Phylogeography of Organophosphate Resistant ace Alleles in Spanish Olive Fruit Fly Populations: A Mediterranean Perspective in the Global Change Context
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Exon IV
2.2.2. Exons VII and X
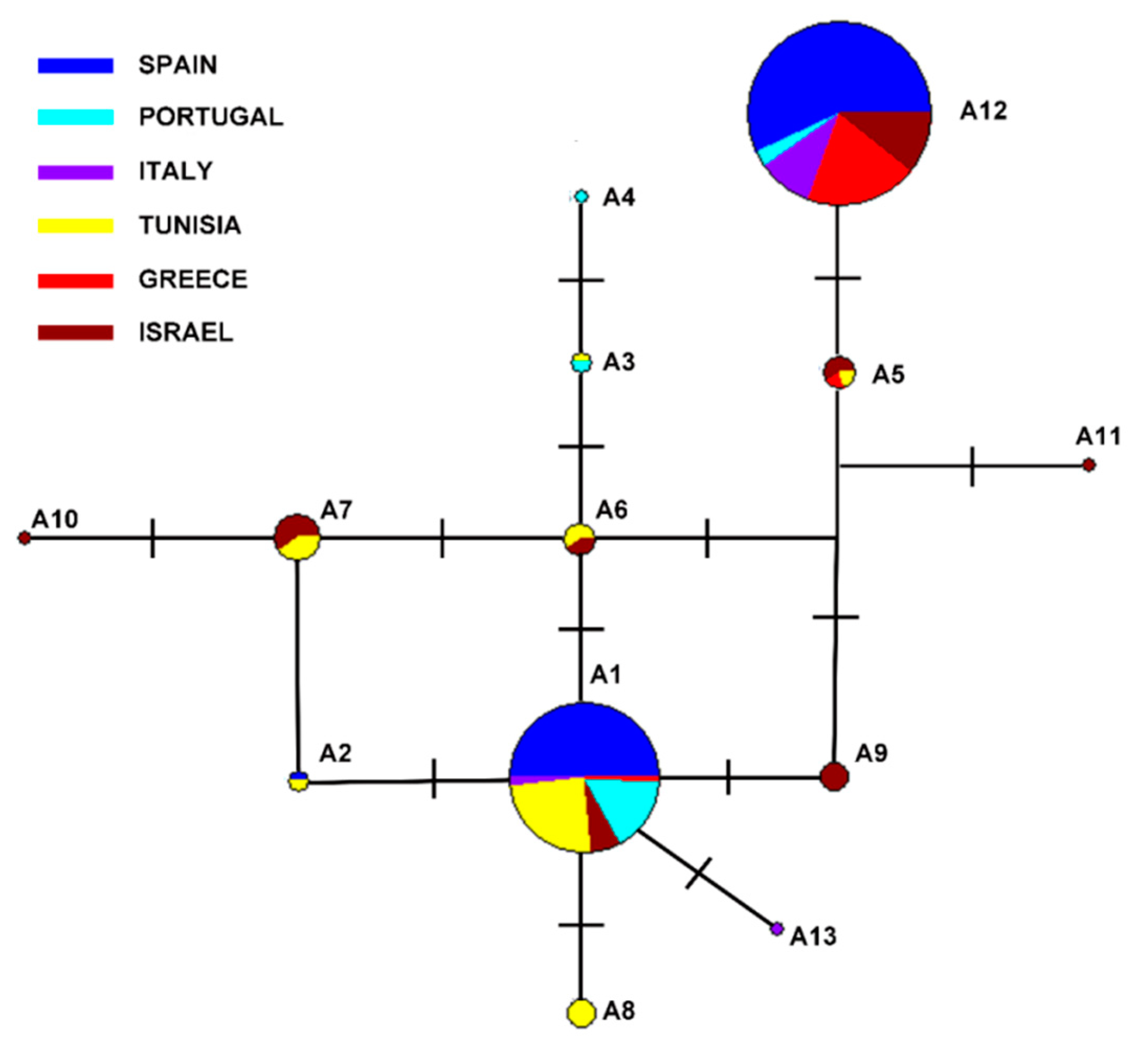
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- PROSODOL. Available online: http://www.prosodol.gr (accessed on 1 January 2015).
