Natural Product Medicines for Honey Bees: Perspective and Protocols
Abstract
:1. Introduction
1.1. Key Agents of Disease
1.2. Disease Costs for Beekeeping
1.3. Registered Bee Disease Treatments
1.4. Why Natural Products?
1.5. Current Research on Natural Products for Bee Disease
1.6. RNA Interference
1.7. High-Throughput Screening in the Laboratory
1.8. Limits of Approach
1.9. Goal of This Review and Protocol Description
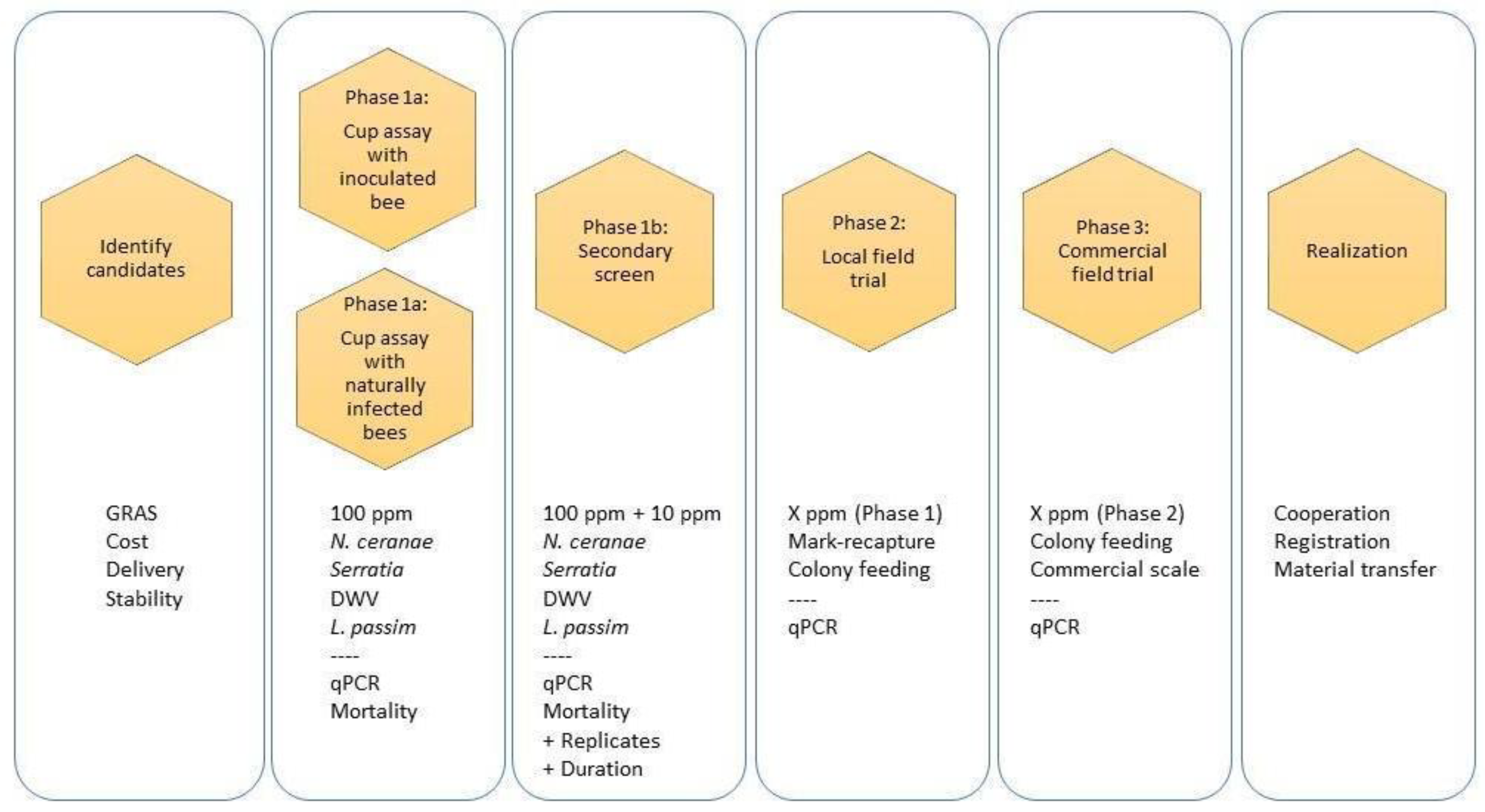
2. Protocols
2.1. Selection Criteria
- Plant nectar products [24];
- Plant resins (and compounds derived from plant resins), especially those collected by bees in the field [23];
- Natural products proposed by community members and tested under material transfer and research agreements (e.g., polypore mushroom extracts developed by Fungi Perfecti [31] and commercialized probiotics);
- FDA GRAS database and lists of natural extracts vetted against peer-reviewed literature (known antiviral and antiparasitic activity);
- Compounds known from the insect literature to be antiviral/antimicrobial/antiparasitic;
- Compounds known from general literature to be antiviral/antimicrobial/antiparasitic;
- Herbs with potential health benefits in other organisms [25];
- The NCI Natural Products Repository (https://dtp.cancer.gov/organization/npb/introduction.htm) that includes extracts from 80,000 plants that are native to Africa and Madagascar, Central and South America, and Southeast Asia; 20,000 specimens from marine invertebrates and algae from the southern oceans; and specimens from 16,000 microbes.
