Evaluation of RNA Interference for Control of the Grape Mealybug Pseudococcus maritimus (Hemiptera: Pseudococcidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. RNA Extraction
2.3. Design of Primers for Amplification of Ps. maritimus and Pl. ficus Genes
2.4. cDNA Synthesis and Sequencing
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Synthesis of dsRNA
2.7. RNAi Experiments
2.8. Species Specificity of dsRNA Reagents
2.9. Statistical Analysis
3. Results
3.1. Identification of Target Genes in Pseudococcus maritimus and Design of dsRNA Sequences
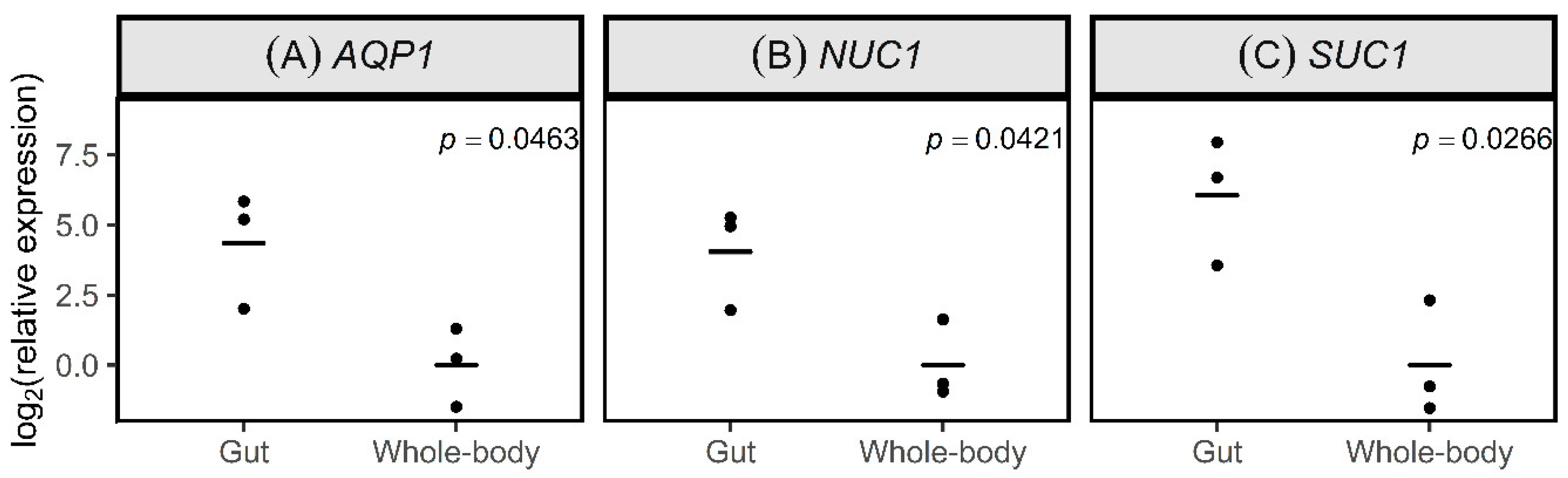
3.2. Expression of Target Genes in the Gut of Ps. maritimus
3.3. Response of Ps. maritimus to Orally Delivered RNAi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Daane, K.M.; Almeida, R.P.P.; Bell, V.A.; Walker, J.T.S.; Botton, M.; Fallahzadeh, M.; Mani, M.; Miano, J.L.; Sforza, R.; Walton, V.M.; et al. Biology and Management of Mealybugs in Vineyards; Bostanian, N.J., Isaacs, R., Vincent, C., Eds.; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Atallah, S.S.; Gomez, M.I.; Fuchsberger, C.; Martinson, T.E. Economic impact of grapevine leafroll disease on Vitis vinifera cv. Cabernet franc in Finger Lakes vineyards of New York. Am. J. Enol. Viticult. 2012, 63, 73–79. [Google Scholar] [CrossRef]
- Cooper, M.L.; Daugherty, M.P.; Jeske, D.R.; Almeida, R.P.P.; Daane, K.M. Incidence of grapevine leafroll disease: Effects of grape mealybug (Pseudococcus maritimus) abundance and pathogen supply. J. Econ. Entomol. 2018, 111, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Wallingford, A.K.; Fuchs, M.F.; Martinson, T.; Hesler, S.; Loeb, G.M. Slowing the Spread of Grapevine Leafroll-Associated Viruses in Commercial Vineyards With Insecticide Control of the Vector, Pseudococcus maritimus (Hemiptera: Pseudococcidae). J. Insect. Sci. 2015, 15, 112. [Google Scholar] [CrossRef]
- Weigle, T.H.; Muza, A.J. (Eds.) New York and Pennsylvania Pest Management Guidelines for Grapes; A Cornell and Penn State Cooperative Extension Publication: Ithaca, NY, USA, 2020. [Google Scholar]
- Cooper, A.M.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2019, 10, 1912. [Google Scholar] [CrossRef] [PubMed]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Zhu, K.Y.; Palli, S.R. Mechanisms, Applications, and Challenges of Insect RNA Interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Cagliari, D.; Dias, N.P.; Galdeano, D.M.; Dos Santos, E.A.; Smagghe, G.; Zotti, M.J. Management of Pest Insects and Plant Diseases by Non-Transformative RNAi. Front. Plant Sci. 2019, 10, 1319. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E. Strategies for Enhanced Crop Resistance to Insect Pests. Annu. Rev. Plant Biol. 2018, 69, 637–660. [Google Scholar] [CrossRef] [PubMed]
- Zotti, M.; Dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [PubMed]
