Discovery of Aphid Lethal Paralysis Virus in Vespa velutina and Apis cerana in China
Abstract
1. Introduction
2. Materials and Methods
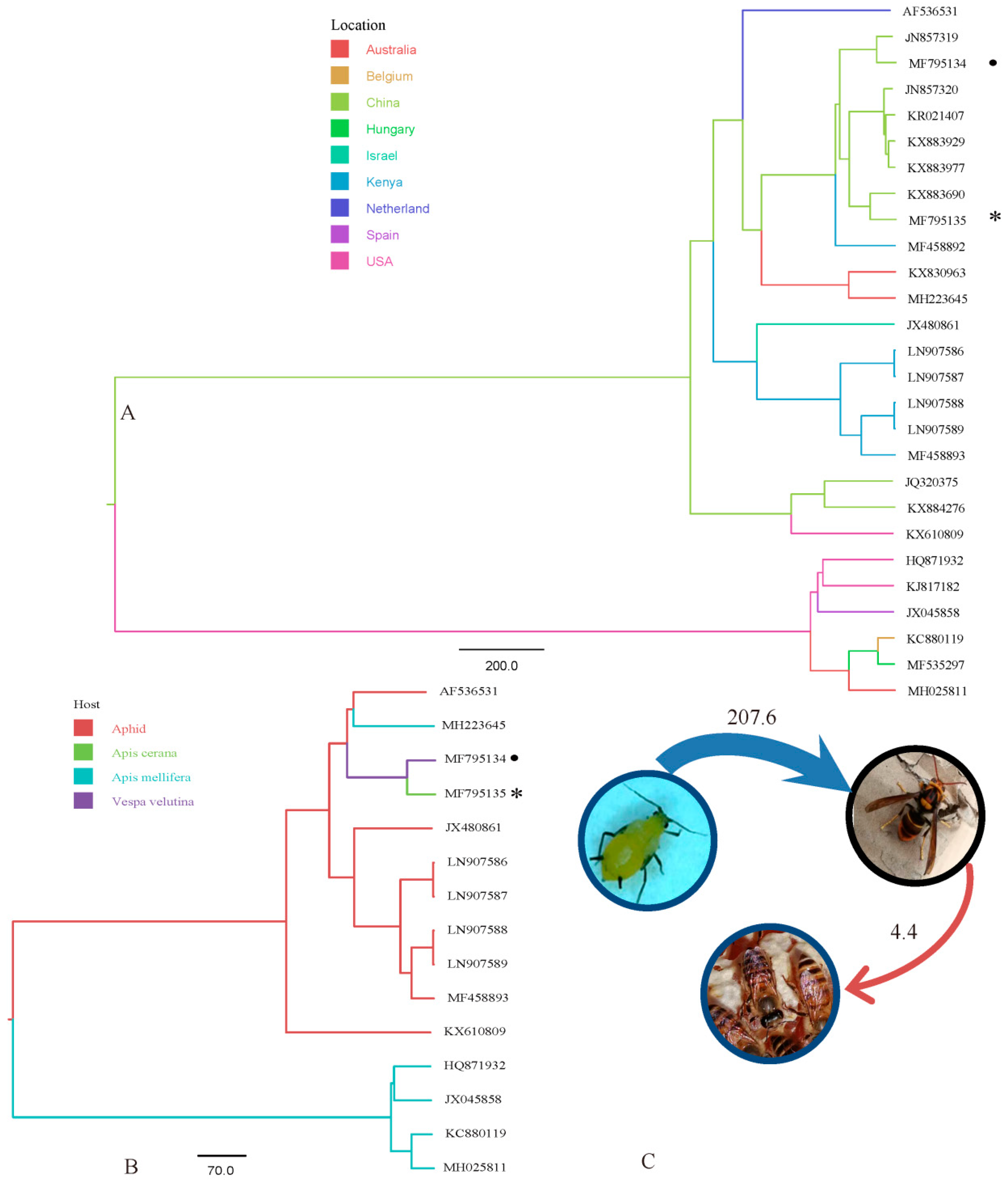
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chauzat, M.P.; Antoine, J.; Marion, L.; Stéphanie, B.; Pascal, H.; Magali, R.C. Risk indicators affecting honeybee colony survival in Europe: One year of surveillance. Apidologie 2016, 3, 348–378. [Google Scholar] [CrossRef]
- Liu, Z.G.; Chen, C.; Niu, Q.S.; Qi, W.Z.; Yuan, C.Y.; Su, S.K.; Liu, S.D.; Zhang, Y.S.; Zhang, X.W.; Ji, T.; et al. Survey results of honey bee (Apis mellifera) colony losses in China (2010–2013). J. Apicul. Res. 2016, 1, 1–8. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Z.; Luo, Y.; Zheng, X.; Wang, S.H.; Zhang, X.W.; Dai, R.G.; Gao, J.L.; Chen, X.; Guo, H.K.; et al. Managed honeybee colony losses of the Eastern honeybee (Apis cerana) in China (2011–2014). Apidologie 2017, 48, 692–702. [Google Scholar] [CrossRef]
- Degrandi-Hoffman, G.; Chen, Y. Nutrition, immunity and viral infections in honey bees. Curr. Opin. Insect Sci. 2015, 10, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Diao, Q.Y.; Li, B.B.; Zhao, H.X.; Wu, Y.Y.; Guo, R.; Dai, P.L.; Chen, D.F.; Wang, Q.; Hou, C.S. Enhancement of chronic bee paralysis virus levels in honeybees acute exposed to imidacloprid: A Chinese case study. Sci. Total Environ. 2018, 630, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Yañez, O.; Zheng, H.Q.; Hu, F.L.; Neumann, P.; Dietemann, V. A scientific note on Israeli acute paralysis virus infection of Eastern honeybee Apis cerana and vespine predator Vespa velutina. Apidologie 2012, 43, 587–589. [Google Scholar] [CrossRef]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F.; Evans, J.D.; Chen, Y. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011, 1, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Monceau, K.; Thiéry, D. Vespa velutina nest distribution at a local scale: An eight-year survey of the invasive honeybee predator. Insect Sci. 2017, 4, 663–674. [Google Scholar] [CrossRef]
- Choi, M.B.; Martin, S.J.; Lee, J.W. Distribution, spread, and impact of the invasive hornet Vespa velutina in South Korea. J. Asia-Pac. Entomol. 2012, 15, 473–477. [Google Scholar] [CrossRef]