- Projected maximum product cost of $5 USD per treatment (approximately half the cost of one current treatment);
- ○
- The natural product in question may end up being more expensive, and then the choice of which treatment to apply will be based on other criteria;
- ○
- Reliability of production (e.g., synthetic compounds inspired from naturally produced compounds);
- Chemoinformatic properties that suggest favorable target interaction:
- ○
- Chemical diversity across terpenoids, carbohydrates, sterols, alkaloids, lipids, small-molecule secondary metabolites and extracts, especially when compounds lack a report of a molecular target [17];
- ○
- Stability at circa 35 °C with high relative humidity (50%–90%), and solubility in high-osmolarity sugar-water solution for administration;
- Consideration of cross-contamination into honey bee products and to flowers and other commercial and wild insects;
- Crude extracts may in fact contain unwanted animal toxins and it is then possible that the compound of interest cannot be confidently removed from the malefactor. For example, many plants, including rhododendrons and others, are known to carry compounds that are toxic to bees (https://bee-health.extension.org/are-there-plants-that-produce-nectar-that-is-poisonous-to-either-honey-bees-or-humans/).
2.2. Compound Qualification
2.3. Metrics of Success
2.4. Testing Compounds Without Artificial Pathogen Inoculation (Phase 1a)
2.4.1. Apiary Collection of Worker Honey Bees for Cup Trials
- Choose two to three healthy honey bee colonies which, based on prior genetic screens have an appropriate abundance of pathogens for which treatments are to be tested.;
- Take the outermost honey frames which tend to have older worker honey bees and are less likely to host queen bees;
- Shake a frame covered with worker bees forcefully into a plastic bucket or alternate container with steep sides >25 cm tall;
- Immediately shake bees from the bucket into ventilated plastic cups and cap these cups, with about 600 bees/16 oz. (ca. 473 mL) cup. Keep cups of bees in the shade after collection, or ideally in a cooler with an adjacent ice pack on hot days;
- After transporting the bee cups into the laboratory, anesthetize the cupped bees with CO2 for ca. 30 s using a 20 × 20 × 20 cm Styrofoam™ shipping container into which CO2 is provided from a tank using surgical tubing. CO2 is preferred to placing the bees on ice. In our experience, recovery is >99% versus 80% when bees were chilled;
- From the cup of anesthetized bees, quickly distribute bees into rearing cups, allocating 30 to 40 bees per rearing cup;
- A video of this process is provided in the supplement.
- Choose multiple healthy honey bee colonies;
- Take one or two brood frames from each colony. Select frames that show signs of emerging worker bees to ensure a supply of young bees within one to two days. Place the frames in a screened frame-holding box or a container with a mesh lid;
- Transport the frames to a laboratory incubator that has been pre-set to 34° C with high relative humidity (50%);
- Allow worker bees to naturally emerge from the comb within the wire mesh frame cage. One can expect at least 1000 bees from a half-filled brood frame with many, but not all, of the bees emerging within two days. By having multiple combs, one can ensure a sufficient number of bees within one day and the remaining brood can be put back into the colony;
- Once young bees have emerged, distribute by hand into single-use, semi-sterile plastic rearing cups with 15 to 30 bees per cup.
2.4.2. Experimental Chambers
2.4.3. Treatment Conditions
2.4.4. Example Trial
2.4.5. Preserving Specimens
2.4.6. Output
2.5. Testing Compounds with Artificial Pathogen Inoculation (Phase 1a)
2.5.1. Nosema as a Targeted Parasite
- To acquire Nosema from local apiary, collect 30–50 live foragers from the entrance of colonies that one knows or suspects to carry Nosema;
- Nosema infects the honey bee gut; therefore, after collection, the next step to isolate Nosema spores is to eviscerate honey bee abdomens. Perform whole intestine extractions as described in [54]. The simplest method is to sedate bees (not by freezing, ideally using CO2), and then to pull the entire intestine out by holding the last abdominal segment with a pair of tweezers and slowly pulling until the entire intestine is removed from the abdomen;
- Combine ten guts in a 1.5 mL sterile centrifuge tube. Add 400 μL of sterile dH20 and grind using a sterile pestle;
- From this mixture, aliquot 5–10 μL onto a standard glass microscope slide;
- Place the slide on a microscope and use 400× magnification with phase contrast to look for spores. With phase contrast, if Nosema is present, one will observe ovate structures that are distinguishable from any other irregular-shaped debris (Figure S5);
- Once you identify spores, wash the crude suspension using a modified triangulation protocol 2.2.4.2.1.2. described in [53]. Centrifuge the crude suspension at 300× g for 2 min, remove the supernatant, suspend the pellet in 400 μL of sterile dH20, do a very quick spin of this in a small tabletop centrifuge, and remove the supernatant into a fresh tube. This tube will now contain a more purified spore suspension. Repeat this process at least two more times. In our experience, the quick tabletop spin allows for additional debris to pellet and the spores to remain in the supernatant. Count the spores using a hemocytometer. Keep the spores in water and do not dilute the entire extraction in sugar water;
- Calculate 10,000 spores per bee and add this number of spores to a volume of 50% sucrose solution when ready to inoculate honey bees. For example, in a cup of 30 bees, 300,000 spores would be suspended in 0.5–1 mL sugar water (cup top feeder):
- If there are enough spores, then one can proceed to the feeding stage;
- If one does not have enough spores, one can survey more bees and repeat the protocol;
- If time allows, feed the suspension created from step 2 to newly emerged bees. Rear the bees using the protocol described above for at least 12 days and then repeat steps 2 through 7 to isolate and count the spores from the bees that were artificially fed Nosema. This should generate tens to hundreds of millions of spores depending on how many bees were inoculated, which is definitely enough for a typical trial.
