Comparative Transcriptome Analysis Reveals the Mechanism Related to Fluazinam Stress of Panonychus citri (Acarina: Tetranychidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. P. citri Samples
2.2. RNA Extraction and Library Construction
2.3. Assembly Sequencing and Functional Annotation
2.4. Differential Expression Analysis
2.5. Enrichment and KEGG Enrichment Analysis
3. Results
3.1. Splicing Transcript Length Distribution
3.2. Gene Annotation Success Rate Statistics
3.3. Unigene Homology Analysis
3.4. GO Function Analysis
3.5. KOG Function Analysis
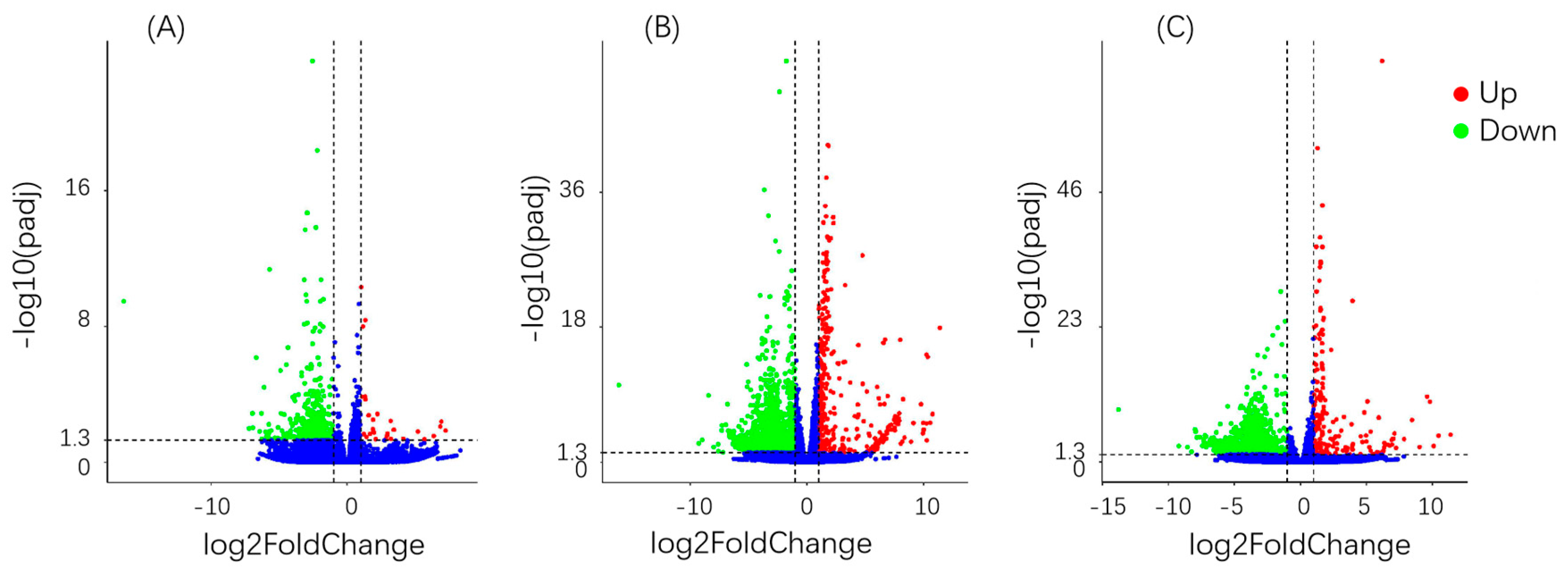
3.6. Differential Gene Analysis
3.7. GO Enriched Gene Analysis
3.8. KEGG Enrichment Pathway Analysis
3.9. Genetic Verification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Livingston, G.; Hack, L.; Steinmann, K.P.; Grafton-Cardwell, E.; Rosenheim, J. An Ecoinformatics Approach to Field-Scale Evaluation of Insecticide Effects in California Citrus: Are Citrus Thrips and Citrus Red Mite Induced Pests? J. Econ. Entomol. 2018, 111, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Ouyang, G.; Lu, H.; Guo, M.; Meng, X.; Liu, H. The effects of different control measures on Panonychus citri and arthropod enemies in citrus orchards. Chin. J. Appl. Entomol. 2013, 50, 413–420. [Google Scholar]
- Jiang, Q. Study on Control Efficiency of the Recommended Concentration of Seven Acaricides on Panonychus citri (McGregor). Pestic. Sci. Admin. 2012, 33, 48–51. (In Chinese) [Google Scholar]
- Hu, J.; Wang, C.; Wang, J.; You, Y.; Chen, F. Monitoring of resistance to spirodiclofen and five other acaricides in Panonychus citri collected from Chinese citrus orchards. Pest Manag. Sci. 2010, 66, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Montez, G.H.; Liu, L.; Grafton-Cardwell, E. Spirodiclofen and spirotetramat bioassays for monitoring resistance in citrus red mite, Panonychus citri (Acari: Tetranychidae). Pest Manag. Sci. 2011, 68, 781–787. [Google Scholar] [CrossRef]
- Niu, J.-Z.; Liu, G.-Y.; Dou, W.; Wang, J. Susceptibility and Activity of GlutathioneS-Transferases in Nine Field Populations of Panonychus citri (Acari: Tetranychidae) to Pyridaben and Azocyclotin. Fla. Entomol. 2011, 94, 321–329. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Van Nieuwenhuyse, P.; Vanholme, B.; Dermauw, W.; Nauen, R.; Tirry, L. Parallel evolution of cytochrome b mediated bifenazate resistance in the citrus red mite Panonychus citri. Insect Mol. Biol. 2010, 20, 135–140. [Google Scholar] [CrossRef]
- Ran, C.; Chen, Y.; Wang, J. Susceptibility and carboxylesterase activity of five field populations of Panonychus citri (McGregor) (Acari: Tetranychidae) to four acaricides. Int. J. Acarol. 2009, 35, 115–121. [Google Scholar] [CrossRef]
- Liao, C.-Y.; Feng, Y.-C.; Li, G.; Shen, X.-M.; Liu, S.-H.; Dou, W.; Wang, J. Antioxidant Role of PcGSTd1 in Fenpropathrin Resistant Population of the Citrus Red Mite, Panonychus citri (McGregor). Front. Physiol. 2018, 9, 314. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Duan, Y.-B.; Zhou, M.-G. Baseline sensitivity and efficacy of fluazinam in controlling Sclerotinia stem rot of rapeseed. Eur. J. Plant Pathol. 2015, 144, 337–343. [Google Scholar] [CrossRef]