- Garigliany, M.; Taminiau, B.; El Agrebi, N.; Cadar, D.; Gilliaux, G.; Hue, M.; Desmecht, D.; Daube, G.; Linden, A.; Farnir, F.; et al. Moku virus in invasive Asian hornets, Belgium, 2016. Emerg. Infect. Dis. 2017, 23, 2109–2112. [Google Scholar] [CrossRef]
- Villemant, C.; Zuccon, D.; Rome, Q.; Muller, F.; Poinar, G.O.; Justine, J.L. Can parasites halt the invader? Mermithid nematodes parasitizing the yellow-legged Asian hornet in France. PeerJ 2015, 3, e947. [Google Scholar] [CrossRef] [PubMed]
- Demichelis, S.; Manino, A.; Minuto, G.; Mariotti, M.; Porporato, M. Social wasp trapping in north west Italy: Comparison of different bait-traps and first detection of Vespa velutina. Bull. Insectol. 2014, 2, 307–317. [Google Scholar]
- Dombrovsky, A.; Luria, N. The Nerium oleander aphid Aphis neriiis tolerant to a local isolate of Aphid lethal paralysis virus (ALPV). Virus Genes 2013, 2, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Granberg, F.; Vicente-Rubiano, M.; Rubio-Guerri, C.; Karlsson, O.E.; Kukielka, D.; Belák, S.; Sánchez-Vizcaíno, J.M. Metagenomic detection of viral pathogens in Spanish honeybees: Co-infection by Aphid Lethal Paralysis, Israel Acute Paralysis and Lake Sinai Viruses. PLoS ONE 2013, 2, e57459. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.X.; Yan, X.; Han, R. Occurrence and prevalence of seven bee viruses in Apis mellifera and Apis cerana apiaries in China. J. Invertebr. Pathol. 2012, 109, 160–164. [Google Scholar] [CrossRef]
- Nakashima, N.; Uchiumi, T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009, 2, 137–147. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 8, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. j Model Test: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 7, 1253–1256. [Google Scholar] [CrossRef]
- Darrouzet, E.; Gévar, J.; Dupont, S. A scientific note about a parasitoid that can parasitize the yellow-legged hornet, Vespa velutina nigrithorax, in Europe. Apidologie 2015, 1, 130–132. [Google Scholar] [CrossRef]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; Vanengelsdorp, D.; Lipkin, W.I.; Depamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef]
