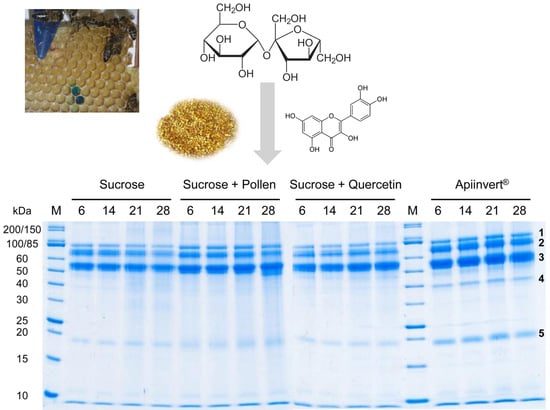
The Effect of Diet on the Composition and Stability of Proteins Secreted by Honey Bees in Honey
Abstract
1. Introduction
2. Materials and Methods
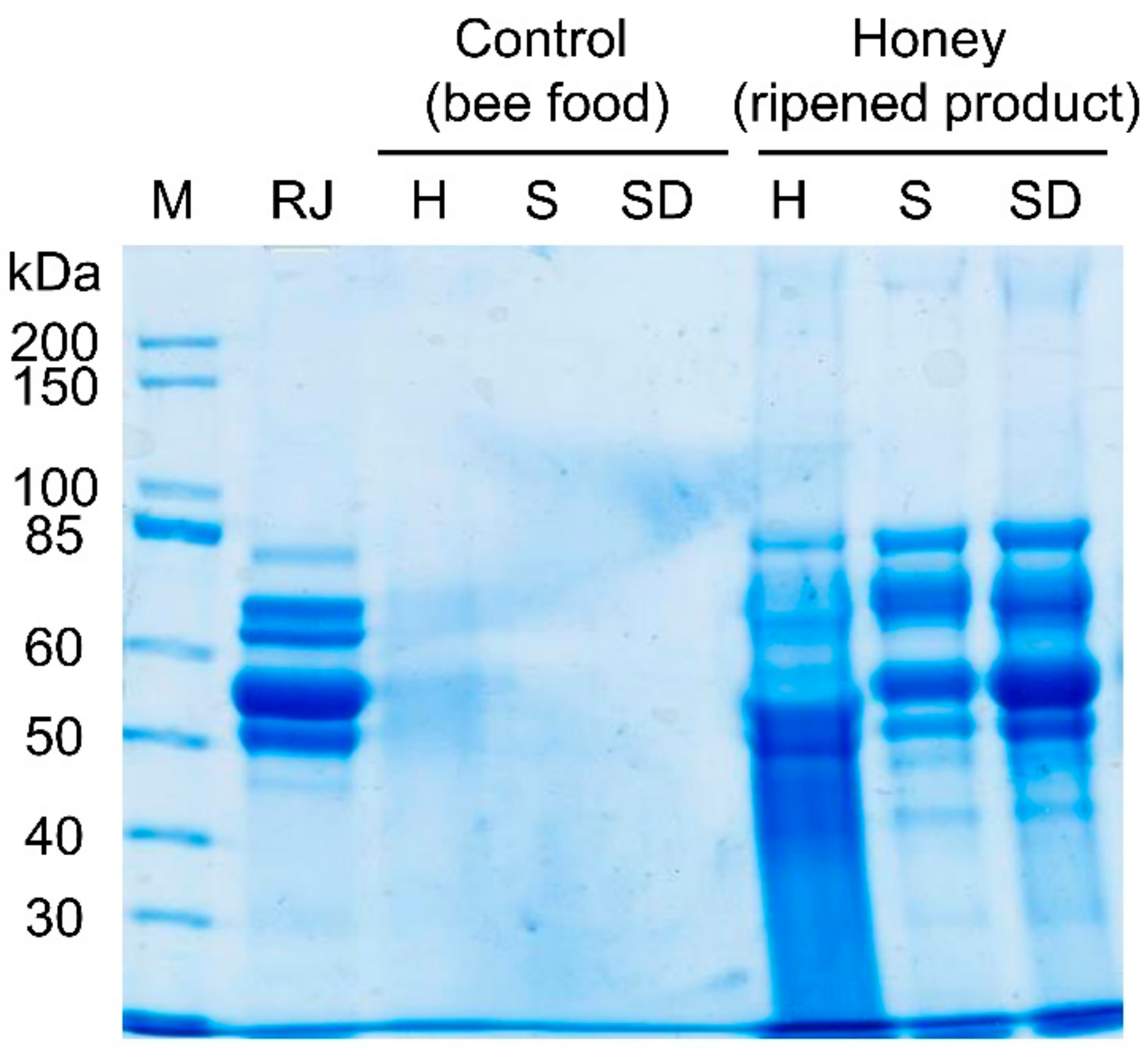
2.1. Bee Feeding Regime; Honey Ripening, Storage, and Protein Analysis
2.2. Protein Precipitation, Dye Removal, and Identification via Mass-Spectrometry
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maurizio, A. How bees make honey. In Honey: A Comprehensive Survey; Crane, E., Ed.; Heinemann: London, UK, 1975; pp. 77–105. [Google Scholar]
- Park, W. The storing and ripening of honey by honeybees. J. Econ. Entomol. 1925, 18, 405–410. [Google Scholar] [CrossRef]
- White, J.W. Honey. Adv. Food Res. 1978, 24, 287–374. [Google Scholar] [CrossRef] [PubMed]
- Chua, L.S.; Lee, J.Y.; Chan, G.F. Honey protein extraction and determination by mass spectrometry. Anal. Bioanal. Chem. 2013, 405, 3063–3074. [Google Scholar] [CrossRef] [PubMed]
- Schepartz, A.I.; Subers, M.H. The glucose oxidase of honey I. Purification and some general properties of the enzyme. Biochim. Biophys. Acta 1964, 85, 228–237. [Google Scholar] [CrossRef]
- Kubota, M.; Tsuji, M.; Nishimoto, M.; Wongchawalit, J.; Okuyama, M.; Mori, H.; Matsui, H.; Surarit, R.; Svasti, J.; Kimura, A.; et al. Localization of α-glucosidases I, II, and III in organs of European honeybees, Apis mellifera L., and the origin of α-glucosidase in honey. Biosci. Biotech. Biochem. 2004, 68, 2346–2352. [Google Scholar] [CrossRef] [PubMed]
- White, J.W.; Kushnir, I. The enzymes of honey: examination by ionexchange chromatography, gel filtration, and starch-gel electrophoresis. J. Apicult. Res. 1967, 6, 69–89. [Google Scholar] [CrossRef]
- Bergner, K.-G.; Diemair, S. Proteine des Bienenhonigs: II. Gelchromatographie, enzymatische Aktivität und Herkunft von Bienenhonig-Proteinen. Z. Lebensm. Unters. Forsch. 1975, 157, 7–13. [Google Scholar] [CrossRef]
- Bucekova, M.; Valachova, I.; Kohutova, L.; Prochazka, E.; Klaudiny, J.; Majtan, J. Honeybee glucose oxidase-its expression in honeybee workers and comparative analyses of its content and H2O2-mediated antibacterial activity in natural honeys. Naturwissenschaften 2014, 101, 661–670. [Google Scholar] [CrossRef]
- Simúth, J.; Bíliková, K.; Kovácová, E.; Kuzmová, Z.; Schroder, W. Immunochemical approach to detection of adulteration in honey: physiologically active royal jelly protein stimulating TNF-α release is a regular component of honey. J. Agric. Food Chem. 2004, 52, 2154–2158. [Google Scholar] [CrossRef]
- Kwakman, P.H.S.; te Velde, A.A.; de Boer, L.; Speijer, D.; Vandenbroucke-Grauls, C.M.J.E.; Zaat, S.A.J. How honey kills bacteria. FASEB J. 2010, 24, 2576–2582. [Google Scholar] [CrossRef]
- Rossano, R.; Larocca, M.; Polito, T.; Perna, A.M.; Padula, M.C.; Martelli, G.; Riccio, P. What are the proteolytic enzymes of honey and what they do tell us? A fingerprint analysis by 2-D zymography of unifloral honeys. PLoS ONE 2012, 7, e49164. [Google Scholar] [CrossRef]