2.5.2. Live Bee Nosema Screening
- Prepare the Nosema ceranae suspension (above);
- Collect adult worker bees or newly emerged bees as described in ‘Apiary Collection of Worker Honey Bees for Cup Trials’;
- After collection of the bees, feed the bees with sterile 50% sucrose solution. Incubate at 34 °C with 50% relative humidity (as above) for one day;
- On the second day, inoculate the adult bees “cage-style” with the suspension from step 1. Using a low volume (likely 500 μL) that contains 10,000 spores for each bee in the cup ensures complete infection and that the bees consume the entire solution within one day [53]:
- (a)
- If using newly emerged bees, one has the option to do “cage-style” feeding or hand inoculations (see Table S1 in the supplement for cage-style and hand-style feeding success). The food source should be removed in the morning and in the late afternoon, feed 5 μL to each bee using a 10–20 μL pipettor and by holding the bee by its wings and letting the bee drink from the inoculum droplet;
- (b)
- On the other hand, foragers will consume this amount of food quickly and the feeder might need to be topped off with extra food;
- After complete consumption of the inoculum, start the compound-sucrose solution feeding at 100 ppm for half of the cups and maintain ad libitum feeding throughout the trial. Replenish as needed;
- Incubate and dose the honey bees for at least twelve more days to ensure complete infection. It takes at most six days for Nosema to complete its life cycle and after twelve days, high mortality is then observed in cup-reared honey bees;
- Count and record the number of deaths daily and/or at the end of the trial;
- At the end of the experiment, remove dead bees (which are left in the cups throughout), freeze the remaining live bees at −80 °C, and then transfer the bees into marked extraction bags or tubes to prepare for total RNA isolation;
- Store bags in a freezer at −80 °C until ready to perform RNA extraction.
2.6. Serratia
- Isolate strains of Serratia from honey bee intestines as in [55]. Honey bee intestines may be fresh or macerated and stored in 20% glycerol at −20 °C. There is no Serratia-type strain for honey bee testing, and the precaution is biosafety level 1;
- In preparation for the trial, swab the glycerol stock of the isolated and verified Serratia strain in LB broth in the evening. Grow overnight, dilute and then incubate to an OD600 of about 1.0. Centrifuge the bacteria, wash the pellet with 1X PBS, centrifuge again and suspend in 1× PBS. Combine 20 mL of the bacterial PBS suspension with 50% sucrose solution to make a 25% bacteria-sucrose feed;
- Collect adult worker bees as described in ‘Apiary Collection of Worker Honey Bees for Cup Trials.’ As above, capture honey bees and then incubate the bees for one day with plain sterile 50% sucrose solution;
- On day two, feed to the bees 1 mL of the Serratia-infused syrup and let the bees consume the solution for one day like “cage-style” feeding for Nosema ceranae;
- The next day, start feeding 100 ppm of the test compound in 50% sucrose solution ad libitum to half of the cups;
- Incubate the honey bees for eight more days. One could expect a sharp increase in mortality after six to seven days;
- Refer to steps 7–9 in ‘Live Bee Nosema Screening’.
2.7. Deformed Wing Virus
- Identify an apiary or colony with relatively high titers of DWV;
- One may assess colony virus load by collecting 50 adult honey bees and measuring virus load using qPCR as described: “4.3.2. Bulk extraction of RNA from 50–100 whole bees using the acid-phenol method” (omitting the hot phenol step) in [60]. There is no standard for what classifies as a very high or, conversely, very low hive infection to be used for testing; however, by screening various colonies at the apiary, the researchers can determine which hives have relatively higher and relatively lower titers of DWV. This survey need not only be for this trial: the survey provides additional information to the researchers on which hives to use for other experimentation, from which hives to avoid using brood frames, possible recombination trends, and survey data can be passed on to interested parties such as the USDA-ARS BRL;
- Alternatively, one may feed filtered bee hemolymph containing DWV to bees using the cage-style method (see Table S2 in the supplement);
- Once a colony with relatively high DWV titers is identified, collect and rear bees as described in ‘Apiary Collection of Worker Honey Bees for Cup Trials’;
- Feed with sterile 50% sucrose solution and incubate for one day;
- The next day, start feeding 100 ppm of the test compound in 50% sucrose solution ad libitum to half of the cups;
- Incubate and treat the honey bees for twelve days;
- Refer to steps 7–9 in ‘Live Bee Nosema Screening’.
