RNA Interference-Based Silencing of the Chitin Synthase 1 Gene for Reproductive and Developmental Disruptions in Panonychus citri
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Rearing of Panonychus citri
2.2. Expression Profiles of the Target Gene
2.3. Preparation of Target Gene Double-Stranded RNA (dsRNA)
2.4. Sample Preparation for RNAi Bioassays
2.5. Impacst of the Target Gene dsRNA on the Expression Patterns of Chitin Synthase 1 Gene Regulation in Different Stages of Panonychus citri
2.6. Time–Mortality Response of the Panonychus citri to Target Gene dsRNAs
2.7. Effects of Target Gene dsRNA on the Reproduction Potential of the Panonychus citri
2.8. Impact of the Target Gene dsRNA on the Chitin Contents of Panonychus citri
2.9. Histological Analysis to Observe Alterations
3. Results
3.1. Expression Profile of the PcCHS1Gene in Different Developmental Stages of the Panonychus citri
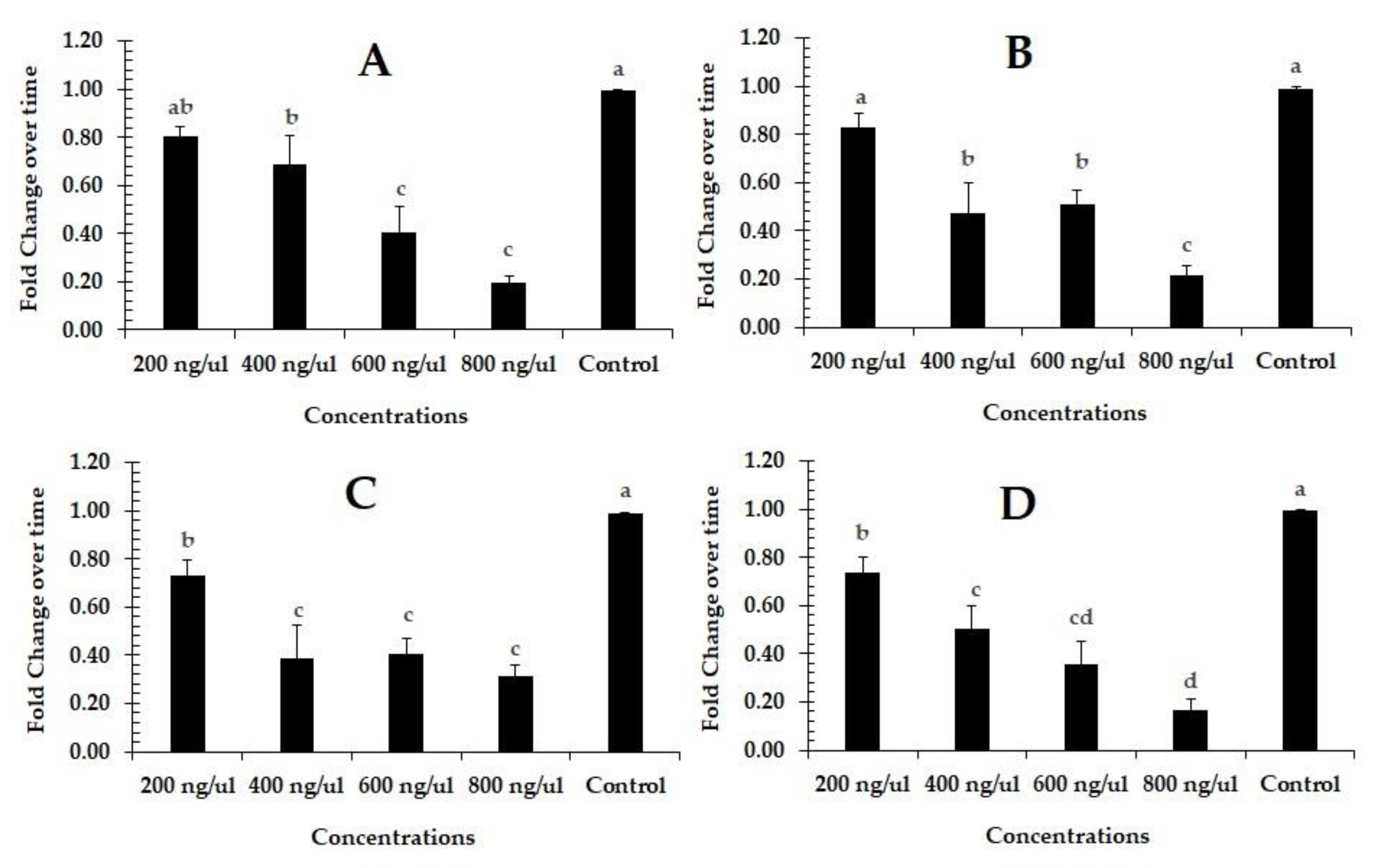
3.2. RNAi-Based Silencing of PcCHS1 in Different Developmental Stages of the Panonychus citri
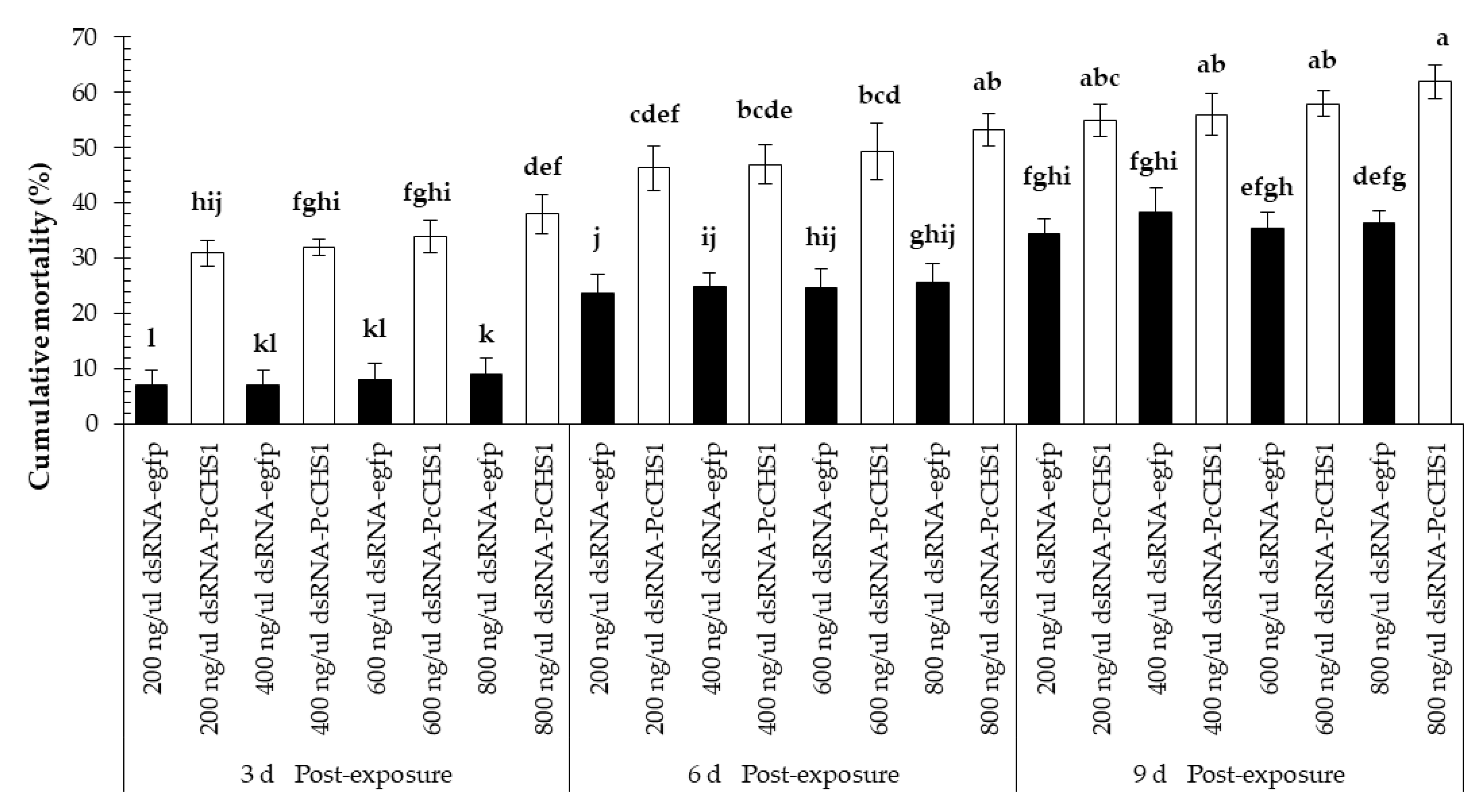
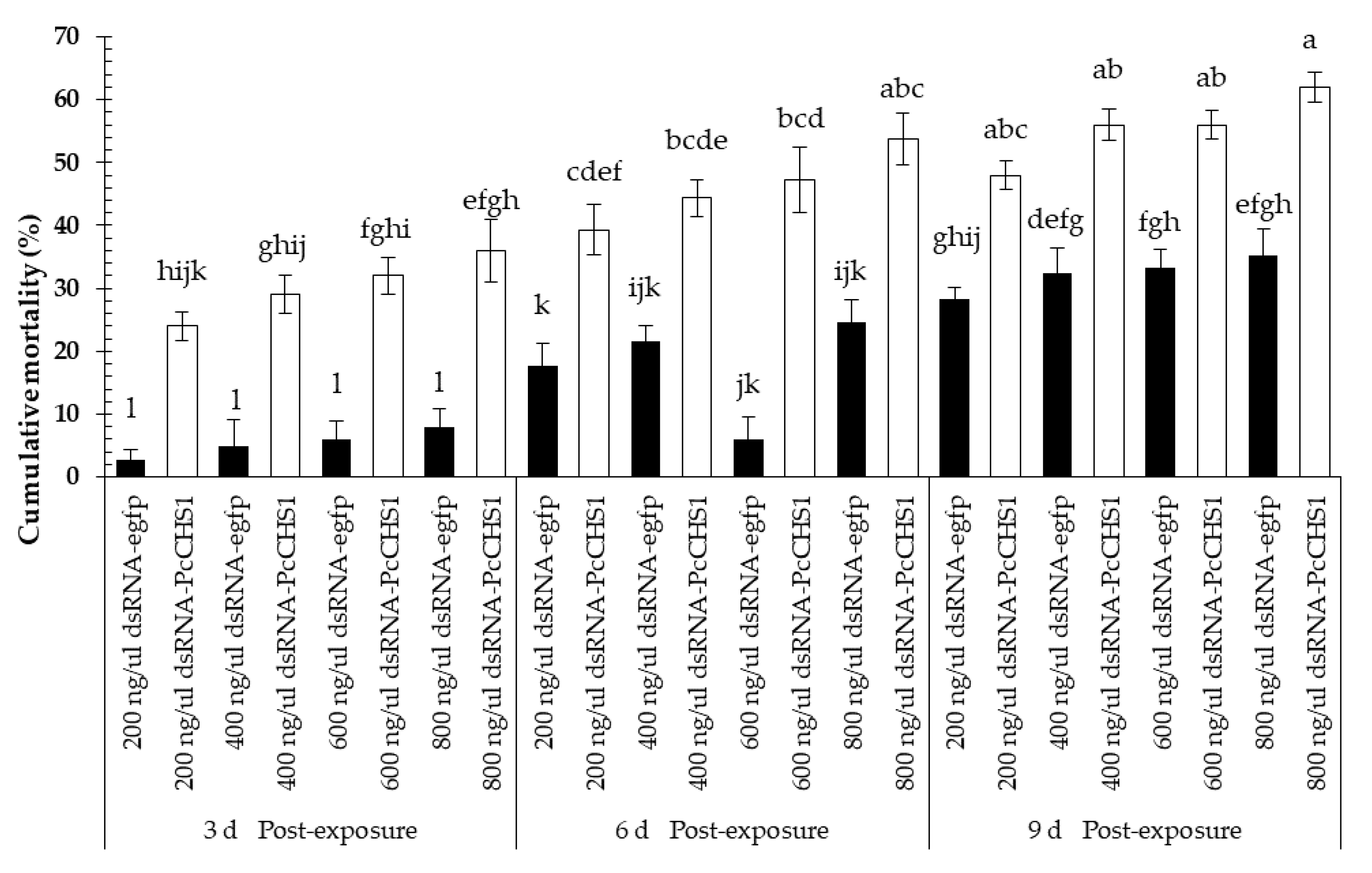
3.3. Time–Mortality Response in Different Developmental Stages of the Panonychus citri
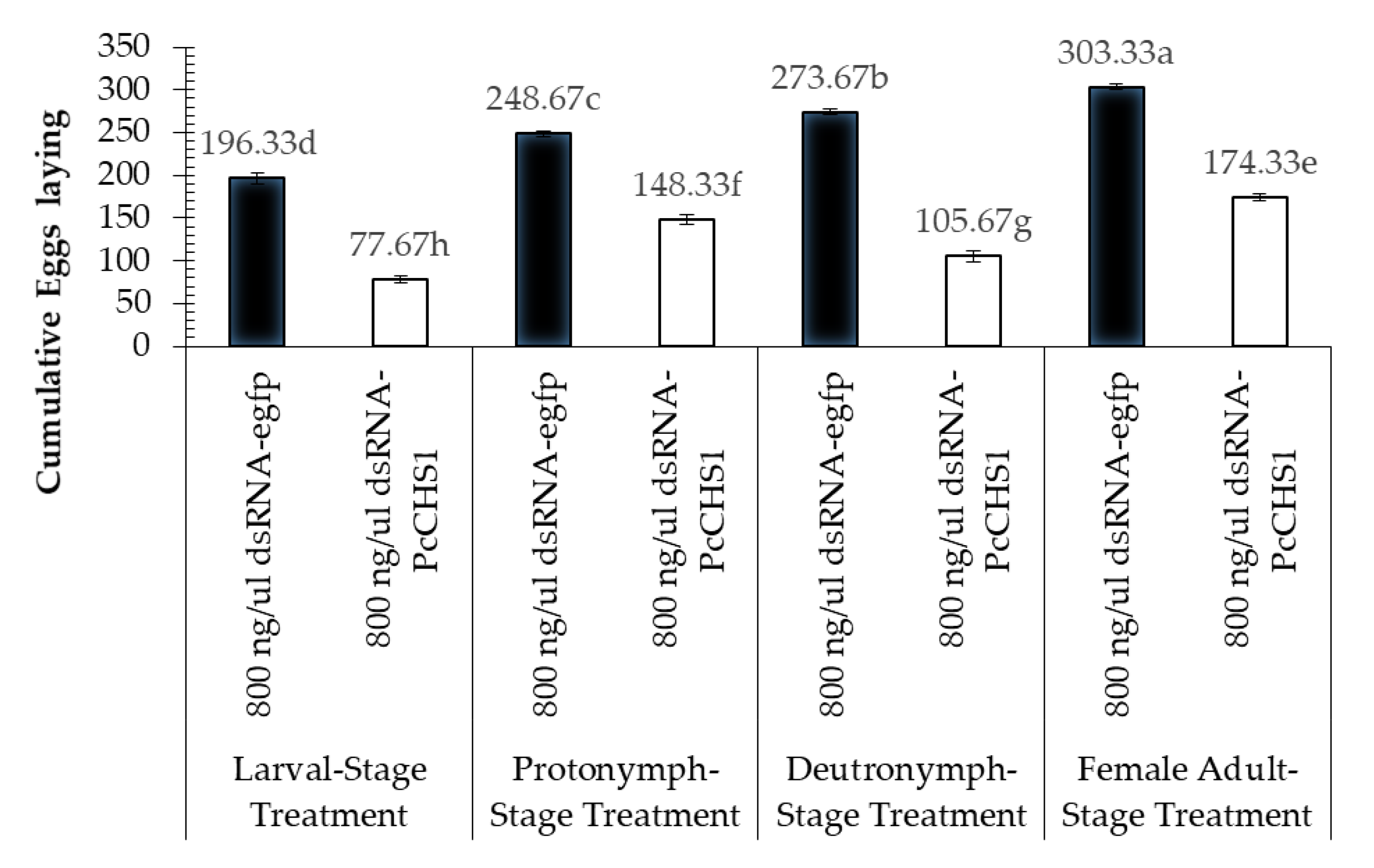
3.4. Impact of Target Gene Silencing on the Reproduction Potential of Panonychus citri
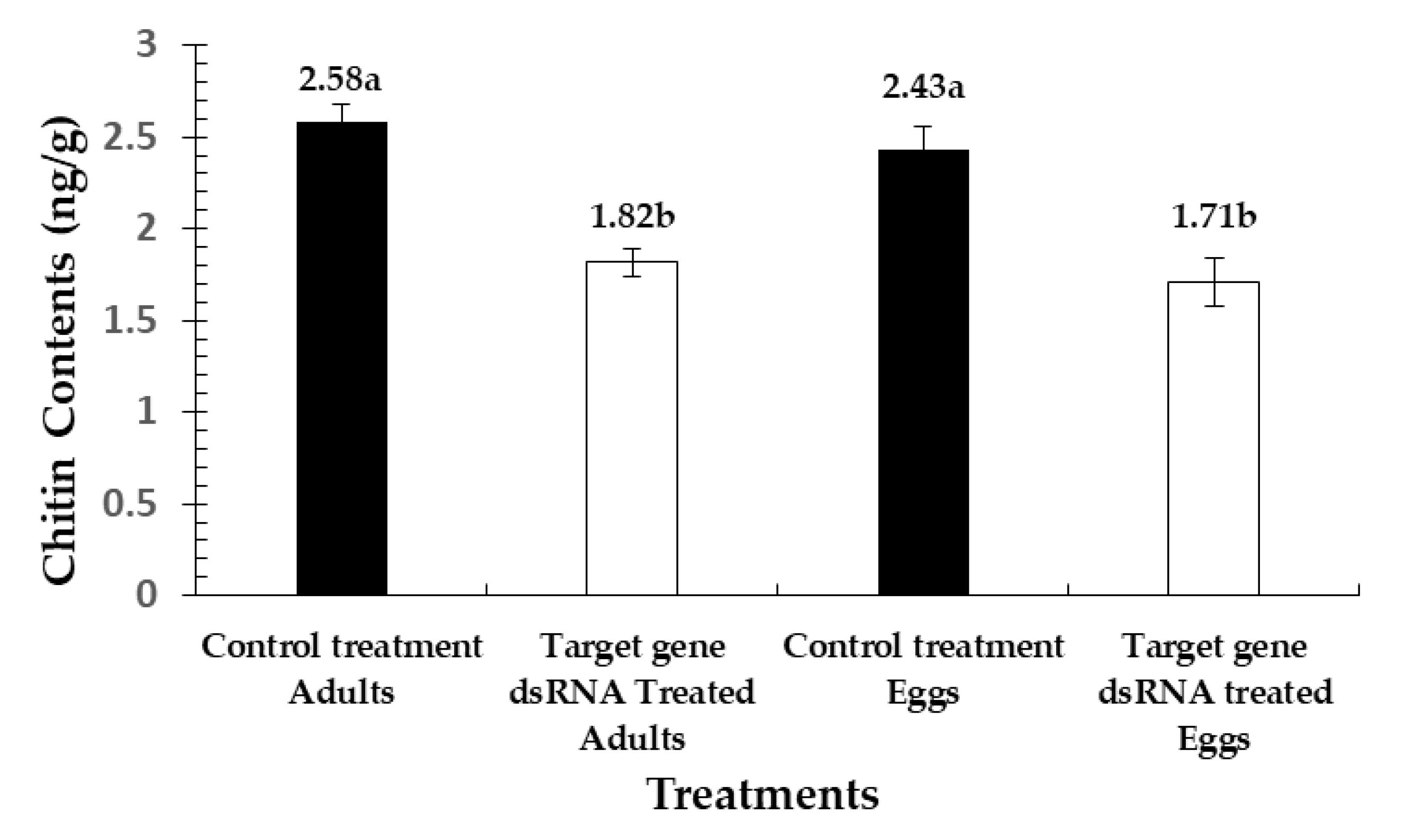
3.5. Disruptions in Chitin Contents
3.6. Cuticular Disruption and Cellular Abnormalities
3.7. Chitin Reduction in the Cuticles of Panonychus citri
3.8. Transmission Electron Microscopic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Vassiliou, V.A.; Papadoulis, G. First record of the citrus red mite Panonychus citri in Cyprus. Phytoparasitica 2009, 37, 99–100. [Google Scholar] [CrossRef]
- Takafuji, A.; Fujimoto, H. Winter Survival of the Non-Diapausing Population of the Citrus Red Mite, Panonychus citri (Mcgregor) (Acarina: Tetranychidae) on Pear and Citrus. Appl. Entomol. Zool. 1986, 21, 467–473. [Google Scholar] [CrossRef]
