The Gut–Brain–Microbiome Axis in Bumble Bees
Abstract
1. Introduction
2. Methods
2.1. Bumble Bee Rearing and Microbe-Inoculation Treatments
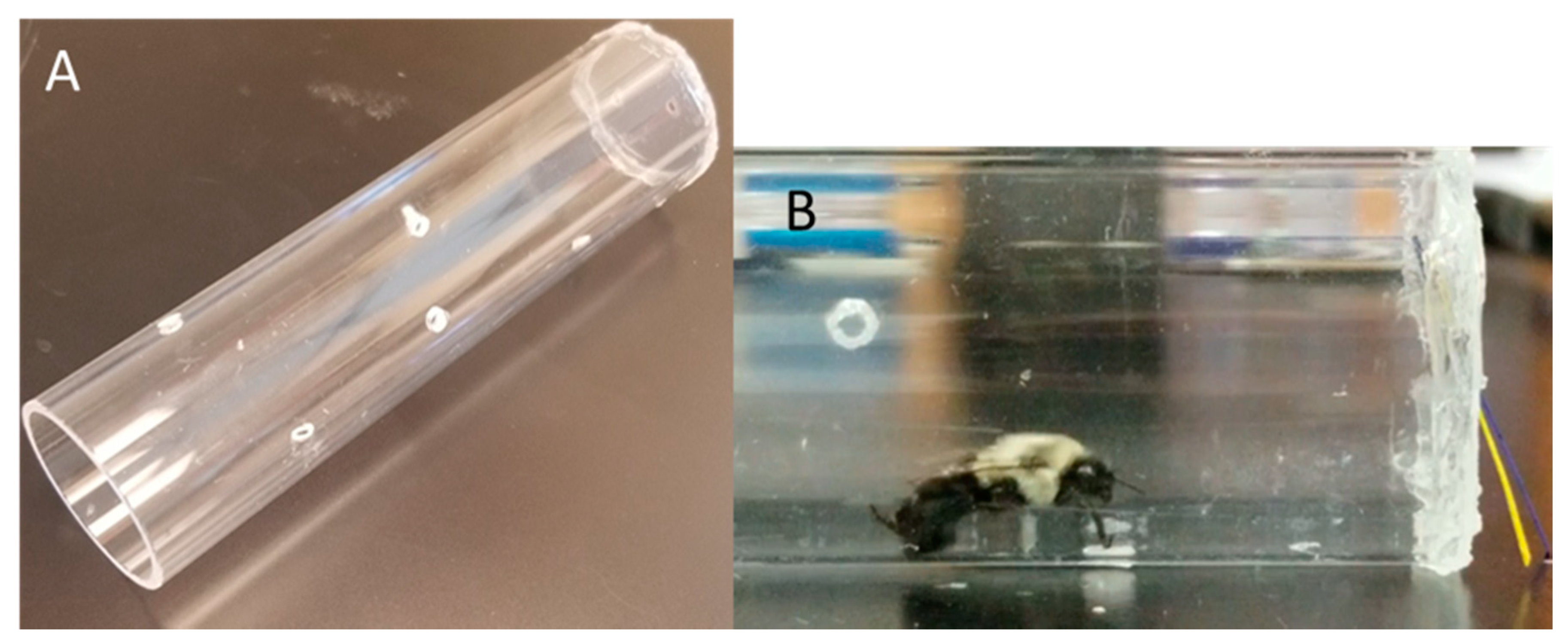
2.2. Free Moving Proboscis Extension Response (FMPER) Assays
2.3. Statistical Analyses
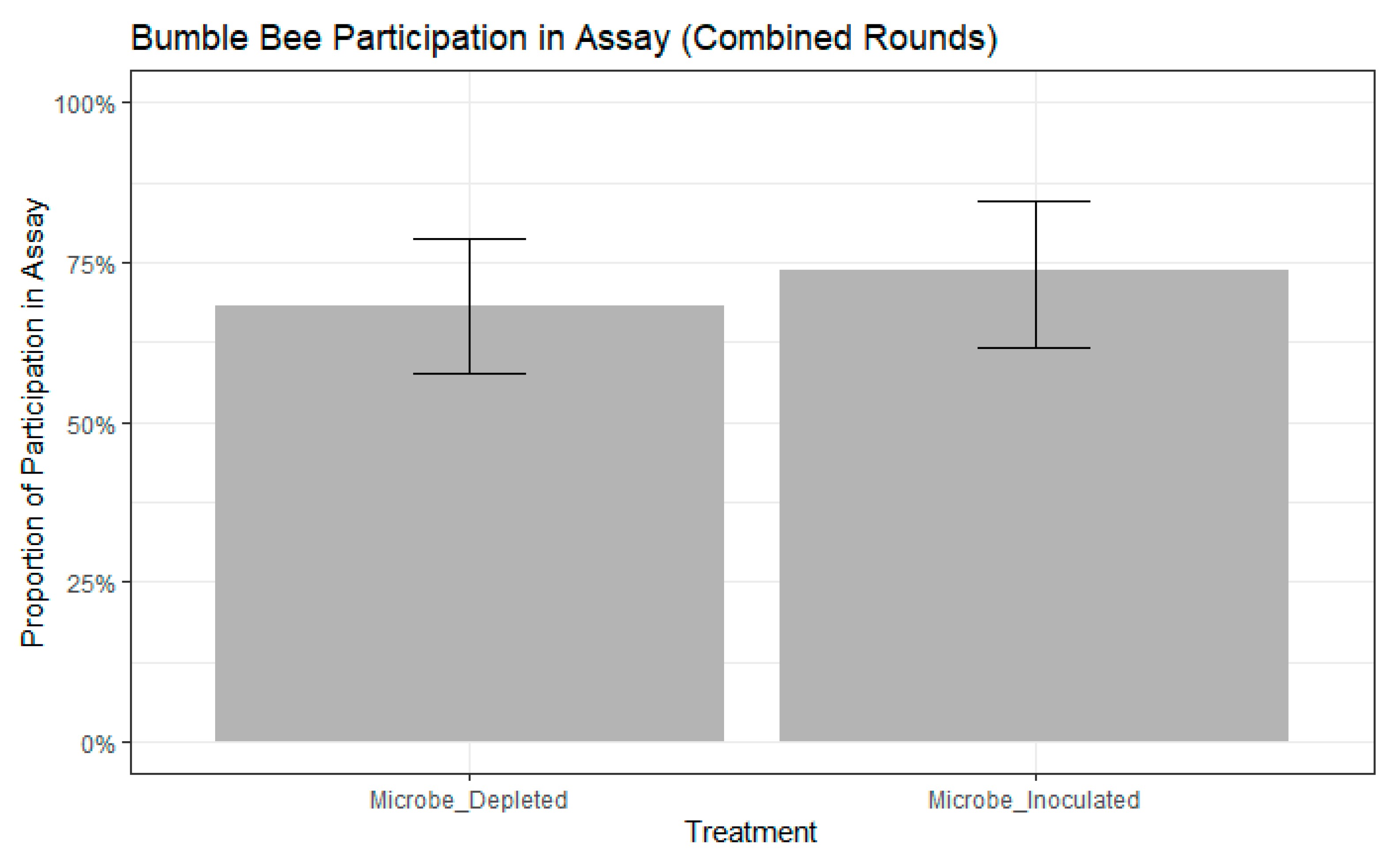
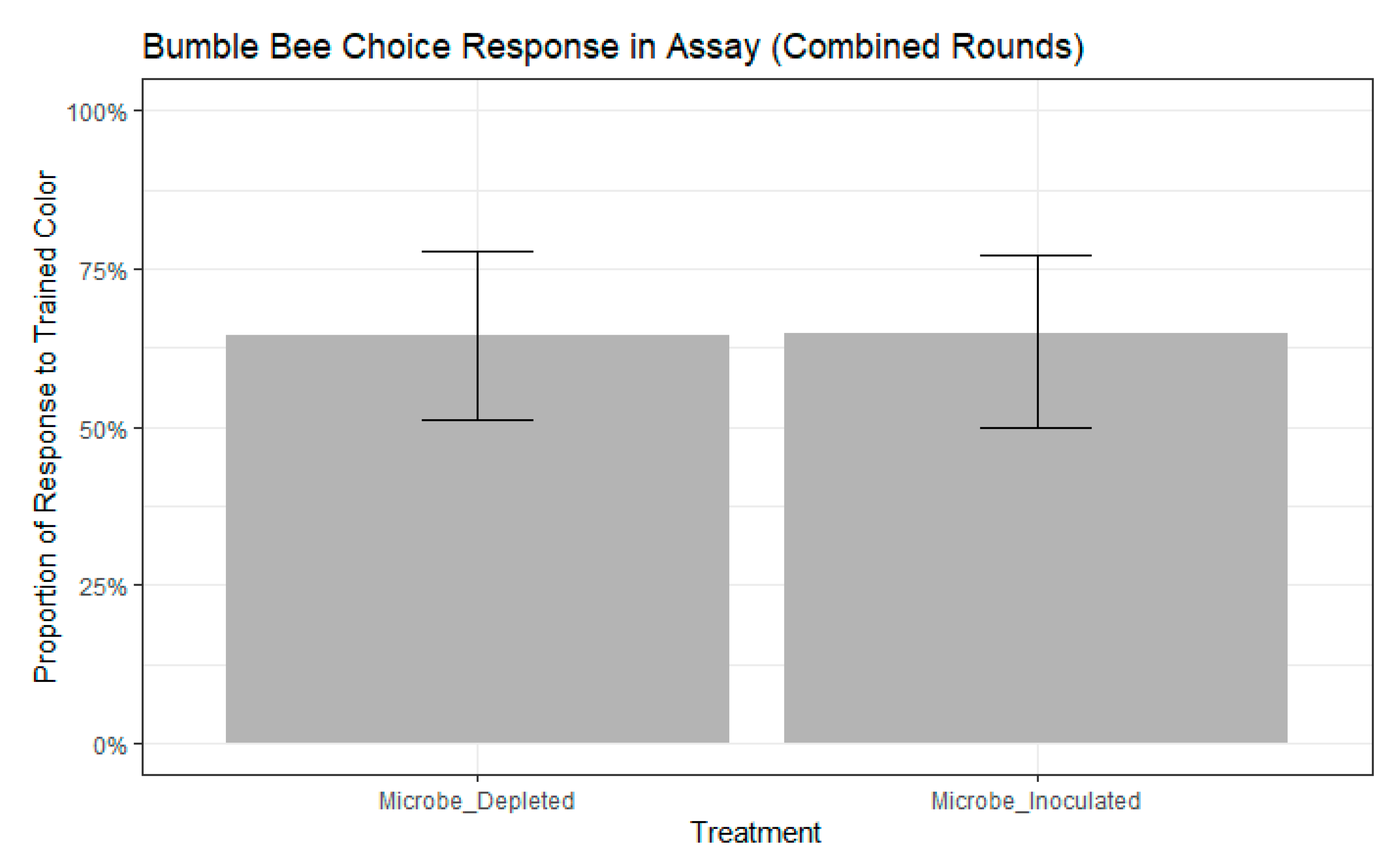
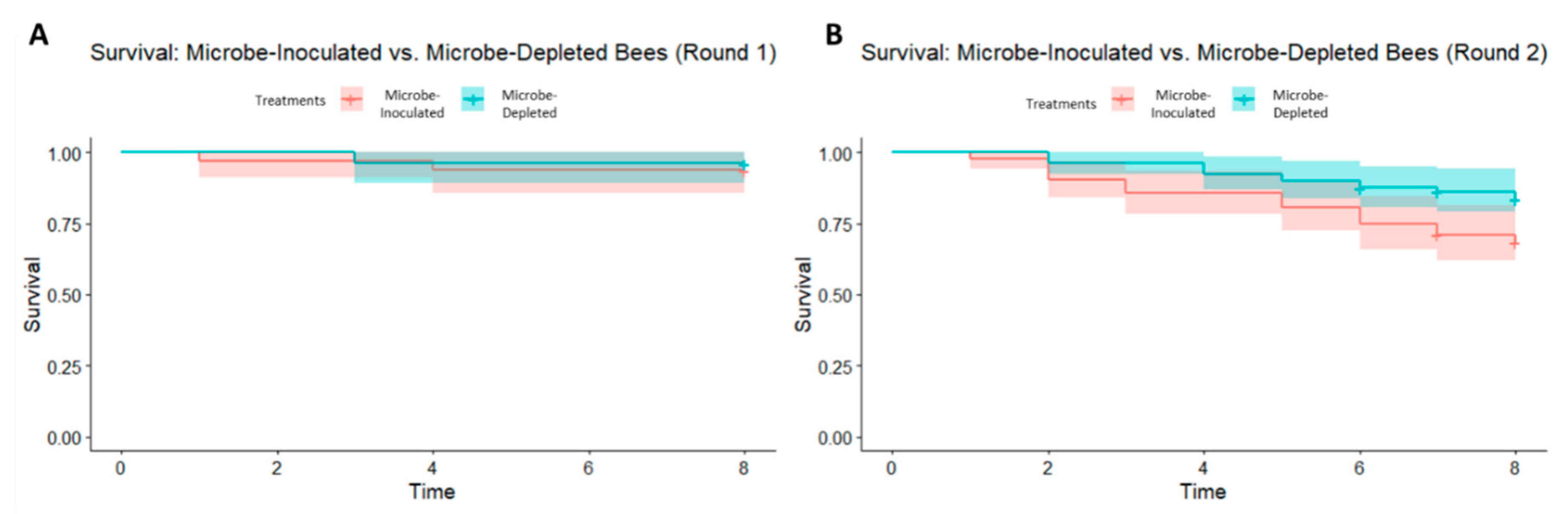
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cryan, J.F.; Dinan, T.G. Mind-Altering Microorganisms: The Impact of the Gut Microbiota on Brain and Behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.M.; Bercik, P. The Relationship Between Intestinal Microbiota and the Central Nervous System in Normal Gastrointestinal Function and Disease. Gastroenterology 2009, 136, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Wine, E.; Rodrigues, D.M.; Cho, J.H.; Whary, M.T.; Philpott, D.J.; Macqueen, G.; Sherman, P.M. Bacterial infection Causes Stress-Induced Memory Dysfunction in Mice. Gut 2011, 60, 307–317. [Google Scholar] [CrossRef]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated with Neurodevelopmental Disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef]
- Sankar, S.A.; Lagier, J.C.; Pontarotti, P.; Raoult, D.; Fournier, P.E. The Human Gut Microbiome, a Taxonomic Conundrum. Syst. Appl. Microbiol. 2015, 38, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Moran, N.A. The Gut Microbiota of Insects—Diversity in Structure and Function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Liberti, J.; Engel, P. The Gut Microbiota—Brain Axis of Insects. Curr. Opin. Insect Sci. 2020, 39, 6–13. [Google Scholar] [CrossRef]