2.8. Lotmaria Passim
- Process brood frames as described in ‘Apiary Collection of Worker Honey Bees for Cup Trials’ to generate newly emerged worker honey bees;
- Three days before the experiment, thaw the stock of L. passim and inoculate into supplemented DS2 media that additionally contains 5% (v/v) fetal bovine serum and antibiotics [8];
- When ready to harvest on the day of inoculation, pellet the dense culture of L. passim in the medium for 10 min at 425× g, and suspend it in 1× PBS;
- Dilute further by 1:100 and count active promastigotes in a Neubauer hemocytometer at 400× magnification;
- Fix the suspension to a 2000 cells per μL inoculum using 20% sucrose solution (1:1 vol/vol) in 1× PBS;
- After the newly emerged bees are incubated in the cups for one day, feed the newly emerged bees 5 μL of the L. passim suspension within one day after removing their food source that morning (discard any bee that does not consume the inoculum);
- Immediately start the treatment of 50% sucrose solution with 100 ppm of the testing compound to half of the cups;
- Continue rearing the bees until day twelve, whereby after six days, there will be a complete infection detectable by qPCR, at least in the control group;
- Refer to steps 7–9 in ‘Live Bee Nosema Screening’.
2.9. Interpretation from Phase 1
2.9.1. Product Toxicity
2.9.2. Bee Collections
2.9.3. RNA Extraction
2.9.4. Measuring Pathogen Load and Honey Bee Health
2.9.5. Analysis of Treatment Effect
3. Secondary Screen (Phase 1b)
3.1. Compound Selection
3.2. Expanded Protocol
- Repeat the original trial with a new set of rearing cups and source colonies in order to increase the number of biological replicates and statistical power. Use the same dose as the initial trial but add a dose treatment that is ten times lower to get a range of efficacy. A dose-dependent response curve can eventually be assessed;
- (a)
- For certain natural products, the dose window might be shifted to address concerns about solubility, toxicity or weak impact at a low dose, as determined from in vitro testing;
- In parallel, run the sucrose-only and compound-only untreated control in order to have additional controls. For example, in the inoculation studies, the first screen had two conditions where all the bees were inoculated with the pathogen yet only one condition was with the test compound. Now, we add two conditions where bees are fed either an unsoiled food source, as well as the compound without the pathogen;
- As noted above, endpoint survival for the first trial may be sufficient, especially when a large number of bee cups are running in parallel. However, in the second round, it is necessary to monitor survival by counting deaths daily or every other day for three to four weeks (time already incubated included) in order to produce a proper survival curve and to monitor the longevity of the worker bee (e.g., a decline in the Nosema-infected population will likely only be seen 12 to 14 days after the start of infection);
- Feeding the compound from the onset of infection for pathogen-inoculated honey bees follows a similar protocol set up in our laboratory, as described in Palmer-Young et al. [24]. However, variations in the experimental design can include waiting three to six days post-infection before administering the compound of interest. This would allow the pathogen in question to establish itself in the host by giving it time to start its reproductive life cycle.
- All candidate substances that might end up as a bee treatment should be verified for stability in 50% sucrose solution by subjecting it to liquid or gas chromatography coupled with mass spectrometry (LC/GS-MS). This will partially confirm that the natural product in question is still present and whether there was extensive degradation. Additionally, crude materials bought from different companies and under different storage conditions, in which one expects there to be the compound of interest, may in fact have degraded or be nonexistent which would severely reduce or eliminate the material’s efficacy; this can also be confirmed by spectroscopy and mass analyses. If there is an extract is of interest, then one should consider purchasing it as fresh as possible and to monitor its composition overtime;
- Since this protocol ensures the availability of stable cDNA from each specimen, it may also be informative and economical to measure additional RNA viruses that are detrimental to bee health (such as Lake Sinai virus 2).
4. Small, Local Field Trials (Phase 2)
4.1. Mark-Recapture Trials
- For source colonies, select three that have a queen (queenright), are of near-equal strength and do not show overt signs of disease:
- (a)
- This is a judgement call on behalf of the beekeeper. At the simplest level, a queenright colony, with an equal number of Langstroth frame boxes and with healthy brood patterns can be considered similar enough for colony tests. Alternatively, weighing a colony is possible, but this is more intensive;
- Collect several hundred very young worker bees from each colony in a similar manner and from similar frames;
- Very young bees on brood frames can be collected gently and quickly by hand or by a modified “insect” vacuum (Figures S3 and S4). In general, collected bees will only differ a couple days in age from each other. The easiest method for identification is whether the bees are attending to the brood and if they are a bit gray in color;
- In the lab, distinctly mark 50 worker bees per condition using non-toxic paint pens (e.g., ‘Pro-Painter’ pens, generally sold for queen marking);
- Place marked bees into single-use plastic cups with each cup having a distinct color scheme (using two color spots it is simple to produce up to 16 distinct marking patterns) (Figure S2);
- Feed bees for 48 h with a 50% sucrose solution treatment containing a natural product or controls using the determined dosage from the second trial in Phase 1b:
- Compounds can be tested in bees that have pathogens that they already acquired from the colony; or
- Bees can also be fed Nosema spores (10,000 cumulatively per bee) or a suspension of viruses (e.g., 109 copies per bee) at this stage, as desired;
- The 50% sucrose solution with and without the compound should be immediately fed to the collected and painted bees (optionally, artificially inoculated bee);
- Mix bees of all colors into a single cup for at least two hours. This improves equal acceptance, we feel, while not offering bees too long to share microbes or carried treatments;
- Select source colonies and introduce marked bees to this hive via the top frames of the top box. We do not find high mortality in these introductions, perhaps because bees are introduced away from the guarded nest entrance. It is not uncommon to see >95% survivorship at one week;
- After ten days, collect all marked bees from the colony using a modified portable vacuum (e.g., the BioQuip bug vacuum). Each hive box, if two, must be separated and each frame pulled and inspected on both sides by two people, one of whom is handling the vacuum. Frames are set outside and then returned to the clean box one at a time, again checking carefully for all marked bees;
- Freeze collected bees at −80 °C. Once frozen, bees can be sorted by color for counting, and placed into color-coded groups for RNA extraction and analyses for disease levels and honey bee transcripts as above.