- Lee, M.H.; Cho, S.H.; Park, H.S.; Bahn, J.W.; Lee, B.J.; Son, J.W.; Kim, Y.K.; Koh, Y.Y.; Min, K.U.; Kim, Y.Y. Citrus red mite (Panonychus citri) is a common sensitizing allergen among children living around citrus orchards. Ann. Allergy Asthma Immunol. 2000, 85, 200–204. [Google Scholar] [CrossRef]
- Ding, T.B.; Niu, J.Z.; Yang, L.H.; Zhang, K.; Dou, W.; Wang, J.J. Transcription profiling of two cytochrome P450 genes potentially involved in acaricide metabolism in citrus red mite Panonychus citri. Pestic. Biochem. Physiol. 2013, 106, 28–37. [Google Scholar] [CrossRef]
- Yamamoto, A.; Yoneda, H.; Hatano, R.; Asada, M. Genetic Analysis of Hexythiazox Resistance in the Citrus Red Mite, Panonychus citri (McGregor). J. Pestic. Sci. 1995, 20, 513–519. [Google Scholar] [CrossRef]
- Hu, J.; Wang, C.; Wang, J.; You, Y.; Chen, F. Monitoring of resistance to spirodiclofen and five other acaricides in Panonychus citri collected from Chinese citrus orchards. Pest Manag. Sci. 2010, 66, 1025–1030. [Google Scholar] [CrossRef]
- Arakane, Y.; Hogenkamp, D.G.; Zhu, Y.C.; Kramer, K.J.; Specht, C.A.; Beeman, R.W.; Kanost, M.R.; Muthukrishnan, S. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem. Mol. Biol. 2004, 34, 291–304. [Google Scholar] [CrossRef]
- Merzendorfer, H. Insect chitin synthases: A review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2006, 176, 1–15. [Google Scholar] [CrossRef]
- Merzendorfer, H. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef]
- Moreira, M.F.; dos Santos, A.S.; Marotta, H.R.; Mansur, J.F.; Ramos, I.B.; Machado, E.A.; Souza, G.H.M.F.; Eberlin, M.N.; Kaiser, C.R.; Kramer, K.J.; et al. A chitin-like component in Aedes aegypti eggshells, eggs and ovaries. Insect Biochem. Mol. Biol. 2007, 37, 1249–1261. [Google Scholar] [CrossRef]