- Miller, W.J.; Ehrman, L.; Schneider, D. Infectious Speciation Revisited: Impact of Symbiont-Depletion on Female Fitness and Mating Behavior of Drosophila Paulistorum. PLoS Pathog. 2010, 6, e1001214. [Google Scholar] [CrossRef]
- Andersen, S.B.; Gerritsma, S.; Yusah, K.M.; Mayntz, D.; Hywel-Jones, N.L.; Billen, J.; Boomsma, J.J.; Hughes, D.P. The Life of a Dead Ant: The Expression of an Adaptive Extended Phenotype. Am. Nat. 2009, 174, 424–433. [Google Scholar] [CrossRef]
- Matsuura, K. Nestmate Recognition Mediated by Intestinal Bacteria in a Termite, Reticulitermes Speratus. Oikos 2001, 92, 20–26. [Google Scholar] [CrossRef]
- Teseo, S.; van Zweden, J.S.; Pontieri, L.; Kooij, P.W.; Sørensen, S.J.; Wenseleers, T.; Poulsen, M.; Boomsma, J.J.; Sapountzis, P. The Scent of Symbiosis: Gut Bacteria may Affect Social Interactions in Leaf-Cutting Ants. Anim. Behav. 2019, 150, 239–254. [Google Scholar] [CrossRef]
- DeNieu, M.; Mounts, K.; Manier, M. Two Gut Microbes are Necessary and Sufficient for Normal Cognition in Drosophila Melanogaster. bioRxiv 2019, 593723. [Google Scholar] [CrossRef]
- Sharon, G.; Segal, D.; Ringo, J.M.; Hefetz, A.; Zilber-Rosenberg, I.; Rosenberg, E. Commensal Bacteria Play a Role in Mating Preference of Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 2010, 107, 20051–20056. [Google Scholar] [CrossRef]
- Klein, S.; Cabirol, A.; Devaud, J.M.; Barron, A.B.; Lihoreau, M. Why Bees Are So Vulnerable to Environmental Stressors. Trends Ecol. Evol. 2017, 32, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Michener, C.D. The Bees of the World; The John Hopkins University Press: Baltimore, MD, USA, 2000. [Google Scholar]