4.2. Small-Scale Colony Trials
- Select a minimum of 16 colonies (one treatment condition and one control set), which have been equalized, paired by size (worker number, brood area, and stored nutrition, [65]) and randomly placed into either set;
- Use standard top feeders to deliver products and nutrients. These trials work best when colonies are actively receiving sugar supplements;
- For each colony, expect to use six liters of 50% sucrose solution in total, three liters per week for two weeks. This provides each bee with about 150 μL of 50% sucrose solution on average, assuming a colony population of 40,000 bees. The bees will store much of the treated 50% sucrose solution, but it remains accessible and the colony should consume it over the next two months;
- Collect and freeze 300 worker bees per colony after two weeks, one month, two months and finally, on the third month after the start of the trial. This is in addition to collected bees from day 0;
- At each collection point, additionally screen colonies for colony metrics and any overt signs of disease;
- Extract RNA from 50 collected bees using a bulk extraction method: e.g., “4.3.2. Bulk extraction of RNA from 50–100 whole bees using the acid-phenol method (omitting the hot phenol step)” [60];
- Perform qPCR to measure pathogen load and bee health using the set of standard pathogen and host gene assays as described above. Add assays to measure additional pathogen or host genes as desired.
5. Large-Scale Field Trials (Phase 3)
6. Epilogue
6.1. Cooperation
6.2. Registration
6.3. Promise of the Natural Product Approach
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- vanEngelsdorp, D.; Meixner, M.D. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 2010, 103, S80–S95. [Google Scholar] [CrossRef] [PubMed]
- Cornman, R.S.; Tarpy, D.R.; Chen, Y.; Jeffreys, L.; Lopez, D.; Pettis, J.S.; vanEngelsdorp, D.; Evans, J.D. Pathogen webs in collapsing honey bee colonies. PLoS ONE 2012, 7, e43562. [Google Scholar] [CrossRef] [PubMed]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Predictive markers of honey bee colony collapse. PLoS ONE 2012, 7, e32151. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of Deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef]
- Schwarz, R.S.; Huang, Q.; Evans, J.D. Hologenome theory and the honey bee pathosphere. Curr. Opin. Insect Sci. 2015, 10, 1–7. [Google Scholar] [CrossRef]
- Schwarz, R.S.; Moran, N.A.; Evans, J.D. Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 2016, 113, 9345–9350. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.E.; Ricigliano, V.A. Honey bee gut dysbiosis: A novel context of disease ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef]
- Burritt, N.L.; Foss, N.J.; Neeno-Eckwall, E.C.; Church, J.O.; Hilger, A.M.; Hildebrand, J.A.; Warshauer, D.M.; Perna, N.T.; Burritt, J.B. Sepsis and hemocyte loss in honey bees (Apis mellifera) Infected with serratia marcescens strain sicaria. PLoS ONE 2016, 11, e0167752. [Google Scholar] [CrossRef]
- Raymann, K.; Shaffer, Z.; Moran, N.A. Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol. 2017, 15, e2001861. [Google Scholar] [CrossRef] [PubMed]
- DeGrandi-Hoffman, G.; Ahumada, F.; Danka, R.; Chambers, M.; DeJong, E.W.; Hidalgo, G. Population growth of Varroa destructor (Acari: Varroidae) in colonies of Russian and unselected honey bee (Hymenoptera: Apidae) stocks as related to numbers of foragers with mites. J. Econ. Entomol. 2017, 110, 809–815. [Google Scholar] [CrossRef]
- Johnson, R.M.; Pollock, H.S.; Berenbaum, M.R. Synergistic interactions between in-hive miticides in Apis mellifera. J. Econ. Entomol. 2009, 102, 474–479. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, D.C.; Alippi, A.M.; Brown, M.; Evans, J.D.; Feldlaufer, M.; Gregorc, A.; Hornitzky, M.; Pernal, S.F.; Schuch, D.M.T.; Tiťra, D.; et al. Diagnosis of American foulbrood in honey bees: A synthesis and proposed analytical protocols. Lett. Appl. Microbiol. 2006, 43, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.F.; Solter, L.F.; Yau, P.M.; Imai, B.S. Nosema ceranae escapes fumagillin control in honey bees. PLoS Pathog. 2013, 9, e1003185. [Google Scholar] [CrossRef]
- Barry, C. Snider total synthesis of (+-)-fumagillin. J. Am. Chem. Soc. 1972, 94, 2549–2550. [Google Scholar]