- Souza-Ferreira, P.S.; Mansur, J.F.; Berni, M.; Moreira, M.F.; Dos Santos, R.E.; Araújo, H.M.M.; De Souza, W.; Ramos, I.B.; Masuda, H. Chitin deposition on the embryonic cuticle of Rhodnius prolixus: The reduction of CHS transcripts by CHS-dsRNA injection in females affects chitin deposition and eclosion of the first instar nymph. Insect Biochem. Mol. Biol. 2014, 51, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Specht, C.A.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2008, 38, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; AlJabr, A.M.; Al-Ayedh, H. Development-Disrupting Chitin Synthesis Inhibitor, Novaluron, Reprogramming the Chitin Degradation Mechanism of Red Palm Weevils. Molecules 2019, 24, 4304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, J.; Zhu, K.Y. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Gagou, M.E.; Kapsetaki, M.; Turberg, A.; Kafetzopoulos, D. Stage-specific expression of the chitin synthase DmeChSA and DMeChSB genes during the onset of drosophila metamorphosis. Insect Biochem. Mol. Biol. 2002, 32, 141–146. [Google Scholar] [CrossRef]
- Bolognesi, R.; Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Terra, W.R.; Ferreira, C. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2005, 35, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Specht, C.A.; Tomoyasu, Y.; Lorenzen, M.D.; Kanost, M.; Beeman, R.W. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol. Biol. 2005, 14, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Zimoch, L.; Merzendorfer, H. Immunolocalization of chitin synthase in the tobacco hornworm. Cell Tissue Res. 2002, 308, 287–297. [Google Scholar] [CrossRef]
- Lehane, M.J. Peritrophic matrix structure and function. Annu. Rev. Entomol. 1997, 42, 525–550. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next-Generation Insect-Resistant Plants: RNAi-Mediated Crop Protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef]