- Ministerio de Agricultura Pesca y Alimentación (MAPA) 2017. Available online: https://www.mapa.gob.es/es/estadistica/temas/estadisticas-agrarias/boletinfrutales2017_tcm30-499069.pdf (accessed on 25 June 2020).
- Food and Agriculture Organization of the United Nations (FAO). Available online: http://www.fao.org/home/es/ (accessed on 25 June 2020).
- Agencia de Información y Control Alimentarios (AICA). Available online: http://www.aica.gob.es (accessed on 25 June 2020).
- Sparks, T.C.; Nauen, R. IRAC: Mode of action classification and insecticide resistance management. Pestic. Biochem. Physiol. 2014, 121, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Budzinski, H.; Couderchet, M. Environmental and human health issues related to pesticides: From usage and environmental fate to impact. Environ. Sci. Pollut. R 2018, 25, 14277–14279. [Google Scholar] [CrossRef] [PubMed]
- Brühl, C.A.; Zaller, J.G. Biodiversity decline as a consequence of an inappropriate environmental risk assessment of pesticides. Front. Environ. Sci. 2019. [Google Scholar] [CrossRef]
- Mulé, R.; Sabella, G.; Robba, L.; Manachini, B. Systematic Review of the Effects of Chemical Insecticides on Four Common Butterfly Families. Front. Environ. Sci. 2017. [Google Scholar] [CrossRef]
- Sánchez-Bayo, F.; Goka, K. Pesticide Residues and Bees—A Risk Assessment. PLoS ONE 2014. [Google Scholar] [CrossRef]
- Sanchez-Bayo, F.; Wyckhuys, K.A.G. Worldwide decline of the entomofauna: A review of its drivers. Biol. Conserv. 2019, 232, 8–27. [Google Scholar] [CrossRef]
- Kimbras, C.B.; Tsakas, S. The genetics of Dacus oleae V. Changes of esterase polymorphism in a natural population following insecticide control-selection or drift? Evolution 1971, 25, 454–460. [Google Scholar]
- Vontas, J.G.; Hejazi, M.J.; Hawkes, N.J.; Cosmidis, N.; Loukas, M.; Hemingway, J. Resistance associated point mutations of organophosphate insensitive acetylcholinesterase in the olive fruit fly Bactrocera oleae. Insect Mol. Biol. 2002, 11, 329–336. [Google Scholar] [CrossRef]
- Kakani, E.G.; Trakala, M.; Drosopoulou, E.; Mavragani-Tsipidou, P.; Mathiopoulos, K.D. Genomic structure, organization and localization of the acetylcholinesterase locus of the olive fruit fly, Bactrocera oleae. Bull. Entomol. Res. 2013, 103, 36–47. [Google Scholar] [CrossRef]
- Kakani, E.G.; Ioannides, I.M.; Margaritopoulos, T.J.; Seraphides, N.A.; Skouras, P.J.; Tsitsipis, J.A.; Mathiopoulos, K.D.A. Small delection in the olive fly acetylcholinesterase gene associated with high levels of organophosphate resistance. Insect Biochem. Mol. Biol. 2008, 38, 781–787. [Google Scholar] [CrossRef]
- Vontas, J.G.; Hernández-Crespo, P.; Margaritopoulos, J.T.; Ortego, F.; Feng, H.T.; Mathiopoulos, K.D.; Hsu, J.C. Insecticide resistance in tephritid flies. Pestic. Biochem. Physiol. 2011, 100, 199–205. [Google Scholar] [CrossRef]
- Hawkes, N.J.; Janes, R.W.; Hemingway, J.; Vontas, J. Detection of resistance associated point mutations of organophosphate insensitive acetylcholinesterase in the olive fruit fly Bactrocera oleae (Gmelin). Pestic. Biochem. Physiol. 2005, 81, 154–163. [Google Scholar] [CrossRef]