- Schepers, H.T.A.M.; Kessel, G.J.T.; Lucca, F.; Förch, M.G.; Bosch, G.B.M.V.D.; Topper, C.G.; Evenhuis, A. Reduced efficacy of fluazinam against Phytophthora infestans in The Netherlands. Eur. J. Plant Pathol. 2018, 151, 947–960. [Google Scholar] [CrossRef] [Green Version]
- Walgenbach, J.; Borchert, D.; Palmer, C.J.I.; Tests, A. Apple, Mite Control with Fluazinam. Insectic. Acaric. Tests 1993, 18, 55. [Google Scholar]
- Wang, Y.B.; Cheng, X.; Wang, Z.R.; Wu, K.L.; Liu, X.Y.; Wu, H.M.; Rao, Q. Toxicity test and field efficacy of seven acaricides against Panonychus citri. Plant Prot. 2019, 45, 292–294. (In Chinese) [Google Scholar]
- Shao, J.; Xu, D.; Li, H.; Mu, W.; Liu, F. Toxicity and modes of action of fluazinam and other fungicides against wheat Fusarium graminearum at different life stages. J. Plant Prot. 2016, 43, 314–320. (In Chinese) [Google Scholar]
- Shao, W.; Zhang, Y.; Ren, W.; Chen, C.-J. Physiological and biochemical characteristics of laboratory induced mutants of Botrytis cinerea with resistance to fluazinam. Pestic. Biochem. Physiol. 2015, 117, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.-J.; Miyoshi, H.; Komyoji, T.; Haga, T.; Fujita, T. Uncoupling activity of a newly developed fungicide, fluazinam [3-chloro-N-(3-chloro-2,6-dinitro-4-trifluoromethylphenyl)-5-trifluoromethyl-2-pyridinamine]. Biochim. Biophys. Acta Bioenerg. 1991, 1056, 89–92. [Google Scholar] [CrossRef]
- Lee, J.E.; Kang, J.S.; Ki, Y.-W.; Park, J.H.; Shin, I.C.; Koh, H.C. Fluazinam targets mitochondrial complex I to induce reactive oxygen species-dependent cytotoxicity in SH-SY5Y cells. Neurochem. Int. 2012, 60, 773–781. [Google Scholar] [CrossRef]
- Wang, X.H.; Zheng, S.S.; Huang, T.; Su, L.M.; Zhao, Y.H.; Souders, C.L.; Martyniuk, C.J. Fluazinam impairs oxidative phosphorylation and induces hyper/hypo-activity in a dose specific manner in zebrafish larvae. Chemosphere 2018, 210, 633–644. [Google Scholar] [CrossRef]
- Leroux, P. Recent developments in the mode of action of fungicides. Pestic. Sci. 1996, 47, 191–197. [Google Scholar] [CrossRef]
- He, W.; You, M.; Vasseur, L.; Yang, G.; Xie, M.; Cui, K.; Bai, J.; Liu, C.; Li, X.; Xu, X.; et al. Developmental and insecticide-resistant insights from the de novo assembled transcriptome of the diamondback moth, Plutella xylostella. Genomics 2012, 99, 169–177. [Google Scholar] [CrossRef]
- Oates, C.N.; Külheim, C.; Myburg, A.A.; Slippers, B.; Naidoo, S. The Transcriptome and Terpene Profile of Eucalyptus grandis Reveals Mechanisms of Defense Against the Insect Pest, Leptocybe invasa. Plant Cell Physiol. 2015, 56, 1418–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Jiang, G.; Zhang, Y.; Li, J.; Li, X.; Yue, J.; Chen, J.; Liu, H.; Li, H.; Zhu, S.; et al. Analysis of Transcriptome Differences between Resistant and Susceptible Strains of the Citrus Red Mite Panonychus citri (Acari: Tetranychidae). PLoS ONE 2011, 6, e28516. [Google Scholar] [CrossRef]