- Wang, M.; Thomas, N.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi for powerful innovative pre- and post-harvest plant protection. Curr. Opin. Plant Biol. 2017, 38, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.W.; Zhang, Z.Y.; Xia, S.; Zhang, H. Biofunctional analysis of Vitellogenin and Vitellogenin receptor in citrus red mites, Panonychus citri by RNA interference. Sci. Rep. 2017, 7, 16123. [Google Scholar] [CrossRef] [PubMed]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Tian, M.-Y.; Wen, S.-Y. Exploring the caste-specific multi-layer defense mechanism of Formosan Subterranean Termites, Coptotermes formosanus Shiraki. Int. J. Mol. Sci. 2017, 18, 2694. [Google Scholar] [CrossRef]
- Handler, A.M.; Harrell, R.A. Transformation of the Caribbean fruit fly, Anastrepha suspensa, with a piggyBac vector marked with polyubiquitin-regulated GFP. Insect Biochem. Mol. Biol. 2001, 31, 199. [Google Scholar] [CrossRef]
- Hussain, A.; Ali, M.W.; Aljabr, A.M.; Al-Kahtani, S.N. Insights into the gryllus bimaculatus immunerelated transcriptomic profiling to combat naturally invading pathogens. J. Fungi 2020, 6, 232. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xia, W.K.; Shen, X.M.; Ding, T.B.; Niu, J.Z.; Zhong, R.; Liao, C.Y.; Feng, Y.C.; Dou, W.; Wang, J.J. Functional analysis of a chitinase gene during the larval-nymph transition in Panonychus citri by RNA interference. Exp. Appl. Acarol. 2016, 70, 1–15. [Google Scholar] [CrossRef]
- Shah, S.; Hafeez, M.; Wu, M.Y.; Zhang, S.S.; Ilyas, M.; Wu, G.; Yang, F.L. Down regulation of chitin synthase A gene by diallyl trisulfide, an active substance from garlic essential oil, inhibits oviposition and alters the morphology of adult Sitotroga cerealella. J. Pest Sci. 2020, 93, 1097–1106. [Google Scholar] [CrossRef]