- Husnik, F.; Nikoh, N.; Koga, R.; Ross, L.; Duncan, R.P.; Fujie, M.; Tanaka, M.; Satoh, N.; Bachtrog, D.; Wilson, A.C.; et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 2013, 153, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Gupta, M.; Pandher, S.; Kaur, G.; Goel, N.; Rathore, P. Using de novo transcriptome assembly and analysis to study RNAi in Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae). Sci. Rep. 2019, 9, 13710. [Google Scholar] [PubMed]
- Khan, A.M.; Ashfaq, M.; Khan, A.A.; Naseem, M.T.; Mansoor, S. Evaluation of potential RNA-interference-target genes to control cotton mealybug, Phenacoccus solenopsis (Hemiptera: Pseudococcuidae). Insect Sci. 2018, 25, 778–786. [Google Scholar]
- Khan, A.M.; Ashfaq, M.; Kiss, Z.; Khan, A.A.; Mansoor, S.; Falk, B.W. Use of recombinant tobacco mosaic virus to achieve RNA interference in plants against the citrus mealybug, Planococcus citri (Hemiptera: Pseudococcidae). PLoS ONE 2013, 8, e73657. [Google Scholar]
- Omar, M.A.A.; Ao, Y.; Li, M.; He, K.; Xu, L.; Tong, H.; Jiang, M.; Li, F. The functional difference of eight chitinase genes between male and female of the cotton mealybug, Phenacoccus solenopsis. Insect Mol. Biol. 2019, 28, 550–567. [Google Scholar]
- Mathew, L.G.; Campbell, E.M.; Yool, A.J.; Fabrick, J.A. Identification and characterization of functional aquaporin water channel protein from alimentary tract of whitefly, Bemisia tabaci. Insect Biochem. Mol. Biol. 2011, 41, 178–190. [Google Scholar] [CrossRef]
- Price, D.R.; Karley, A.J.; Ashford, D.A.; Isaacs, H.V.; Pownall, M.E.; Wilkinson, H.S.; Gatehouse, J.A.; Douglas, A.E. Molecular characterisation of a candidate gut sucrase in the pea aphid, Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 2007, 37, 307–317. [Google Scholar]
- Shakesby, A.J.; Wallace, I.S.; Isaacs, H.V.; Pritchard, J.; Roberts, D.M.; Douglas, A.E. A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem. Mol. Biol. 2009, 39, 1–10. [Google Scholar] [CrossRef]
- Ashford, D.A.; Smith, W.A.; Douglas, A.E. Living on a high sugar diet: The fate of sucrose ingested by a phloem-feeding insect, the pea aphid Acyrthosiphon pisum. J. Insect Physiol. 2000, 46, 335–341. [Google Scholar] [CrossRef]
- Karley, A.J.; Ashford, D.A.; Minto, L.M.; Pritchard, J.; Douglas, A.E. The significance of gut sucrase activity for osmoregulation in the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 2005, 51, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.; Ashford, D.; Pritchard, J.; Douglas, A. Honeydew sugars and osmoregulation in the pea aphid Acyrthosiphon pisum. J. Exp. Biol. 1997, 200, 2137–2143. [Google Scholar] [PubMed]
- Douglas, A.E. Nutritional physiology of aphids. Adv. Insect Physiol. 2003, 31, 73–140. [Google Scholar]