- Dogaç, E.; Kandemir, I.; Taskin, V. Geographical distribution and frequencies of organophosphate- resistant ace alleles and morphometric variations in olive fruit fly populations. Pest. Manag. Sci. 2014, 71, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Kakani, E.G.; Sagri, E.; Omirou, M.; Ioannides, I.M.; Mathiopoulos, K.D. Detection and geographical distribution of the organophosphate resistance associated Δ3Q ace mutation in the olive fruit fly, Bactrocera oleae (Rossi). Pest. Manag. Sci. 2014, 70, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Kampouraki, A.; Marianna, S.; Karataraki, A.; Katsikogiannis, G.; Pitika, E.; Varikou, K.; Vlachaki, A.; Chrysargys, A.; Malandraki, E.; Sidiropoulos, N.; et al. Recent evolution and operational impact of insecticide resistance in olive fruit fly Bactrocera oleae populations from Greece. J. Pest. Sci. 2018, 91, 1429–1439. [Google Scholar] [CrossRef]
- Skouras, P.J.; Margaritopoulos, J.; Seraphides, N.; Ioannides, I.; Kakani, E.; Mathiopoulos, K.; Tsitsipis, J. Organophosphate resistance in the olive fruit fly Bactrocera oleae populations in Greece and Cyprus. J. Pest. Sci. 2007, 63, 42–48. [Google Scholar] [CrossRef]
- Phytoma. Noticias de Actualidad. IRAC Alerta del Riesgo de Resistencias de Bactrocera oleae a Resistencias, 317/11/ 2019. Available online: https://www.phytoma.com/ (accessed on 15 November 2019).
- Pereira-Castro, I.; Van-Asch, B.; Trinidade-Rei, F.; Teixeira Da Costa, L. Bactrocera oleae (Diptera: Tephritidae) organophosphate resistance alleles in Iberia: Recent expansion and variable frequencies. Eur. J. Entomol. 2015, 112, 20–26. [Google Scholar] [CrossRef]
- Nobre, T.; Gomes, L.; Trinidade Rei, F. A Re-evaluation of olive fruit fly organophosphate resistant ace alleles in Iberia and field-testing population effects after in-practice dimethoate use. Insects 2018. [Google Scholar] [CrossRef]
- European Commission. Available online: https://ec.europa.eu/info/food-farming-fisheries_es (accessed on 25 June 2020).
- Pu, J.; Wang, Z.; Chung, H. Climate change and the genetics of insecticide resistance. Pest. Manag. Sci. 2020. [Google Scholar] [CrossRef]
- Nardi, F.; Carapelli, A.; Vontas, J.N.; Dallai, R.; Roderick GKFrati, F. Geographical distribution and evolutionary history of organophosphate-resistant Ace alleles in the olive fly (Bactrocera oleae). Insect Biochem. Mol. Biol. 2006, 36, 593–602. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal X window interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 24, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Rozas, J.; Sánchez Del Barrio, J.C.; Messenguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2008, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Vontas, J.G.; Cosmidis, N.; Loukas, M.; Tsakas, S.; Hejazi, M.J.; Ayoutani, A.; Hemingway, J. Altered Acetylcholinesterase confers organophosphate resistance in the olive fruit fly Bactrocera oleae. Pestic. Biochem. Physiol. 2001, 71, 124–132. [Google Scholar] [CrossRef]
- Matallanas, B.; Lantero, E.; M’Saad, M.; Callejas, C.; Ochando, M.D. Genetic polymorphism at the cytochrome oxidase I gene in mediterranean populations of Bactrocera oleae (Diptera: Tephritidae). J. Appl. Entomol. 2013, 137, 624–630. [Google Scholar] [CrossRef]
- Lantero, E. Estudio genético de la plaga del olivo Bactrocera oleae (Rossi 1790) y su aplicación al control biológico. Ph.D. Thesis, Universidad Complutense de Madrid, Madrid, Spain, 2018. [Google Scholar]
- Daane, K.M.; Johnson, M.W. Olive fruit fly: Managing an ancient pest in modern times. Ann. Rev. Entomol. 2010, 55, 151–169. [Google Scholar] [CrossRef] [PubMed]
- ASAJA (Asociación Agraria de Jóvenes Agricultores). Jaén: Portugal Apuesta por la Modernización del Olivar, Aunque sólo el 23% de su Superficie es de Riego, 20 December 2017. Available online: https://www.asajajaen.com/ (accessed on 25 June 2020).