- Wang, X.-W.; Luan, J.-B.; Li, J.; Bao, Y.-Y.; Zhang, C.-X.; Liu, S.-S. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genom. 2010, 11, 400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.; Gao, Y.; Liu, Y.; Li, X.; Liang, Y.; Dai, X.; Xu, Y.; Zhou, Y.; Wang, H.-B. Comparative transcriptome profiling reveals candidate genes related to insecticide resistance of Glyphodes pyloalis. Bull. Entomol. Res. 2019, 110, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA Using TRIzol (TRI Reagent). Cold Spring Harb. Protoc. 2010, 2010, 5439. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Lu, L.; Liu, H.; Li, Y.; Su, X.; Zhang, B.; Chen, X. De novo assembly of the transcriptome for Greenbug (Schizaphis graminum Rondani) and analysis on insecticide resistance-related genes. Entomol. Res. 2019, 49, 363–373. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.E.; Tanaka, T.; Miyata, T. A cytochrome P450 gene involved in methidathion resistance in Amblyseius womersleyi Schicha (Acari: Phytoseiidae). Pestic. Biochem. Physiol. 2007, 88, 337–345. [Google Scholar] [CrossRef]
- Pavlidi, N.; Khalighi, M.; Myridakis, A.; Dermauw, W.; Wybouw, N.; Tsakireli, D.; Stephanou, E.G.; Labrou, N.E.; Vontas, J.; Van Leeuwen, T. A glutathione-S-transferase (TuGSTd05) associated with acaricide resistance in Tetranychus urticae directly metabolizes the complex II inhibitor cyflumetofen. Insect Biochem. Mol. Biol. 2017, 80, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Shi, L.; Shen, G.; Xu, Z.; Liu, J.; Pan, Y.; He, L. Characteristics of carboxylesterase genes and their expression-level between acaricide-susceptible and resistant Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Physiol. 2016, 131, 87–95. [Google Scholar] [CrossRef]
- Ding, T.-B.; Niu, J.-Z.; Yang, L.-H.; Zhang, K.; Dou, W.; Wang, J. Transcription profiling of two cytochrome P450 genes potentially involved in acaricide metabolism in citrus red mite Panonychus citri. Pestic. Biochem. Physiol. 2013, 106, 28–37. [Google Scholar] [CrossRef]
- Bu, C.; Li, J.; Wang, X.-Q.; Shi, G.; Peng, B.; Han, J.; Gao, P.; Wang, Y. Transcriptome Analysis of the Carmine Spider Mite, Tetranychus cinnabarinus (Boisduval, 1867) (Acari: Tetranychidae), and Its Response to β-Sitosterol. BioMed Res. Int. 2015, 2015, 794718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuglik, M.T.; Babik, W.; Prokop, Z.M.; Radwan, J. Alternative reproductive tactics and sex-biased gene expression: The study of the bulb mite transcriptome. Ecol. Evol. 2014, 4, 623–632. [Google Scholar] [CrossRef]
- Cabrera, A.R.; Donohue, K.V.; Khalil, S.M.; Scholl, E.; Opperman, C.; Sonenshine, D.E.; Roe, R.M. New approach for the study of mite reproduction: The first transcriptome analysis of a mite, Phytoseiulus persimilis (Acari: Phytoseiidae). J. Insect Physiol. 2011, 57, 52–61. [Google Scholar] [CrossRef]