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.-W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic Microbiome Evolution in Social Bees. Sci. Adv. 2017, 3, e1600513. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Gut Microbial Communities of Social Bees. Nat. Rev. Microbiol. 2016, 14, 374–384. [Google Scholar] [CrossRef]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee Gut Microbiota Promotes Host Weight Gain via Bacterial Metabolism and Hormonal Signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef]
- Zheng, H.; Nishida, A.; Kwong, W.K.; Koch, H.; Engel, P.; Steele, M.I.; Moran, N.A. Metabolism of Toxic Sugars by Strains of the Bee Gut Symbiont Gilliamella Apicola. MBio 2016, 7, e01326-16. [Google Scholar] [CrossRef]
- Koch, H.; Schmid-Hempel, P. Socially Transmitted Gut Microbiota Protect Bumble Bees against an Intestinal Parasite. Proc. Natl. Acad. Sci. USA 2011, 108, 19288–19292. [Google Scholar] [CrossRef]
- Hammer, T.J.; Moran, N.A. Links between Metamorphosis and Symbiosis in Holometabolous Insects. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190068. [Google Scholar] [CrossRef]
- Rothman, J.A.; Leger, L.; Graystock, P.; Russell, K.; McFrederick, Q.S. The Bumble Bee Microbiome Increases Survival of Bees Exposed to Selenate Toxicity. Environ. Microbiol. 2019, 21, 3417–3429. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Steele, M.I.; Leonard, S.P.; Motta, E.V.S.; Moran, N.A. Honey Bees as Models for Gut Microbiota Research. Lab Anim. (NY) 2018, 47, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Abrol, D.P.; Li, J.; Schmid-Hempel, P. Diversity and Evolutionary Patterns of Bacterial Gut Associates of Corbiculate Bees. Mol. Ecol. 2013, 22, 2028–2044. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; de Miranda, J.R.; Doublet, V.; et al. The Bee Microbiome: Impact on Bee Health and Model for Evolution and Ecology of Host-Microbe Interactions. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Gegear, R.J.; Otterstatter, M.C.; Thomson, J.D. Does Parasitic Infection Impair the Ability of Bumblebees to Learn Flower-Handling Techniques? Anim. Behav. 2005, 70, 209–215. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef]
- Meeus, I.; Mommaerts, V.; Billiet, A.; Mosallanejad, H.; Van de Wiele, T.; Wäckers, F.; Smagghe, G. Assessment of Mutualism between Bombus Terrestris and its Microbiota by Use of Microcolonies. Apidologie 2013, 44, 708–719. [Google Scholar] [CrossRef]
- Muth, F.; Cooper, T.R.; Bonilla, R.F.; Leonard, A.S. A Novel Protocol for Studying Bee Cognition in the Wild. Methods Ecol. Evol. 2017. [Google Scholar] [CrossRef]
- Takeda, K. Classical Conditioned Response in the Honey Bee. J. Insect Physiol. 1961, 6, 168–179. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Menzel, R.; Sandoz, J.C.; Giurfa, M. Revisiting Olfactory Classical Conditioning of the Proboscis Extension Response in Honey Bees: A Step toward Standardized Procedures. J. Neurosci. Methods 2012, 211, 159–167. [Google Scholar] [CrossRef]