- Byler, K.G.; Ogungbe, I.V.; Setzer, W.N. In-silico screening for anti-Zika virus phytochemicals. J. Mol. Graph. Model. 2016, 69, 78–91. [Google Scholar] [CrossRef]
- Ng, Y.C.; Kim, Y.W.; Ryu, S.; Lee, A.; Lee, J.S.; Song, M.J. Suppression of norovirus by natural phytochemicals from Aloe vera and Eriobotryae Folium. Food Control 2017, 73, 1362–1370. [Google Scholar] [CrossRef]
- Rasouli, H.; Farzaei, M.H.; Khodarahmi, R. Polyphenols and their benefits: A review. Int. J. Food Prop. 2017, 20, 1700–1741. [Google Scholar] [CrossRef]
- Vijayasri, S.; Hopper, W. Towards the identification of novel phytochemical leads as macrodomain inhibitors of Chikungunya virus using molecular docking approach. J. Appl. Pharm. Sci. 2017, 7, 74–82. [Google Scholar] [CrossRef]
- Simone-Finstrom, M.; Borba, R.S.; Wilson, M.; Spivak, M. Propolis counteracts some threats to honey bee health. Insects 2017, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Simone-Finstrom, M.D.; Spivak, M. Increased resin collection after parasite challenge: A case of self-medication in honey bees? PLoS ONE 2012, 7, e34601. [Google Scholar] [CrossRef]
- Simone-Finstrom, M.; Spivak, M. Propolis and bee health: The natural history and significance of resin use by honey bees. Apidologie 2010, 41, 295–311. [Google Scholar] [CrossRef]
- Palmer-Young, E.C.; Tozkar, C.O.; Schwarz, R.S.; Chen, Y.; Irwin, R.E.; Adler, L.S.; Evans, J.D. Nectar and pollen phytochemicals stimulate honey bee (Hymenoptera: Apidae) immunity to viral infection. J. Econ. Entomol. 2017, 110, 1959–1972. [Google Scholar] [CrossRef]
- Aurori, A.C.; Bobiş, O.; Dezmirean, D.S.; Mărghitaş, L.A.; Erler, S. Bay laurel (Laurus nobilis) as potential antiviral treatment in naturally BQCV infected honeybees. Virus Res. 2016, 222, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Gherman, B.I.; Denner, A.; Bobiş, O.; Dezmirean, D.S.; Mǎrghitaş, L.A.; Schlüns, H.; Moritz, R.F.A.; Erler, S. Pathogen-associated self-medication behavior in the honeybee Apis mellifera. Behav. Ecol. Sociobiol. 2014, 68, 1777–1784. [Google Scholar] [CrossRef]
- Erler, S.; Moritz, R.F.A. Pharmacophagy and pharmacophory: Mechanisms of self-medication and disease prevention in the honeybee colony (Apis mellifera). Apidologie 2016, 47, 389–411. [Google Scholar] [CrossRef]
- Mihai, C.M.; Mârghitaş, L.A.; Dezmirean, D.S.; Chirilâ, F.; Moritz, R.F.A.; Schlüns, H. Interactions among flavonoids of propolis affect antibacterial activity against the honeybee pathogen Paenibacillus larvae. J. Invertebr. Pathol. 2012, 110, 68–72. [Google Scholar] [CrossRef]
- Wolska, K.; Górska, A.; Adamiak, A. Antibacterial properties of propolis. Postepy Mikrobiol. 2016, 55, 343–350. [Google Scholar]
- Flesar, J.; Havlik, J.; Kloucek, P.; Rada, V.; Titera, D.; Bednar, M.; Stropnicky, M.; Kokoska, L. In vitro growth-inhibitory effect of plant-derived extracts and compounds against Paenibacillus larvae and their acute oral toxicity to adult honey bees. Vet. Microbiol. 2010, 145, 129–133. [Google Scholar] [CrossRef]
- Stamets, P.E.; Naeger, N.L.; Evans, J.D.; Han, J.O.; Hopkins, B.K.; Lopez, D.; Moershel, H.M.; Nally, R.; Sumerlin, D.; Taylor, A.W.; et al. Extracts of polypore mushroom mycelia reduce viruses in honey bees. Sci. Rep. 2018, 8, 13936. [Google Scholar] [CrossRef] [PubMed]
- Burnham, A.J. Scientific Advances in Controlling Nosema ceranae (Microsporidia) Infections in Honey Bees (Apis mellifera). Front. Vet. Sci. 2019, 6. [Google Scholar] [CrossRef] [PubMed]
- Ptaszyńska, A.A.; Trytek, M.; Borsuk, G.; Buczek, K.; Rybicka-Jasińska, K.; Gryko, D. Porphyrins inactivate Nosema spp. microsporidia. Sci. Rep. 2018, 8, 5523. [Google Scholar] [CrossRef] [PubMed]
- Maori, E.; Lavi, S.; Mozes-Koch, R.; Gantman, Y.; Peretz, Y.; Edelbaum, O.; Tanne, E.; Sela, I. Isolation and characterization of Israeli acute paralysis virus, a dicistrovirus affecting honeybees in Israel: Evidence for diversity due to intra- and inter-species recombination. J. Gen. Virol. 2007, 88, 3428–3438. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.; Ellis, J.; vanEngelsdorp, D.; Hayes, J.; Westervelt, D.; Glick, E.; Williams, M.; Sela, I.; Maori, E.; Pettis, J.; et al. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, hymenoptera: Apidae). PLoS Pathog. 2010, 6, e1001160. [Google Scholar] [CrossRef]