- Xu, Z.; Zhu, W.; Liu, Y.; Liu, X.; Chen, Q.; Peng, M.; Wang, X.; Shen, G.; He, L. Analysis of Insecticide Resistance-Related Genes of the Carmine Spider Mite Tetranychus cinnabarinus Based on a De novo Assembled Transcriptome. PLoS ONE 2014, 9, e94779. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W. Analysis of Insecticide Resistance-Related Genes of the Carmme Spider Mite Tetranychus cinnabarlnus Based on High-Throughput Sequencing; Southwest University: Chongqing, China, 2013. (In Chinese) [Google Scholar]
- Brandt, U.; Schubert, J.; Geck, P.; Von Jagow, G. Uncoupling activity and physicochemical properties of derivatives of fluazinam. Biochim. Biophys. Acta BBA Bioenerg. 1992, 1101, 41–47. [Google Scholar] [CrossRef]
- Khailova, L.S.; Firsov, A.M.; Kotova, E.A.; Antonenko, Y.N. Interaction of Potent Mitochondrial Uncouplers with Thiol-Containing Antioxidants. Antioxidants 2019, 8, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; He, J.; Chi, C.; Gu, Y. Identification and analysis of icCu/Zn-SOD, Mn-SOD and ecCu/Zn-SOD in superoxide dismutase multigene family of Pseudosciaena crocea. Fish Shellfish Immunol. 2015, 43, 491–501. [Google Scholar] [CrossRef]
- Zelko, I.N.; Mariani, T.J.; Folz, R.J. Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 2002, 33, 337–349. [Google Scholar] [CrossRef]
- Hassan, H.M. Determination of microbial damage caused by oxygen free radicals, and the protective role of superoxide dismutase. Methods Enzymol. 1984, 105, 404–412. [Google Scholar] [CrossRef]
- Ge, B.; Scheller, F.W.; Lisdat, F. Electrochemistry of immobilized CuZnSOD and FeSOD and their interaction with superoxide radicals. Biosens. Bioelectron. 2003, 18, 295–302. [Google Scholar] [CrossRef]
- Li, J.-M.; Su, Y.-L.; Gao, X.; He, J.; Liu, S.-S.; Wang, X.-W. Molecular characterization and oxidative stress response of an intracellular Cu/Zn superoxide dismutase (CuZnSOD) of the whitefly, Bemisia tabaci. Arch. Insect Biochem. Physiol. 2011, 77, 118–133. [Google Scholar] [CrossRef]
- El-Shenawy, N.S. Effects of insecticides fenitrothion, endosulfan and abamectin on antioxidant parameters of isolated rat hepatocytes. Toxicol. In Vitro 2010, 24, 1148–1157. [Google Scholar] [CrossRef]
- Zhang, G.-H.; Liu, H.; Wang, J.; Wang, Z.-Y. Effects of thermal stress on lipid peroxidation and antioxidant enzyme activities of the predatory mite, Neoseiulus cucumeris (Acari: Phytoseiidae). Exp. Appl. Acarol. 2014, 64, 73–85. [Google Scholar] [CrossRef]
- Yang, L.-H.; Huang, H.; Wang, J. Antioxidant responses of citrus red mite, Panonychus citri (McGregor) (Acari: Tetranychidae), exposed to thermal stress. J. Insect Physiol. 2010, 56, 1871–1876. [Google Scholar] [CrossRef]
- Feng, Y.-C.; Liao, C.-Y.; Xia, W.-K.; Jiang, X.-Z.; Shang, F.; Yuan, G.-R.; Wang, J. Regulation of three isoforms of SOD gene by environmental stresses in citrus red mite, Panonychus citri. Exp. Appl. Acarol. 2015, 67, 49–63. [Google Scholar] [CrossRef]
- Zhu, W.Y. Cloning and Function Exploration of Superoxide Dismutase Gene in Panonychus citri; Southwest University: Chongqing, China, 2016. (In Chinese) [Google Scholar]