- Mutero, A.; Pravalorio, M.; Bride, J.M.; Fournier, D. Resistance associated point mutations in insecticide insensitive acetylcholinesterase. Proc. Natl. Acad. Sci. USA 1994, 91, 5922–5926. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, J.L.; Nardi, F.; Zhang, R.J. Population genetic structure of the melon fly, Bactrocera cucurbitae (Diptera: Tephritidae), from China and Southeast Asia. Genetica 2008, 134, 319–324. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Rural Development of Israel, MoAg. Available online: https://www.moag.gov.il/en/ (accessed on 25 June 2020).
- Elfekih, S.; Shannon, M.; Haran, J.; Vogler, A.P. Detection of the Acetylcholinesterase Insecticide Resistance Mutation (G328A) in Natural Populations of Ceratitis capitata. J. Econ. Entomol. 2014, 107, 1965–1968. [Google Scholar] [CrossRef][Green Version]
- Cheikh, H.B.; Ali-Haouas, Z.B.; Marquine, M.; Pasteur, N. Resistance to Organophosphorus and Pyrethroid Insecticides in Culex pipiens (Diptera: Culicidae) from Tunisia. J. Med. Entomol. 1998, 35, 251–260. [Google Scholar] [CrossRef]
- Ffrench-Constant, R.H. The molecular genetics of insecticide resistance. Genetics 2013, 194, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Hartley, C.J.; Newcomb, R.D.; Russell, R.J.; Yong, C.G.; Stevens, J.R.; Yeates, D.K.; La Salle, J.; Oakeshott, J.G. Amplification of DNA from preserved specimens shows blowflies were preadapted for the rapid evolution of insecticide resistance. Proc. Natl. Acad. Sci. USA 2006, 103, 8757–8762. [Google Scholar] [CrossRef] [PubMed]
- McCart, C.; Buckling, A.; Ffrench-Constant, R.H. DDT resistance in flies carries no cost. Curr. Biol. 2005, 15, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Pavlidi, N.; Kampouraki, A.; Tseliou, V.; Wybouw, N.; Dermauw, W.; Roditakis, E.; Nauen, R.; Van Leeuwen, T.; Vontas, J. Molecular characterization of Pyrethroid resistance in the olive fruit fly Bactrocera oleae. Pestic. Biochem. Physiol. 2018, 148, 1–7. [Google Scholar] [CrossRef] [PubMed]