- Van Ekert, E.; Chauvigne, F.; Finn, R.N.; Mathew, L.G.; Hull, J.J.; Cerda, J.; Fabrick, J.A. Molecular and functional characterization of Bemisia tabaci aquaporins reveals the water channel diversity of hemipteran insects. Insect Biochem. Mol. Biol. 2016, 77, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, Q.; Luan, J.; Chung, S.H.; Van Eck, J.; Turgeon, R.; Douglas, A.E. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol. 2017, 88, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Malik, H.J.; Shafiq, M.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Mansoor, S. RNA Interference based Approach to Down Regulate Osmoregulators of Whitefly (Bemisia tabaci): Potential Technology for the Control of Whitefly. PLoS ONE 2016, 11, e0153883. [Google Scholar] [CrossRef] [PubMed]
- Santos-Ortega, Y.; Killiny, N. Silencing of sucrose hydrolase causes nymph mortality and disturbs adult osmotic homeostasis in Diaphorina citri (Hemiptera: Liviidae). Insect Biochem. Mol. Biol. 2018, 101, 131–143. [Google Scholar] [CrossRef]
- Tzin, V.; Yang, X.; Jing, X.; Zhang, K.; Jander, G.; Douglas, A.E. RNA interference against gut osmoregulatory genes in phloem-feeding insects. J. Insect Physiol. 2015, 79, 105–112. [Google Scholar] [CrossRef]
- Jing, X.; White, T.A.; Luan, J.; Jiao, C.; Fei, Z.; Douglas, A.E. Evolutionary conservation of candidate osmoregulation genes in plant phloem sap-feeding insects. Insect Mol. Biol. 2016, 25, 251–258. [Google Scholar] [CrossRef]
- Scott, J.G.; Michel, K.; Bartholomay, L.C.; Siegfried, B.D.; Hunter, W.B.; Smagghe, G.; Zhu, K.Y.; Douglas, A.E. Towards the elements of successful insect RNAi. J. Insect Physiol. 2013, 59, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.D.; Liu, Z.C.; Huang, S.L.; Chen, Z.Q.; Sun, Y.W.; Duan, P.F.; Ma, Y.Z.; Xia, L.Q. RNAi-mediated plant protection against aphids. Pest Manag. Sci. 2016, 72, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Whyard, S.; Vélez, A.M.; Smagghe, G. Double-stranded RNA technology to control insect pests: Current status and challenges. Front. Plant Sci. 2020, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.H.; Jing, X.; Luo, Y.; Douglas, A.E. Targeting symbiosis-related insect genes by RNAi in the pea aphid-Buchnera symbiosis. Insect Biochem. Mol. Biol. 2018, 95, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Cousins, P. Tiny grape could do big things. Agric. Res. 2007, 55, 23. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Douglas, A.E.; Minto, L.B.; Wilkinson, T.L. Quantifying nutrient production by the microbial symbionts in an aphid. J. Exp. Biol. 2001, 204, 349–358. [Google Scholar]
- Luck, S.; Kreszies, T.; Strickert, M.; Schweizer, P.; Kuhlmann, M.; Douchkov, D. siRNA-Finder (si-Fi) Software for RNAi-Target Design and Off-Target Prediction. Front. Plant Sci. 2019, 10, 1023. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015; Available online: https://www.R-project.org/ (accessed on 7 September 2019).