- Zheng, S.S.; Wang, X.H.; Zhao, Y.H. The Toxic Effect of Fluazinam on Mitochondrial Respiratory Function and Dopamine Neural System in Zebrafish Embryos. Asian J. Ecotoxicol. 2018, 13, 170–178. (In Chinese) [Google Scholar]
- Martelli, F.; Zuo, Z.; Wang, J.; Wong, C.-O.; Karagas, N.E.; Roessner, U.; Rupasinghe, T.; Venkatachalam, K.; Perry, T.; Bellen, H.J.; et al. Low doses of the neonicotinoid insecticide imidacloprid induce ROS triggering neurological and metabolic impairments in Drosophila. Proc. Natl. Acad. Sci. USA 2020, 117, 25840–25850. [Google Scholar] [CrossRef]

| Transcript Length Interval | 200–500 bp | 0.5–1 kbp | 1–2 kbp | >2 kbp | Total |
|---|---|---|---|---|---|
| Number of transcripts | 18,885 | 15,594 | 13,651 | 29,443 | 77,573 |
| Number of genes | 10,492 | 8395 | 4151 | 5793 | 28,831 |
| Databases with Annotation | Number of Genes | Percentage |
|---|---|---|
| NR | 14,734 | 51.1 |
| NT | 9499 | 32.94 |
| KO | 8018 | 27.81 |
| Swiss-Prot | 12,746 | 44.2 |
| GO | 14,277 | 49.51 |
| KOG | 8601 | 29.83 |
| All databases | 4386 | 15.21 |
| At least one database | 18,771 | 65.1 |
| Total unigenes | 28,831 | 100 |
| Time | GO Accession | Description | Term Type | p-Value | DEG Item | DEG List |
|---|---|---|---|---|---|---|
| 6 h | GO: 0009277 | Fungal-type cell wall | cellular component | 8.75 × 10−8 | 6 | 368 |
| GO: 0003824 | Catalytic activity | molecular function | 2.69 × 10−7 | 209 | 368 | |
| GO: 0008152 | Metabolic process | biological process | 1.36 × 10−6 | 255 | 368 | |
| GO: 0055114 | Oxidation–reduction process | biological process | 6.04 × 10−6 | 73 | 368 | |
| GO: 0016491 | Oxidoreductase activity | molecular function | 1.08 × 10−5 | 71 | 368 | |
| GO: 0048037 | Cofactor binding | molecular function | 1.33 × 10−5 | 34 | 368 | |
| GO: 0008610 | Lipid biosynthetic process | biological process | 1.39 × 10−5 | 22 | 368 | |
| GO: 0003857 | 3-Hydroxyacyl-CoA dehydrogenase activity | molecular function | 1.52 × 10−5 | 9 | 368 | |
| GO: 0042430 | Indole-containing compound metabolic process | biological process | 2.48 × 10−5 | 15 | 368 | |
| GO: 0006550 | Isoleucine catabolic process? | biological process | 2.60 × 10−5 | 9 | 368 | |
| 24 h | GO: 0055114 | Oxidation–reduction process | biological process | 4.81 × 10−9 | 263 | 1627 |
| GO: 0016491 | Oxidoreductase activity | molecular function | 3.97 × 10−8 | 253 | 1627 | |
| GO: 0006081 | Cellular aldehyde metabolic process | biological process | 9.11 × 10−6 | 35 | 1627 | |
| GO: 0008152 | Metabolic process | biological process | 1.38 × 10−5 | 1059 | 1627 | |
| GO: 0009277 | Fungal-type cell wall | cellular component | 2.01 × 10−5 | 6 | 1627 | |
| GO: 0044283 | Small molecule biosynthetic process | biological process | 2.72 × 10−5 | 87 | 1627 | |
| GO: 0006550 | Isoleucine catabolic process | biological process | 2.77 × 10−5 | 19 | 1627 | |
| GO: 0006552 | Leucine catabolic process | biological process | 2.77 × 10−5 | 19 | 1627 | |
| GO: 0006574 | valine catabolic process | biological process | 2.77 × 10−5 | 19 | 1627 | |
| GO: 0009083 | Branched-chain amino acid catabolic process | biological process | 2.77 × 10−5 | 19 | 1627 | |