- Li, J.; Qin, H.; Wu, J.; Sadd, B.M.; Wang, X.; Evans, J.D.; Peng, W.; Chen, Y. The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PLoS ONE 2012, 11, e47955. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.Z.; Wang, Y.Z. Wasps Fauna of Yunna; Henan Science and Technology Press: Zhengzhou, China, 2017; pp. 6–7. [Google Scholar]
- Chen, Y.; Zhao, Y.; Hammond, J.; Hsu, H.T.; Evans, J.; Feldlaufer, M. Multiple virus infections in the honey bee and genome divergence of honey bee viruses. J. Invertebr. Pathol. 2004, 87, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Vijayendran, D.; Carrillo-Tripp, J.; Miller, W.A.; Bonning, B.C. Analysis of new aphid lethal paralysis virus (ALPV) isolates suggests evolution of two ALPV species. J. Gen. Virol. 2014, 95, 2809–2819. [Google Scholar] [CrossRef] [PubMed]
- Bowen-Walker, P.L.; Martin, S.J.; Gunn, A. The transmission of deformed wing virus between honey bees (Apis mellifera L.) by the Ectoparasitic mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 1999, 73, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Eyer, M.; Chen, Y.P.; Schäfer, M.O.; Pettis, J.; Neumann, P. Small hive beetle, Aethina tumida, as a potential biological vector of honeybee viruses. Apidologie 2009, 4, 419–428. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; Zhao, H.; Shi, J.; Xu, X.; Wu, Y.; Guo, R.; Chen, D.; Wang, X.; Deng, S.; Yang, S.; et al. Discovery of Aphid Lethal Paralysis Virus in Vespa velutina and Apis cerana in China. Insects 2019, 10, 157. https://doi.org/10.3390/insects10060157
Yang D, Zhao H, Shi J, Xu X, Wu Y, Guo R, Chen D, Wang X, Deng S, Yang S, et al. Discovery of Aphid Lethal Paralysis Virus in Vespa velutina and Apis cerana in China. Insects. 2019; 10(6):157. https://doi.org/10.3390/insects10060157
Chicago/Turabian StyleYang, Dahe, Hongxia Zhao, Junming Shi, Xiang Xu, Yanyan Wu, Rui Guo, Dafu Chen, Xinling Wang, Shuai Deng, Sa Yang, and et al. 2019. "Discovery of Aphid Lethal Paralysis Virus in Vespa velutina and Apis cerana in China" Insects 10, no. 6: 157. https://doi.org/10.3390/insects10060157
APA StyleYang, D., Zhao, H., Shi, J., Xu, X., Wu, Y., Guo, R., Chen, D., Wang, X., Deng, S., Yang, S., Diao, Q., & Hou, C. (2019). Discovery of Aphid Lethal Paralysis Virus in Vespa velutina and Apis cerana in China. Insects, 10(6), 157. https://doi.org/10.3390/insects10060157