- Desai, S.D.; Eu, Y.J.; Whyard, S.; Currie, R.W. Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect Mol. Biol. 2012, 21, 446–455. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Childers, A.K.; Lopez, L.; Grubbs, K.; Posada, F.; Weaver, D.; vanEngelsdorp, D.; Chen, Y.P.; Evans, J.D. Dynamic evolution in the key honey bee pathogen Deformed wing virus. PLoS Biol. 2019, in press. [Google Scholar] [CrossRef]
- Wiese, N.; Fischer, J.; Heidler, J.; Lewkowski, O.; Degenhardt, J.; Erler, S. The terpenes of leaves, pollen, and nectar of thyme (Thymus vulgaris) inhibit growth of bee disease-associated microbes. Sci. Rep. 2018, 8, 14634. [Google Scholar] [CrossRef]
- Kim, J.; Park, S.; Shin, Y.K.; Kang, H.; Kim, K.Y. In vitro antibacterial activity of macelignan and corosolic acid against the bacterial bee pathogens Paenibacillus larvae and Melissococcus plutonius. Acta Vet. Brno 2018, 87, 277–284. [Google Scholar] [CrossRef]
- Isidorov, V.A.; Buczek, K.; Segiet, A.; Zambrowski, G.; Swiecicka, I. Activity of selected plant extracts against honey bee pathogen Paenibacillus larvae. Apidologie 2018, 49, 687–704. [Google Scholar] [CrossRef]
- Alippi, A.M.; Reynaldi, F.J. Inhibition of the growth of Paenibacillus larvae, the causal agent of American foulbrood of honeybees, by selected strains of aerobic spore-forming bacteria isolated from apiarian sources. J. Invertebr. Pathol. 2006, 91, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Armstrong, T.N. Inhibition of the American foulbrood bacterium, Paenibacillus larvae larvae, by bacteria isolated from honey bees. J. Apic. Res. 2005, 44, 168–171. [Google Scholar] [CrossRef]
- Beims, H.; Wittmann, J.; Bunk, B.; Spröer, C.; Rohde, C.; Günther, G.; Rohde, M.; von der Ohe, W.; Steinert, M. Paenibacillus larvae-directed bacteriophage HB10c2 and its application in American foulbrood-affected honey bee larvae. Appl. Environ. Microbiol. 2015, 81, 5411–5419. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.B.; Oliveira, A.; Melo, L.D.R.; Azeredo, J. Identification of the first endolysin Cell Binding Domain (CBD) targeting Paenibacillus larvae. Sci. Rep. 2019, 9, 2568. [Google Scholar] [CrossRef]
- Goblirsch, M.J.; Spivak, M.S.; Kurtti, T.J. A cell line resource derived from honey bee (Apis mellifera) embryonic tissues. PLoS ONE 2013, 8, e69831. [Google Scholar] [CrossRef]
- Carrillo-Tripp, J.; Dolezal, A.G.; Goblirsch, M.J.; Miller, W.A.; Toth, A.L.; Bonning, B.C. In vivo and in vitro infection dynamics of honey bee viruses. Sci. Rep. 2016, 6, 22265. [Google Scholar] [CrossRef]
- Gisder, S.; Mockel, N.; Linde, A.; Genersch, E. A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environ. Microbiol. 2011, 13, 404–413. [Google Scholar] [CrossRef]
- Procházková, M.; Füzik, T.; Skubník, K.; Moravcová, J.; Ubiparip, Z.; Pridal, A.; Plevka, P. Virion structure and genome delivery mechanism of sacbrood honeybee virus. Proc. Natl. Acad. Sci. USA 2018, 115, 7759–7764. [Google Scholar] [CrossRef] [Green Version]
- Organtini, L.J.; Shingler, K.L.; Ashley, R.E.; Capaldi, E.A.; Durrani, K.; Dryden, K.A.; Makhov, A.M.; Conway, J.F.; Pizzorno, M.C.; Hafenstein, S. Honey bee Deformed wing virus structures reveal that conformational changes accompany genome release. J. Virol. 2017, 91, e01795-16. [Google Scholar] [CrossRef]
- Wilke, R.; Reif, D.; Moore, J. Combinatorial pharmacogenetics. Nat. Rev. Drug Discov. 2005, 4, 911–918. [Google Scholar] [CrossRef]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Chen, Y.P.; Di Prisco, G.; Pettis, J.; Williams, V. Bee cups: Single-use cages for honey bee experiments. J. Apic. Res. 2009, 48, 300–302. [Google Scholar] [CrossRef]
- Fries, I.; Chauzat, M.P.; Chen, Y.P.; Doublet, V.; Genersch, E.; Gisder, S.; Higes, M.; McMahon, D.P.; Martin-Hernandez, R.; Natsopoulou, M.; et al. Standard methods for Nosema research. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Carreck, N.L.; Andree, M.; Brent, C.S.; Cox-Foster, D.; Dade, H.A.; Ellis, J.D.; Hatjina, F.; vanEnglesdorp, D. Standard methods for Apis mellifera anatomy and dissection. J. Apic. Res. 2013, 52, 1–40. [Google Scholar] [CrossRef]
- Raymann, K.; Coon, K.L.; Shaffer, Z.; Salisbury, S.; Moran, N.A. Pathogenicity of Serratia marcescens Strains in Honey Bees. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Ryabov, E.V.; Childers, A.K.; Chen, Y.; Madella, S.; Nessa, A.; vanEngelsdorp, D.; Evans, J.D. Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci. Rep. 2017, 7, 17447. [Google Scholar] [CrossRef]