- Bilikova, K.; Kristof Krakova, T.; Yamaguchi, K.; Yamaguchi, Y. Major royal jelly proteins as markers of authenticity and quality of honey. Arh. Hig. Rada Toksikol. 2015, 66, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Baroni, M.V.; Chiabrando, G.A.; Costa, C.; Wunderlin, D.A. Assessment of the floral origin of honey by SDS-page immunoblot techniques. J. Agric. Food Chem. 2002, 50, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kliks, M.M.; Qu, W.; Jun, S.; Shi, G.; Li, Q.X. Rapid determination of the geographical origin of honey based on protein fingerprinting and barcoding using MALDI TOF MS. J. Agric. Food Chem. 2009, 57, 10081–10088. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-Z.; Chen, Y.-F.; Wu, Y.-Q.; Si, J.-J.; Zhang, C.-P.; Zheng, H.-Q.; Hu, F.-L. Discrimination of the entomological origin of honey according to the secretions of the bee (Apis cerana or Apis mellifera). Food Res. Int. 2019, 116, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Borutinskaitė, V.; Treigytė, G.; Matuzevičius, D.; Zaikova, I.; Čeksterytė, V.; Navakauskas, D.; Kurtinaitienė, B.; Navakauskienė, R. Proteomic analysis of pollen and blossom honey from rape seed Brassica napus L. J. Apic. Sci. 2017, 61, 73–92. [Google Scholar] [CrossRef]
- Erban, T.; Shcherbachenko, E.; Talacko, P.; Harant, K. The unique protein composition of honey revealed by comprehensive proteomic analysis: Allergens, venom-like proteins, antibacterial properties, royal jelly proteins, serine proteases, and their inhibitors. J. Nat. Prod. 2019, 82, 1217–1226. [Google Scholar] [CrossRef]
- Di Girolamo, F.; D’Amato, A.; Righetti, P.G. Assessment of the floral origin of honey via proteomic tools. J. Proteomics 2012, 75, 3688–3693. [Google Scholar] [CrossRef] [PubMed]
- Borutinskaite, V.; Treigyte, G.; Čeksteryte, V.; Kurtinaitiene, B.; Navakauskiene, R. Proteomic identification and enzymatic activity of buckwheat (Fagopyrum esculentum) honey based on different assays. J. Food Nutr. Res. 2018, 57, 57–69. [Google Scholar]
- Rinderer, T.E.; Baxter, J.R. Effect of empty comb on hoarding behavior and honey production of the honey bee. J. Econ. Entomol. 1978, 71, 757–759. [Google Scholar] [CrossRef]
- Park, O.W. Further studies on the evaporation of nectar. J. Econ. Entomol. 1928, 21, 882–887. [Google Scholar] [CrossRef]
- Park, O.W. Studies on the rate at which honeybees ripen honey. J. Econ. Entomol. 1933, 26, 188–193. [Google Scholar] [CrossRef]
- Erler, S.; Moritz, R.F.A. Pharmacophagy and pharmacophory: Mechanisms of self-medication and disease prevention in the honeybee colony (Apis mellifera). Apidologie 2016, 47, 389–411. [Google Scholar] [CrossRef]