- Xia, W.K.; Ding, T.B.; Niu, J.Z.; Liao, C.Y.; Zhong, R.; Yang, W.J.; Liu, B.; Dou, W.; Wang, J.J. Exposure to diflubenzuron results in an up-regulation of a chitin synthase 1 gene in citrus red mite, Panonychus citri (Acari: Tetranychidae). Int. J. Mol. Sci. 2014, 15, 253–281. [Google Scholar] [CrossRef]
- Beeman, R.W. Recent Advances in Mode of Action of Insecticides. Annu. Rev. Entomol. 1982, 27, 253–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, K.Y. Characterization of a chitin synthase cDNA and its increased mRNA level associated with decreased chitin synthesis in Anopheles quadrimaculatus exposed to diflubenzuron. Insect Biochem. Mol. Biol. 2006, 36, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Koehler, P.G.; Patterson, R.S. Effects of Chitin Synthesis Inhibitors on German Cockroach (Orthoptera: Blattellidae) Mortality and Reproduction. J. Econ. Entomol. 1989, 82, 143–148. [Google Scholar] [CrossRef]
- Mothes-Wagner, U. Effects of the chitin synthesis inhibitor complex nikkomycin on oogenesis in the mite Tetranychus urticae. Pestic. Sci. 1984, 15, 455–461. [Google Scholar] [CrossRef]
- Miura, T.; Schaefer, C.H.; Takahashi, R.M.; Mulligan, F.S. Effects of the insect growth inhibitor, Dimilin, on hatching of mosquito eggs. J. Econ. Entomol. 1976, 69, 655–658. [Google Scholar] [CrossRef]
- Wilson, T.G.; Cryan, J.R. Lufenuron, a chitin-synthesis inhibitor, interrupts development of Drosophila melanogaster. J. Exp. Zool. 1997, 278, 37–44. [Google Scholar] [CrossRef]
- Sáenz-de-cabezón, F.J.; Pérez-moreno, I.; Zalom, F.G.; Marco, V. Effects of Lufenuron on Lobesia botrana (Lepidoptera: Tortricidae) Egg, Larval, and Adult Stages. J. Econ. Entomol. 2009, 99, 427–431. [Google Scholar] [CrossRef]
- Braga, I.A.; Mello, C.B.; Peixoto, A.A.; Valle, D. Evaluation of methoprene effect on Aedes aegypti (Diptera: Culicidae) development in laboratory conditions. Mem. Inst. Oswaldo Cruz 2005, 100, 435–440. [Google Scholar] [CrossRef]