- Bates, D.; Machler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme-4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, K.; Chen, J.; Wang, J.; Zhang, H.; Ze, L.; Zhu, G.; Zhao, C.; Xiao, H.; Han, Z. Identification of a double-stranded RNA-degrading nuclease influencing both ingestion and injection RNA interference efficiency in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 2020, 125, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, K.; Fu, W.; Sheng, C.; Han, Z. Biochemical Comparison of dsRNA Degrading Nucleases in Four Different Insects. Front. Physiol. 2018, 9, 624. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Li, D.; Cooper, A.M.W.; Silver, K.; Li, T.; Liu, X.; Ma, E.; Zhu, K.Y.; Zhang, J. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem. Mol. Biol. 2017, 86, 68–80. [Google Scholar] [CrossRef]
- Spit, J.; Philips, A.; Wynant, N.; Santos, D.; Plaetinck, G.; Vanden Broeck, J. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2017, 81, 103–116. [Google Scholar] [CrossRef]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef]
- Wynant, N.; Santos, D.; Verdonck, R.; Spit, J.; Van Wielendaele, P.; Vanden Broeck, J. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2014, 46, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tayler, A.; Heschuk, D.; Giesbrecht, D.; Park, J.Y.; Whyard, S. Efficiency of RNA interference is improved by knockdown of dsRNA nucleases in tephritid fruit flies. Open Biol. 2019, 9, 190198. [Google Scholar] [CrossRef]
- Chung, S.H.; Parker, B.J.; Blow, F.; Brisson, J.A.; Douglas, A.E. Host and symbiont genetic determinants of nutritional phenotype in a natural population of the pea aphid. Mol. Ecol. 2020, 29, 848–858. [Google Scholar] [CrossRef]
- Huang, J.H.; Jing, X.; Douglas, A.E. The multi-tasking gut epithelium of insects. Insect Biochem. Mol. Biol. 2015, 67, 15–20. [Google Scholar] [CrossRef]
- Ohlstein, B.; Spradling, A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 2006, 439, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Kragler, F. Long distance RNA movement. New Phytol. 2018, 218, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Flenniken, M.L.; Andino, R. Non-Specific dsRNA-mediated antiviral response in the honey bee. PLoS ONE 2013, 8, e77263. [Google Scholar] [CrossRef] [PubMed]
- Nunes, F.M.F.; Aleixo, A.C.; Barchuk, A.R.; Bomtorin, A.D.; Grozinger, C.M.; Simões, Z.L.P. Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assays. Insects 2013, 4, 90–103. [Google Scholar] [CrossRef]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef]
- Wuriyanghan, H.; Rosa, C.; Falk, B.W. Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli. PLoS ONE 2011, 6, e27736. [Google Scholar] [CrossRef]