- Palmer-Young, E.C.; Farrell, I.W.; Adler, L.S.; Milano, N.J.; Egan, P.A.; Junker, R.R.; Irwin, R.E.; Stevenson, P.C. Chemistry of floral rewards: Intra- and interspecific variability of nectar and pollen secondary metabolites across taxa. Ecol. Monogr. 2019, 89, e01335. [Google Scholar] [CrossRef]
- Liao, L.-H.; Wu, W.-Y.; Berenbaum, M.R. Behavioral responses of honey bees (Apis mellifera) to natural and synthetic xenobiotics in food. Sci. Rep. 2017, 7, 15924. [Google Scholar] [CrossRef]
- Ehrenberg, S.; Lewkowski, O.; Erler, S. Dyeing but not dying: Colourful dyes as a non-lethal method of food labelling for in vitro-reared honey bee (Apis mellifera) larvae. J. Insect Physiol. 2019, 113, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mureşan, C.I.; Schierhorn, A.; Buttstedt, A. The fate of Major Royal Jelly Proteins during proteolytic digestion in the human gastrointestinal tract. J. Agric. Food Chem. 2018, 66, 4164–4170. [Google Scholar] [CrossRef]
- Helm, D.; Vissers, J.P.C.; Hughes, C.J.; Hahne, H.; Ruprecht, B.; Pachl, F.; Grzyb, A.; Richardson, K.; Wildgoose, J.; Maier, S.K.; et al. Ion mobility tandem mass spectrometry enhances performance of bottom-up proteomics. Mol. Cell Proteom. 2014, 13, 3709–3715. [Google Scholar] [CrossRef]
- Dobritzsch, D.; Aumer, D.; Fuszard, M.; Erler, S.; Buttstedt, A. The rise and fall of major royal jelly proteins during a honey bee (Apis mellifera) workers’ life. Ecol. Evol. 2019, 9, 8771–8782. [Google Scholar] [CrossRef]
- Buttstedt, A.; Moritz, R.F.A.; Erler, S. Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family. Biol. Rev. 2014, 89, 255–269. [Google Scholar] [CrossRef]
- Stadelmeier, M.; Bergner, K.G. The proteins of honey VII. Behaviour and origin of honey amylase. Zeitschrift für Lebensmittel-Untersuchung und Forschung 1986, 182, 196–199. [Google Scholar] [CrossRef]
- Moritz, B.; Crailsheim, K. Physiology of protein digestion in the midgut of the honeybee (Apis mellifera L.). J. Insect Physiol. 1987, 33, 923–931. [Google Scholar] [CrossRef]
- Brudzynski, K.; Sjaarda, C.; Maldonado-Alvarez, L. A new look on protein-polyphenol complexation during honey storage: Is this a random or organized event with the help of dirigent-like proteins? PLoS ONE 2013, 8, e72897. [Google Scholar] [CrossRef] [PubMed]
- Abou-Shaara, H.F.; Owayss, A.A.; Ibrahim, Y.Y.; Basuny, N.K. A review of impacts of temperature and relative humidity on various activities of honey bees. Insectes Soc. 2017, 64, 455–463. [Google Scholar] [CrossRef]
- Jones, J.C.; Oldroyd, B.P. Nest thermoregulation in social insects. Adv. Insect Physiol. 2006, 33, 153–191. [Google Scholar] [CrossRef]
- Ohashi, K.; Natori, S.; Kubo, T. Expression of amylase and glucose oxidase in the hypopharyngeal gland with an age-dependent role change of the worker honeybee (Apis mellifera L.). Eur. J. Biochem. 1999, 265, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Valachová, I.; Bučeková, M.; Majtán, J. Quantification of bee-derived peptide defensin-1 in honey by competitive enzyme-linked immunosorbent assay, a new approach in honey quality control. Czech J. Food Sci. 2016, 34, 233–243. [Google Scholar] [CrossRef]