- Lanzi, G.; De Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef]
- Kevill, J.L.; Highfield, A.; Mordecai, G.J.; Martin, S.J.; Schroeder, D.C. ABC assay: Method development and application to quantify the role of three DWV master variants in overwinter colony losses of European honey bees. Viruses 2017, 9, 314. [Google Scholar] [CrossRef]
- Evans, J.D.; Schwarz, R.S.; Chen, Y.P.; Budge, G.; Cornman, R.S.; De La Rua, P.; De Miranda, J.R.; Foret, S.; Foster, L.; Gauthier, L.; et al. Standard methods for molecular research in Apis mellifera. J. Apic. Res. 2013, 52, 1–54. [Google Scholar] [CrossRef]
- Tauber, J.; Nguyen, V.; Lopez, D.; Evans, J.D. Effects of a resident yeast from the honeybee gut on immunity, microbiota, and nosema disease. Insects 2019, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Vejnovic, B.; Stevanovic, J.; Schwarz, R.S.; Aleksic, N.; Mirilovic, M.; Jovanovic, N.M.; Stanimirovic, Z. Quantitative PCR assessment of Lotmaria passim in Apis mellifera colonies co-infected naturally with Nosema ceranae. J. Invertebr. Pathol. 2018, 151, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Bradford, E.L.; Christie, C.R.; Campbell, E.M.; Bowman, A.S. A real-time PCR method for quantification of the total and major variant strains of the deformed wing virus. PLoS ONE 2017, 12, e0190017. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D. Beepath: An ordered quantitative-PCR array for exploring honey bee immunity and disease. J. Invertebr. Pathol. 2006, 93, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Delaplane, K.S.; Van Der Steen, J.; Guzman-Novoa, E. Standard methods for estimating strength parameters of Apis mellifera colonies. J. Apic. Res. 2013, 52, 1–12. [Google Scholar] [CrossRef]
| Disease/Host | Target | Forward Primer | Reverse Primer | Reference |
|---|---|---|---|---|
| Lotmaria passim-specific | Lotmaria passim | AGTATGAGCAGTAGGTTTTATTATA | GCCAAACACCAATAACTGGTACT | [62] |
| Deformed wing virus- | DWV-A,-B | ACGCAACCCCAGGAAT | GTAGCTAATTTTACCCAATCTTTAAA | [63] |
| Nosemosis | Nosema ceranae | TATTGTAGAGAGGTGGGAGATT | GTCGCTATGATCGCTTGCC | [53] |
| Bacterial infection | Bacteria (all), including Serratia | AGAGTTTGATCCTGGCTCAG | CTGCTGCCTCCCGTAGGAGT | [8] |
| Reference gene (host) | Ribosomal protein S5a (Rps5a) | AATTATTTGGTCGCTGGAATTG | TAACGTCCAGCAGAATGTGGTA | [8] |
| Reference gene (host) | Actin related protein 1 (Arp1) | CCAAAGACCCAAGCTCCCTA | TGGCTTATTGGTTTATGTTTTTCGT | [8] |
| Immunity gene (host) | Hymenoptaecin (Hym) | CTCTTCTGTGCCGTTGCATA | GCGTCTCCTGTCATTCCATT | [64] |
| Age/nutrition/immunity (host) | Vitellogenin (Vg) | TCGACAACTGCGATCAAAGGA | TGGTCACCGACGATTGGATG | [8] |
| Sample ID# | Treatment | Replicate | Rps5a1st | Rps5a2nd | Average | Replicate Difference | CoV | |
| 1 | +Nosema+NP | 1 | 20.19 | 19.75 | 19.97 | 0.44 | 1.557971876 | |
| 2 | +Nosema-NP | 1 | 21.1 | 20.03 | 20.565 | 1.07 | 3.67908707 | |
| Sample ID# | Treatment | Hym1st | Hym2nd | Average | Replicate Difference | CoV | ΔCq | |
| 1 | +Nosema+NP | 1 | 25.2 | 26.1 | 25.65 | −0.9 | 2.481076425 | −5.68 |
| 2 | +Nosema-NP | 1 | 28.2 | 28.9 | 28.55 | −0.7 | 1.733711898 | −7.985 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tauber, J.P.; Collins, W.R.; Schwarz, R.S.; Chen, Y.; Grubbs, K.; Huang, Q.; Lopez, D.; Peterson, R.; Evans, J.D. Natural Product Medicines for Honey Bees: Perspective and Protocols. Insects 2019, 10, 356. https://doi.org/10.3390/insects10100356
Tauber JP, Collins WR, Schwarz RS, Chen Y, Grubbs K, Huang Q, Lopez D, Peterson R, Evans JD. Natural Product Medicines for Honey Bees: Perspective and Protocols. Insects. 2019; 10(10):356. https://doi.org/10.3390/insects10100356
Chicago/Turabian StyleTauber, James P., William R. Collins, Ryan S. Schwarz, Yanping Chen, Kyle Grubbs, Qiang Huang, Dawn Lopez, Raymond Peterson, and Jay D. Evans. 2019. "Natural Product Medicines for Honey Bees: Perspective and Protocols" Insects 10, no. 10: 356. https://doi.org/10.3390/insects10100356
APA StyleTauber, J. P., Collins, W. R., Schwarz, R. S., Chen, Y., Grubbs, K., Huang, Q., Lopez, D., Peterson, R., & Evans, J. D. (2019). Natural Product Medicines for Honey Bees: Perspective and Protocols. Insects, 10(10), 356. https://doi.org/10.3390/insects10100356