- Ge, L.-Q.; Xia, T.; Huang, B.; Gu, H.-T.; Song, Q.-S.; Yang, G.-Q.; Liu, F.; Wu, J.-C. PHF7, a novel male gene influences female fecundity and population growth in Nilaparvata lugens Stål (Hemiptera: Delphacidae). Sci. Rep. 2017, 7, 11611. [Google Scholar] [CrossRef]
- Al-Ayedh, H.; Rizwan-ul-Haq, M.; Hussain, A.; Aljabr, A.M. Insecticidal potency of RNAi-based catalase knockdown in Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae). Pest Manag. Sci. 2016, 72, 2118–2127. [Google Scholar] [CrossRef]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA Interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, J.; Yao, Q.; Pan, Z.; Chen, J.; Zhang, W. Functional analysis of two chitinase genes during the pupation and eclosion stages of the beet armyworm Spodoptera exigua by rna interference. Arch. Insect Biochem. Physiol. 2012, 79, 220–234. [Google Scholar] [CrossRef]
- Ye, C.; Jiang, Y.D.; An, X.; Yang, L.; Shang, F.; Niu, J.; Wang, J.J. Effects of RNAi-based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum). Sci. Rep. 2019, 9, 3694. [Google Scholar] [CrossRef] [PubMed]
- Ioriatti, C.; Pasqualini, E.; Pasquier, D.; Tomasi, C. Efficacy baselines of seven insecticides against larvae of Pandemis heparana (Lepidoptera: Tortricidae). J. Pest Sci. 2006, 79, 163–168. [Google Scholar] [CrossRef]
- Acheuk, F.; Cusson, M.; Doumandji-Mitiche, B. Effects of a methanolic extract of the plant Haplophyllum tuberculatum and of teflubenzuron on female reproduction in the migratory locust, Locusta migratoria (Orthoptera: Oedipodinae). J. Insect Physiol. 2012, 58, 335–341. [Google Scholar] [CrossRef]
- Farnesi, L.C.; Brito, J.M.; Linss, J.G.; Pelajo-Machado, M.; Valle, D.; Rezende, G.L. Physiological and morphological aspects of Aedes aegypti developing larvae: Effects of the chitin synthesis inhibitor novaluron. PLoS ONE 2012, 7, e30363. [Google Scholar] [CrossRef]