- Brudzynski, K.; Maldonado-Alvarez, L. Polyphenol-protein complexes and their consequences for the redox activity, structure and function of honey. A current view and new hypothesis—A review. Pol. J. Food Nutr. Sci. 2015, 65, 71–80. [Google Scholar] [CrossRef]
- Won, S.R.; Lee, D.C.; Ko, S.H.; Kim, J.W.; Rhee, H.I. Honey major protein characterization and its application to adulteration detection. Food Res. Int. 2008, 41, 952–956. [Google Scholar] [CrossRef]
- Baracchi, D.; Francese, S.; Turillazzi, S. Beyond the antipredatory defence: Honey bee venom function as a component of social immunity. Toxicon 2011, 58, 550–557. [Google Scholar] [CrossRef]
- Maori, E.; Navarro, I.C.; Boncristiani, H.; Seilly, D.J.; Rudolph, K.L.M.; Sapetschnig, A.; Lin, C.C.; Ladbury, J.E.; Evans, J.D.; Heeney, J.L.; et al. A secreted RNA binding protein forms RNA-stabilizing granules in the honeybee royal jelly. Mol. Cell. 2019, 74, 598–608. [Google Scholar] [CrossRef] [PubMed]

| Accession Number | Description | MW | Quantitative Data (Normalized to Total Spectra) | |||
|---|---|---|---|---|---|---|
| (NCBI) | (All Apis mellifera) | (kDa) | Apiinvert | Sucrose (S) | S + Pollen | S + Quercetin |
| NP_001011579.1 | Major royal jelly protein 1 | 48.886 | 21 | 9 | 21 | 12 |
| NP_001011601.1 | Major royal jelly protein 3 | 61.662 | 12 | 5 | 7 | 7 |
| NP_001011580.1 | Major royal jelly protein 2 | 51.074 | 9 | 7 | 8 | 5 |
| NP_001011574.1 | Glucose oxidase | 67.938 | 6 | 1 | 6 | 3 |
| NP_001011608.1 | Alpha-glucosidase Hbg3 precursor | 65.565 | 4 | 3 | 5 | 3 |
| NP_001011599.1 | Major royal jelly protein 5 | 70.236 | 2 | 0 | 2 | 1 |
| NP_001014429.1 | Major royal jelly protein 7 | 50.541 | 1 | 0 | 1 | 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewkowski, O.; Mureșan, C.I.; Dobritzsch, D.; Fuszard, M.; Erler, S. The Effect of Diet on the Composition and Stability of Proteins Secreted by Honey Bees in Honey. Insects 2019, 10, 282. https://doi.org/10.3390/insects10090282
Lewkowski O, Mureșan CI, Dobritzsch D, Fuszard M, Erler S. The Effect of Diet on the Composition and Stability of Proteins Secreted by Honey Bees in Honey. Insects. 2019; 10(9):282. https://doi.org/10.3390/insects10090282
Chicago/Turabian StyleLewkowski, Oleg, Carmen I. Mureșan, Dirk Dobritzsch, Matthew Fuszard, and Silvio Erler. 2019. "The Effect of Diet on the Composition and Stability of Proteins Secreted by Honey Bees in Honey" Insects 10, no. 9: 282. https://doi.org/10.3390/insects10090282
APA StyleLewkowski, O., Mureșan, C. I., Dobritzsch, D., Fuszard, M., & Erler, S. (2019). The Effect of Diet on the Composition and Stability of Proteins Secreted by Honey Bees in Honey. Insects, 10(9), 282. https://doi.org/10.3390/insects10090282