- Prentice, K.; Smagghe, G.; Gheysen, G.; Christiaens, O. Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. Biol. 2019, 110, 80–89. [Google Scholar] [CrossRef]
- Fellmann, C.; Lowe, S.W. Stable RNA interference rules for silencing. Nat. Cell. Biol. 2014, 16, 10–18. [Google Scholar] [CrossRef]
- Doench, J.G.; Petersen, C.P.; Sharp, P. A siRNAs can function as miRNAs. Genes Dev. 2003, 17, 438–442. [Google Scholar] [CrossRef]
- Zeng, Y.; Yi, R.; Cullen, B.R. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784. [Google Scholar] [CrossRef]
- Roberts, A.F.; Devos, Y.; Lemgo, G.N.; Zhou, X. Biosafety research for non-target organism risk assessment of RNAi-based GE plants. Front. Plant Sci. 2015, 6, 958. [Google Scholar]
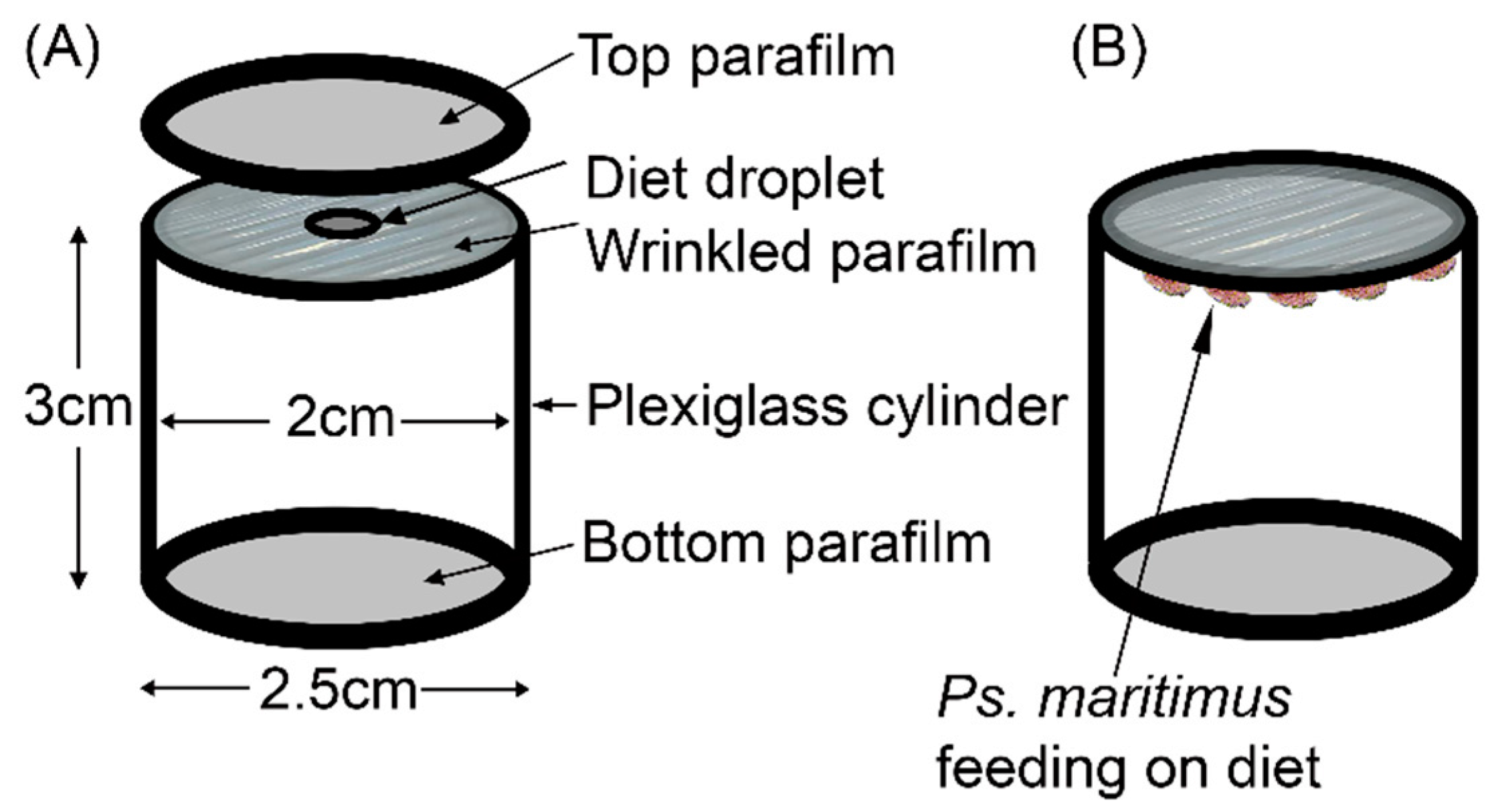


| Treatment | Double Stranded RNA (dsRNA) in Diet | |
|---|---|---|
| Day 0 to Day 2 Larvae | Day 2 to Day 7 Larvae | |
| Diet | dsRNA free | dsRNA free |
| G | dsRNA free | dsGFP (0.4 µg µL−1) |
| N+G | dsNUC1 (0.2 µg µL−1) | dsGFP (0.2 µg µL−1) + dsNUC1 (0.2 µg µL−1) |
| A+S | dsRNA free | dsAQP1 (0.2 µg µL−1) + dsSUC1 (0.2 µg µL−1) |
| N+A+S | dsNUC1 (0.2 µg µL−1) | dsAQP1 (0.1 µg µL−1) + dsSUC1 (0.1 µg µL−1) + dsNUC1 (0.2 µg µL−1) |
| dsRNA Sequence of Ps. maritimus | Insect Species | Number of Predicted Targets of dsRNA-Derived 21 nt Predicted Target Sequences |
|---|---|---|
| dsAQP1 | Planococcus citri | 27 |
| Planococcus ficus | 17 | |
| Acyrthosiphon pisum | 0 | |
| Myzus persicae | 0 | |
| dsSUC1 | Planococcus citri | 0 |
| Planococcus ficus | 0 | |
| Acyrthosiphon pisum | 0 | |
| Myzus persicae | 0 | |
| dsNUC1 | Planococcus citri | 9 |
| Planococcus ficus | 1 | |
| Acyrthosiphon pisum | 0 | |
| Myzus persicae | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arora, A.K.; Clark, N.; Wentworth, K.S.; Hesler, S.; Fuchs, M.; Loeb, G.; Douglas, A.E. Evaluation of RNA Interference for Control of the Grape Mealybug Pseudococcus maritimus (Hemiptera: Pseudococcidae). Insects 2020, 11, 739. https://doi.org/10.3390/insects11110739
Arora AK, Clark N, Wentworth KS, Hesler S, Fuchs M, Loeb G, Douglas AE. Evaluation of RNA Interference for Control of the Grape Mealybug Pseudococcus maritimus (Hemiptera: Pseudococcidae). Insects. 2020; 11(11):739. https://doi.org/10.3390/insects11110739
Chicago/Turabian StyleArora, Arinder K., Noah Clark, Karen S. Wentworth, Stephen Hesler, Marc Fuchs, Greg Loeb, and Angela E. Douglas. 2020. "Evaluation of RNA Interference for Control of the Grape Mealybug Pseudococcus maritimus (Hemiptera: Pseudococcidae)" Insects 11, no. 11: 739. https://doi.org/10.3390/insects11110739
APA StyleArora, A. K., Clark, N., Wentworth, K. S., Hesler, S., Fuchs, M., Loeb, G., & Douglas, A. E. (2020). Evaluation of RNA Interference for Control of the Grape Mealybug Pseudococcus maritimus (Hemiptera: Pseudococcidae). Insects, 11(11), 739. https://doi.org/10.3390/insects11110739