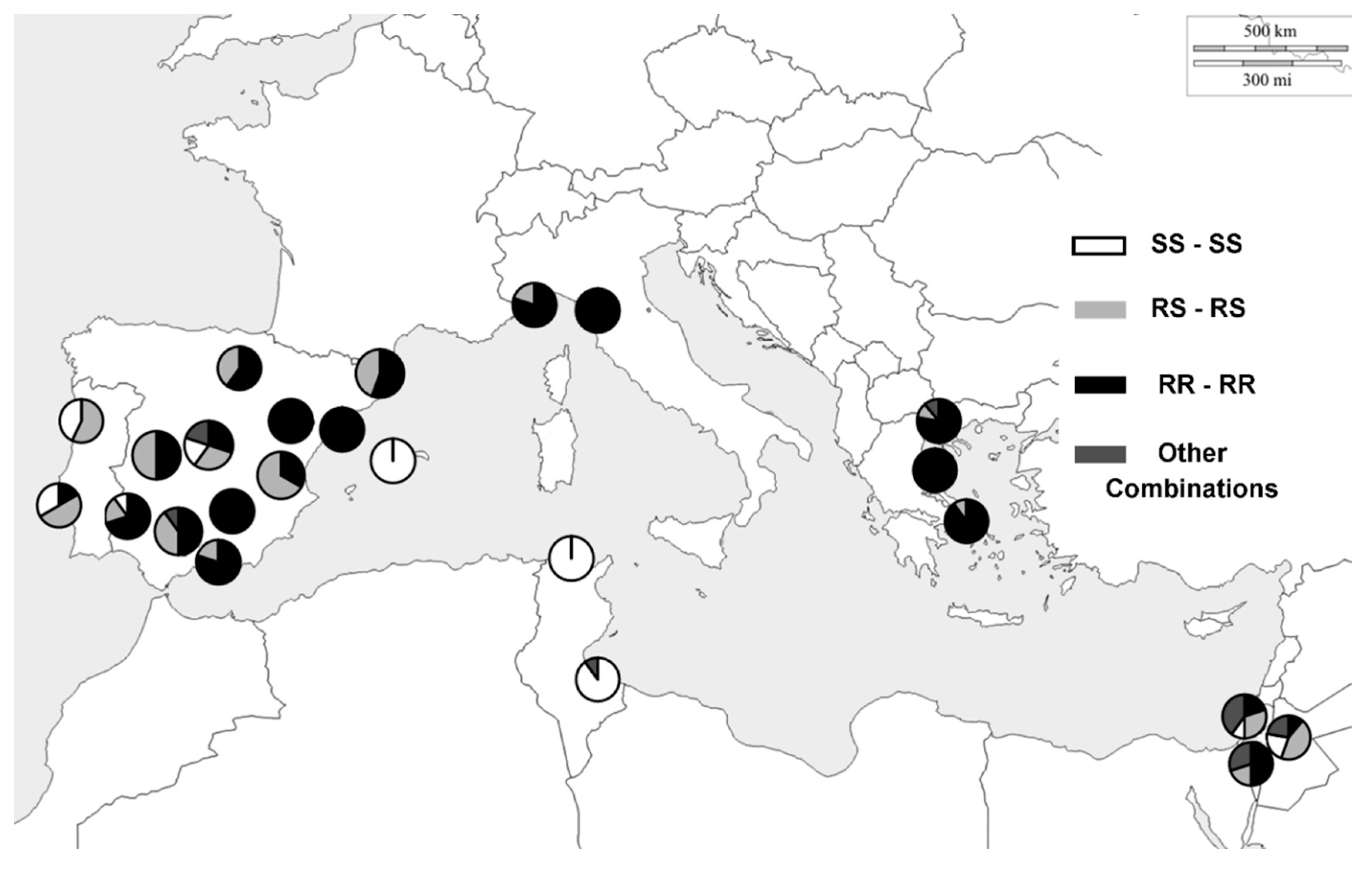
| CODE | LOCALITY/COUNTRY | LAT | LONG | N |
|---|---|---|---|---|
| SPA1 | Morata de Tajuña, Madrid, ES | 40.2275 | −3.4369 | 10 |
| SPA2 | Arróniz, Navarra, ES | 42.4222 | −2.0913 | 11 |
| SPA3 | Tortosa, Tarragona, ES | 40.811 | 0.5209 | 10 |
| SPA4 | Montemolín, Badajoz, ES | 38.1552 | −6.2069 | 10 |
| SPA5 | Mallorca, Baleares Isles, ES | 39.6952 | 3.0175 | 10 |
| SPA6 | Castañar de Ibor, Cáceres, ES | 39.6277 | −5.4166 | 10 |
| SPA7 | Campus Rabanales, Córdoba, ES | 37.2647 | −4.6327 | 10 |
| SPA8 | El Cortalet, Gerona, ES | 42.2253 | 3.0970 | 10 |
| SPA9 | Íllora, Granada, ES | 37.3461 | −3.8727 | 9 |
| SPA10 | La Iruela, Jaén, ES | 37.9469 | −2.9583 | 10 |
| SPA11 | La Portellada, Teruel, ES | 40.89 | −0.0336 | 9 |
| SPA12 | Requena, Valencia, ES | 39.4878 | −1.1003 | 6 |
| POR1 | Fundao, Castelo Branco, PT | 40.1369 | −7.4994 | 7 |
| POR2 | Lisbon, Lisbon, PT | 38.7069 | −9.1356 | 6 |
| ITA1 | Diana Marina, Liguria, IT | 43.9098 | 8.0818 | 10 |
| ITA2 | Pisa, Toscana, IT | 43.7498 | 10.5497 | 10 |
| TUN1 | Sidi Thabet, Ariana, TN | 36.9081 | 10.0222 | 10 |
| TUN3 | Zarzis, Médenine, TN | 33.523 | 11.0852 | 10 |
| GRE1 | Agiá, Tesalia, GR | 39.7188 | 22.7550 | 10 |
| GRE2 | Salónica, Tesalónica, GR | 40.6393 | 22.9446 | 9 |
| GRE3 | Atenas, Central Atenas, GR | 37.9791 | 23.7166 | 10 |
| ISR1 | Jerusalem, Jerusalem District, IL | 31.7383 | 35.2137 | 9 |
| ISR2 | Rehovot, Central District, IL | 31.8927 | 34.8112 | 10 |
| ISR3 | Lahav Forest, Southern District, IL | 31.3725 | 34.8408 | 10 |
| 73 | 88 | 122 | 259 | 274 | 316 | 406 | 412 | 415 | 484 | Freq | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | T | T | A | G | A | C | G | G | G | T | 24.1% |
| A2 | * | * | * | * | * | T | * | * | * | * | 0.44% |