- Kassambara, A.; Kosinski, M.; Biecek, P. Drawing Survival Curves Using “ggplot2” [R package survminer version 0.4.6]. Available online: https://cran.r-project.org/web/packages/survminer/index.html. (accessed on 19 February 2020).
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 1548-7660, 1–48. [Google Scholar]
- Vincent Dorie, A.; Vincent Dorie, M.; Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Package “Blme” Title Bayesian Linear Mixed-Effects Models Description Maximum a Posteriori Estimation for Linear and Generalized Linear Mixed-Effects Models in a Bayesian Setting. Extends “lme4” by. 2016. Available online: https://rdrr.io/cran/blme/ (accessed on 5 August 2020).
- Giurfa, M. Cognition with Few Neurons: Higher-Order Learning in Insects. Trends Neurosci. 2013, 36, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.J.F.; Schmid-Hempel, R.; Schmid-Hempel, P. Strong Context-Dependent Virulence in a Host-Parasite System: Reconciling Genetic Evidence with theory. J. Anim. Ecol. 2003, 72, 994–1002. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leger, L.; McFrederick, Q.S. The Gut–Brain–Microbiome Axis in Bumble Bees. Insects 2020, 11, 517. https://doi.org/10.3390/insects11080517
Leger L, McFrederick QS. The Gut–Brain–Microbiome Axis in Bumble Bees. Insects. 2020; 11(8):517. https://doi.org/10.3390/insects11080517
Chicago/Turabian StyleLeger, Laura, and Quinn S. McFrederick. 2020. "The Gut–Brain–Microbiome Axis in Bumble Bees" Insects 11, no. 8: 517. https://doi.org/10.3390/insects11080517
APA StyleLeger, L., & McFrederick, Q. S. (2020). The Gut–Brain–Microbiome Axis in Bumble Bees. Insects, 11(8), 517. https://doi.org/10.3390/insects11080517