- Zhuo, W.; Fang, Y.; Kong, L.; Li, X.; Sima, Y.; Xu, S. Chitin synthase A: A novel epidermal development regulation gene in the larvae of Bombyx mori. Mol. Biol. Rep. 2014, 41, 4177–4186. [Google Scholar] [CrossRef]
- Ostrowski, S.; Dierick, H.A.; Bejsovec, A. Genetic control of cuticle formation during embryonic development of Drosophila melanogaster. Genetics 2002, 161, 171–182. [Google Scholar]
- Belinato, T.A.; Martins, A.J.; Lima, J.B.P.; de Lima-Camara, T.N.; Peixoto, A.A.; Valle, D. Effect of the chitin synthesis inhibitor triflumuron on the development, viability and reproduction of Aedes aegypti. Mem. Inst. Oswaldo Cruz 2009, 104, 43–47. [Google Scholar] [CrossRef]
- López, J.D.; Latheef, M.A.; Hoffmann, W.C. Effect of Hexaflumuron on Feeding Response and Reproduction of Bollworm, Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae) 1. Southwest. Entomol. 2011, 36, 247–259. [Google Scholar] [CrossRef]
- Ampasala, D.R.; Zheng, S.; Zhang, D.; Ladd, T.; Doucet, D.; Krell, P.J.; Retnakaran, A.; Feng, Q. An epidermis-specific chitin synthase cDNA in Choristoneura fumiferana: Cloning, characterization, developmental and hormonal-regulated expression. Arch. Insect Biochem. Physiol. 2011, 76, 83–96. [Google Scholar] [CrossRef]
- Matsumura, F. Studies on the action mechanism of benzoylurea insecticides to inhibit the process of chitin synthesis in insects: A review on the status of research activities in the past, the present and the future prospects. Pestic. Biochem. Physiol. 2010, 97, 133–139. [Google Scholar] [CrossRef]
- Tang, B.; Chen, X.; Liu, Y.; Tian, H.; Liu, J.; Hu, J.; Xu, W.; Zhang, W. Characterization and expression patterns of a membrane-bound trehalase from Spodoptera exigua. BMC Mol. Biol. 2008, 9, 51. [Google Scholar] [CrossRef] [PubMed]