| A3 | * | * | * | * | * | * | * | * | T | C | 0.44% |
| A4 | * | * | * | * | C | * | * | * | T | C | 0.22% |
| A5 | C | * | * | C | * | * | * | A | T | * | 1.11% |
| A6 | * | * | * | * | * | * | * | * | T | * | 1.11% |
| A7 | * | * | * | * | * | T | * | * | T | * | 2.23% |
| A8 | * | G | * | * | * | * | * | * | * | * | 0.88% |
| A9 | C | * | * | * | * | * | * | * | * | * | 0.88% |
| A10 | * | * | * | T | * | T | * | * | T | * | 0.22% |
| A11 | C | * | * | C | * | * | A | * | T | * | 0.22% |
| A12 | C | * | G | C | * | * | * | A | T | * | 67.85% |
| A13 | * | * | G | * | * | * | * | * | * | * | 0.22% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lantero, E.; Matallanas, B.; Pascual, S.; Ochando, M.D.; Callejas, C. Phylogeography of Organophosphate Resistant ace Alleles in Spanish Olive Fruit Fly Populations: A Mediterranean Perspective in the Global Change Context. Insects 2020, 11, 396. https://doi.org/10.3390/insects11060396
Lantero E, Matallanas B, Pascual S, Ochando MD, Callejas C. Phylogeography of Organophosphate Resistant ace Alleles in Spanish Olive Fruit Fly Populations: A Mediterranean Perspective in the Global Change Context. Insects. 2020; 11(6):396. https://doi.org/10.3390/insects11060396
Chicago/Turabian StyleLantero, Esther, Beatriz Matallanas, Susana Pascual, M. Dolores Ochando, and Carmen Callejas. 2020. "Phylogeography of Organophosphate Resistant ace Alleles in Spanish Olive Fruit Fly Populations: A Mediterranean Perspective in the Global Change Context" Insects 11, no. 6: 396. https://doi.org/10.3390/insects11060396
APA StyleLantero, E., Matallanas, B., Pascual, S., Ochando, M. D., & Callejas, C. (2020). Phylogeography of Organophosphate Resistant ace Alleles in Spanish Olive Fruit Fly Populations: A Mediterranean Perspective in the Global Change Context. Insects, 11(6), 396. https://doi.org/10.3390/insects11060396







