly Bemisia tabaci (Homoptera: Aleyrodidae). Plant Dis. 1998, 82, 1147–1151. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Misiewicz-Krzeminska, I.; Krzeminski, P.; Corchete, L.A.; Quwaider, D.; Rojas, E.A.; Herrero, A.B.; Gutiérrez, N.C. Factors regulating microRNA expression and function in multiple myeloma. Non Coding RNA 2019, 5, 9. [Google Scholar] [CrossRef]
- Jin, H.; Tuo, W.; Lian, H.; Liu, Q.; Zhu, X.-Q.; Gao, H. Strategies to identify microRNA targets: New advances. New Biotechnol. 2010, 27, 734–738. [Google Scholar] [CrossRef]

| Length | No. miRNAs | ||
|---|---|---|---|
| Conserved | B. tabaci-Specific | Total | |
| 19 | 1 | 4 | 5 |
| 20 | 0 | 1 | 1 |
| 21 | 22 | 9 | 31 |
| 22 | 32 | 61 | 93 |
| 23 | 10 | 14 | 24 |
| 24 | 1 | 2 | 3 |
| 25 | 0 | 3 | 3 |
| Total | 66 | 94 | 160 |
| Name | Target | Annotation | 24 h | 48 h | 72 h | |||
|---|---|---|---|---|---|---|---|---|
| miRNA | Target Gene * | miRNA | Target Gene * | miRNA | Target Gene * | |||
| Bta-miRn88 | Bta07821 | Zinc finger CCCH domain-containing protein 14 | −1.16 | 1.02 | −1.03 | 1.27 | 1.03 | 1.01 |
| Bta-miRn61 | Bta03447 | Cadherin EGF LAG seven-pass G-type receptor 3 | −1.20 | 1.07 | 1.10 | −1.04 | −1.10 | −1.09 |
| Bta-miRn100 | Bta13203 | FGGY family of carbohydrate kinase | −1.21 | −1.25 | 1.01 | −1.20 | −1.03 | −1.05 |
| Bta-miRn62 | Bta09225 | UPF0472 protein C16orf72 | −1.30 | 1.11 | −1.02 | 1.05 | 1.17 | 1.19 |
| Bta-miRn54 | Bta07082 | Centrosomal protein of 78 kDa | −1.05 | −1.39 | 1.17 | 1.19 | 1.44 | 1.06 |
| Bta-miRn108 | Bta00358 | Dynamin-1-like protein | −1.16 | −1.04 | −1.03 | 1.05 | 1.10 | 1.12 |
| Bta-miRn71 | Bta05170 | Unknown protein | 1.00 | 12.00 | 1.01 | −1.38 | 1.17 | −1.30 |
| Bta-miRn69 | Bta13835 | Unknown protein | −1.19 | 1.10 | −1.03 | −1.01 | 1.02 | 1.31 |
| Bta-miRn72 | Bta08675 | Unknown protein | −1.27 | −1.38 | 1.07 | 1.12 | −1.11 | −1.06 |
| Bta-miRn112 | Bta04487 | Transcription factor BTF3-like protein 4 | 1.23 | −1.08 | −1.15 | −1.06 | 1.05 | −1.12 |
| Bta-miRn58 | Bta04052 | Centrosomal protein of 164 kDa | −1.21 | −1.04 | −1.03 | 1.19 | 1.03 | −1.06 |
| Bta-miRn93 | Bta09366 | Zinc finger protein 250 | −1.14 | 1.12 | 1.25 | −1.19 | −1.13 | 1.12 |
| Bta-miRn23 | Bta05482 | Nuclear receptor | −1.44 | −1.09 | −3.13 | −1.23 | −1.04 | −1.73 |
| Name | Sequence | Length | Target | Annotation | Fold Change | ||
| 24 h | 48 h | 72 h | |||||
| miR-996 | ugacuagaguuacacucguca | 21 | Bta08371 | Zinc finger protein, putative | −1.72 | 1.05 | 1.37 |
| miR-219 | ugauuguccaaacgcaauucuug | 23 | Bta10860 | Beta-1,3-galactosyltransferase, putative | −1.50 | −1.00 | 1.26 |
| Bta-miRn23 | ccccucgccgcgcggagcu | 19 | Bta05482 | Nuclear receptor | −1.44 | −3.13 * | −1.04 |
| miR-1-3p | uggaauguaaagaaguauggag | 22 | Bta10702 | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 2 | −1.20 | 1.25 | −1.52 * |
| miR-iab-4 | acguauacuaaauguauccuga | 22 | None | N/A | −1.27 | 1.05 | 1.53 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasegawa, D.K.; Shamimuzzaman, M.; Chen, W.; Simmons, A.M.; Fei, Z.; Ling, K.-S. Deep Sequencing of Small RNAs in the Whitefly Bemisia tabaci Reveals Novel MicroRNAs Potentially Associated with Begomovirus Acquisition and Transmission. Insects 2020, 11, 562. https://doi.org/10.3390/insects11090562
Hasegawa DK, Shamimuzzaman M, Chen W, Simmons AM, Fei Z, Ling K-S. Deep Sequencing of Small RNAs in the Whitefly Bemisia tabaci Reveals Novel MicroRNAs Potentially Associated with Begomovirus Acquisition and Transmission. Insects. 2020; 11(9):562. https://doi.org/10.3390/insects11090562
Chicago/Turabian StyleHasegawa, Daniel K., Md Shamimuzzaman, Wenbo Chen, Alvin M. Simmons, Zhangjun Fei, and Kai-Shu Ling. 2020. "Deep Sequencing of Small RNAs in the Whitefly Bemisia tabaci Reveals Novel MicroRNAs Potentially Associated with Begomovirus Acquisition and Transmission" Insects 11, no. 9: 562. https://doi.org/10.3390/insects11090562
APA StyleHasegawa, D. K., Shamimuzzaman, M., Chen, W., Simmons, A. M., Fei, Z., & Ling, K.-S. (2020). Deep Sequencing of Small RNAs in the Whitefly Bemisia tabaci Reveals Novel MicroRNAs Potentially Associated with Begomovirus Acquisition and Transmission. Insects, 11(9), 562. https://doi.org/10.3390/insects11090562