- Tatun, N.; Singtripop, T.; Tungjitwitayakul, J.; Sakurai, S. Regulation of soluble and membrane-bound trehalase activity and expression of the enzyme in the larval midgut of the bamboo borer Omphisa fuscidentalis. Insect Biochem. Mol. Biol. 2008, 38, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Zhang, D.; Tang, B.; Chen, J.; Chen, J.; Lu, L.; Zhang, W. Identification of 20-hydroxyecdysone late-response genes in the chitin biosynthesis pathway. PLoS ONE 2010, 5, e14058. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, J.; Zhu, K.Y. Chito-Protein Matrices in Arthropod Exoskeletons and Peritrophic Matrices; Springer International Publishing Imprint, Springer: Cham, Switzerland, 2019; Volume 12. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.W.; Khan, M.M.; Song, F.; Wu, L.; He, L.; Wang, Z.; Zhang, Z.-y.; Zhang, H.; Jiang, Y. RNA Interference-Based Silencing of the Chitin Synthase 1 Gene for Reproductive and Developmental Disruptions in Panonychus citri. Insects 2020, 11, 786. https://doi.org/10.3390/insects11110786
Ali MW, Khan MM, Song F, Wu L, He L, Wang Z, Zhang Z-y, Zhang H, Jiang Y. RNA Interference-Based Silencing of the Chitin Synthase 1 Gene for Reproductive and Developmental Disruptions in Panonychus citri. Insects. 2020; 11(11):786. https://doi.org/10.3390/insects11110786
Chicago/Turabian StyleAli, Muhammad Waqar, Muhammad Musa Khan, Fang Song, Liming Wu, Ligang He, Zhijing Wang, Zhen-yu Zhang, Hongyu Zhang, and Yingchun Jiang. 2020. "RNA Interference-Based Silencing of the Chitin Synthase 1 Gene for Reproductive and Developmental Disruptions in Panonychus citri" Insects 11, no. 11: 786. https://doi.org/10.3390/insects11110786
APA StyleAli, M. W., Khan, M. M., Song, F., Wu, L., He, L., Wang, Z., Zhang, Z.-y., Zhang, H., & Jiang, Y. (2020). RNA Interference-Based Silencing of the Chitin Synthase 1 Gene for Reproductive and Developmental Disruptions in Panonychus citri. Insects, 11(11), 786. https://doi.org/10.3390/insects11110786