| 48 h | GO: 0048037 | Cofactor binding | molecular function | 1.50 × 10−14 | 103 | 1128 |
| GO: 0050662 | Coenzyme binding | molecular function | 1.75 × 10−13 | 88 | 1128 | |
| GO: 0016491 | Oxidoreductase activity | molecular function | 5.25 × 10−13 | 209 | 1128 | |
| GO: 0008152 | Metabolic process | biological process | 4.18 × 10−11 | 770 | 1128 | |
| GO: 0055114 | Oxidation–reduction process | biological process | 6.79 × 10−11 | 204 | 1128 | |
| GO: 0072330 | Monocarboxylic acid biosynthetic process | biological process | 2.45 × 10−9 | 34 | 1128 | |
| GO: 0003824 | Catalytic activity | molecular function | 2.77 × 10−9 | 617 | 1128 | |
| GO: 0032787 | monocarboxylic acid metabolic process | biological process | 5.14 × 10−9 | 67 | 1128 | |
| GO: 0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | molecular function | 9.08 × 10−9 | 43 | 1128 | |
| GO: 0006550 | isoleucine catabolic process | biological process | 1.44 × 10−8 | 20 | 1128 |
| Gene | Annotation | Time | Fold a | Fold b |
|---|---|---|---|---|
| CYP392A22 | Cytochrome P450 | 6 h | −2.08 | −2.43 |
| CYP385C8 | Cytochrome P450 | 6 h | −2.22 | −1.56 |
| Panonychus citri vitellogenin | Vitellogenin | 24 h | 6.6 | 1.19 |
| Catalase | Catalase-like (Tetranychus urticae) | 24 h | 1.24 | 2.43 |
| Superoxide dismutase 3 | Superoxide dismutase 3 (Panonychus citri) | 24 h | 1.02 | 1.5 |
| Cu.Zn-superoxide dismutase | Cytoplasmic Cu.Zn-superoxide dismutase (Ditylenchus destructor) | 24 h | 1.01 | −1.61 |
| Panonychus citri heat shock gene | Panonychus citri heat shock protein 70-2 mRNA | 24 h | 1.65 | 1.5 |
| Superoxide dismutase (Tieghemostelium lacteum) | Superoxide dismutase | 48 h | −33.3 | −20 |
| CYP13A7 | Putative cytochrome P450 CYP13A7 | 48 h | −36.2 | −13.74 |
| Carboxylesterase family protein | Carboxylesterase family protein (Planoprotostelium fungivorum) | 48 h | −8.45 | 1.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, Y.; Wang, Y.; Deng, J.; Liu, X.; Fang, Y.; Rao, Q.; Wu, H. Comparative Transcriptome Analysis Reveals the Mechanism Related to Fluazinam Stress of Panonychus citri (Acarina: Tetranychidae). Insects 2020, 11, 730. https://doi.org/10.3390/insects11110730
Shang Y, Wang Y, Deng J, Liu X, Fang Y, Rao Q, Wu H. Comparative Transcriptome Analysis Reveals the Mechanism Related to Fluazinam Stress of Panonychus citri (Acarina: Tetranychidae). Insects. 2020; 11(11):730. https://doi.org/10.3390/insects11110730
Chicago/Turabian StyleShang, Yi, Yanbo Wang, Jianyu Deng, Xunyue Liu, Yihao Fang, Qiong Rao, and Huiming Wu. 2020. "Comparative Transcriptome Analysis Reveals the Mechanism Related to Fluazinam Stress of Panonychus citri (Acarina: Tetranychidae)" Insects 11, no. 11: 730. https://doi.org/10.3390/insects11110730
APA StyleShang, Y., Wang, Y., Deng, J., Liu, X., Fang, Y., Rao, Q., & Wu, H. (2020). Comparative Transcriptome Analysis Reveals the Mechanism Related to Fluazinam Stress of Panonychus citri (Acarina: Tetranychidae). Insects, 11(11), 730. https://doi.org/10.3390/insects11110730





