Individualized Transcriptional Resolution of Complicated Malaria in a Colombian Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Subject Recruitment
2.2. Blood Sample Collection
2.3. Whole Transcriptome Sequencing (RNAseq) and Preprocessing of the Data
2.4. Analysis of RNAseq Data
2.4.1. Principal Variance Component Analysis
2.4.2. Modular Analysis
2.4.3. Differentially Expressed Gene Calling and Gene Ontology Analysis
3. Results
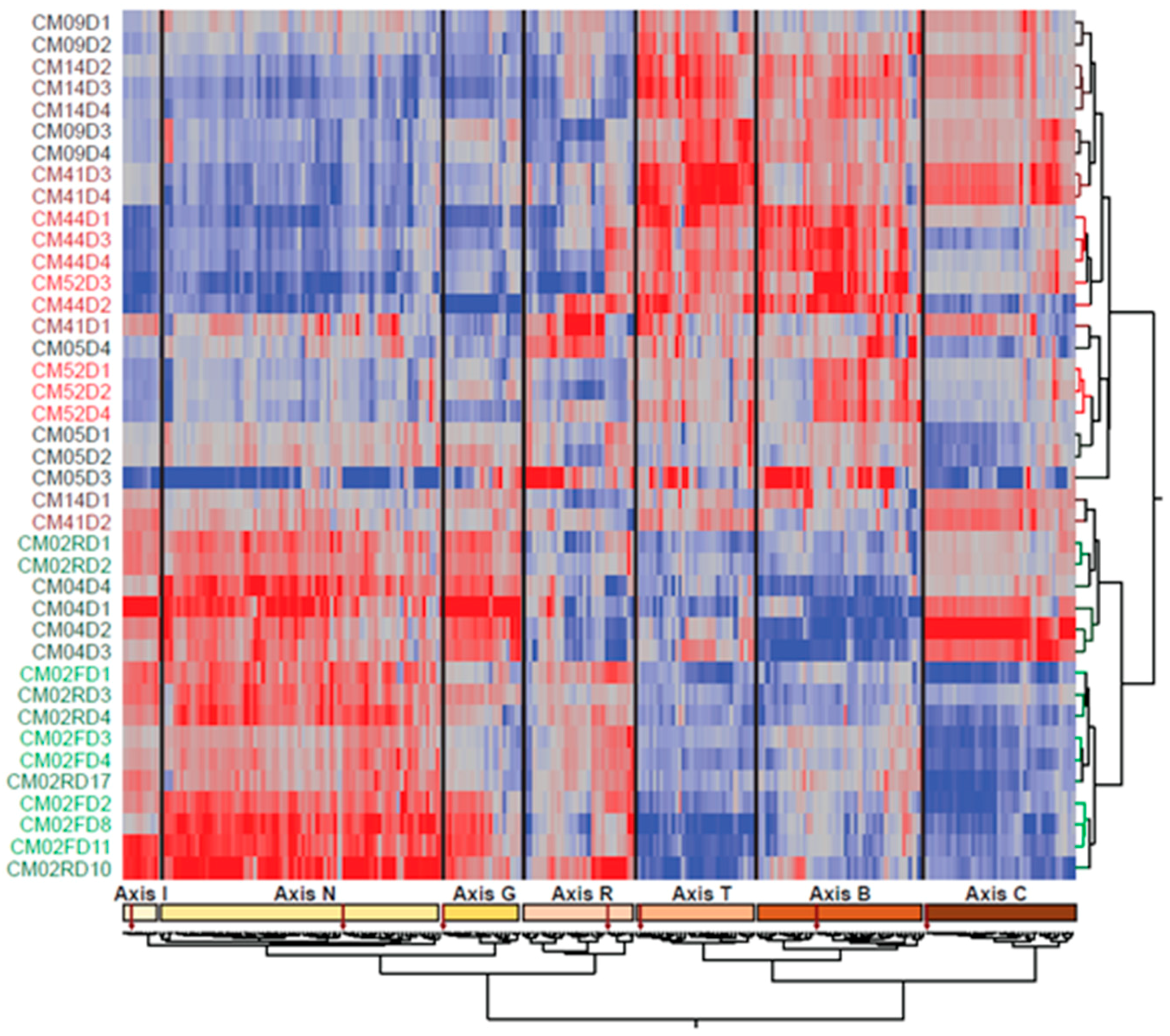
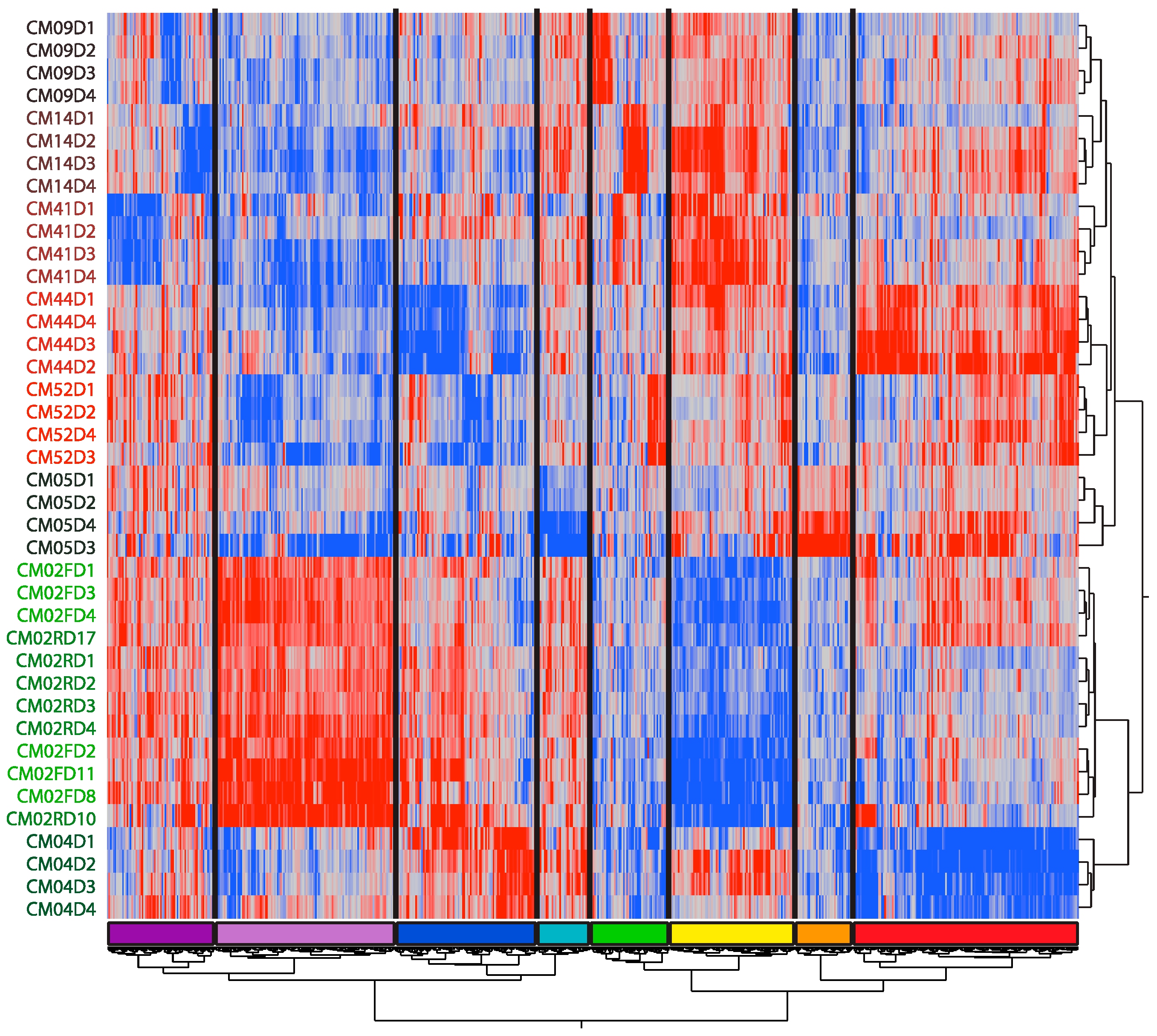
3.1. The Individual Is the Major Source of Variation as Patients Recover from Infection
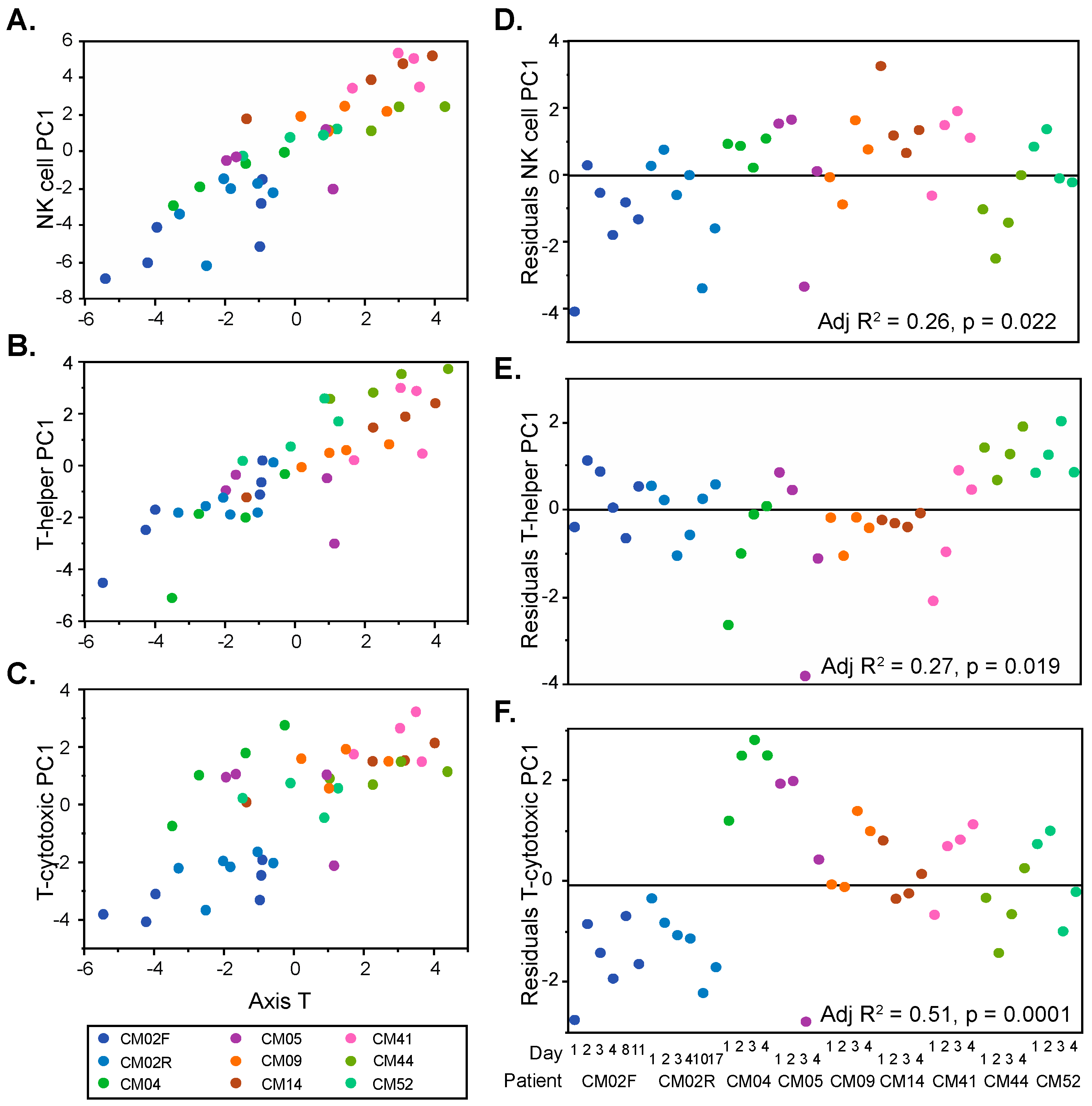
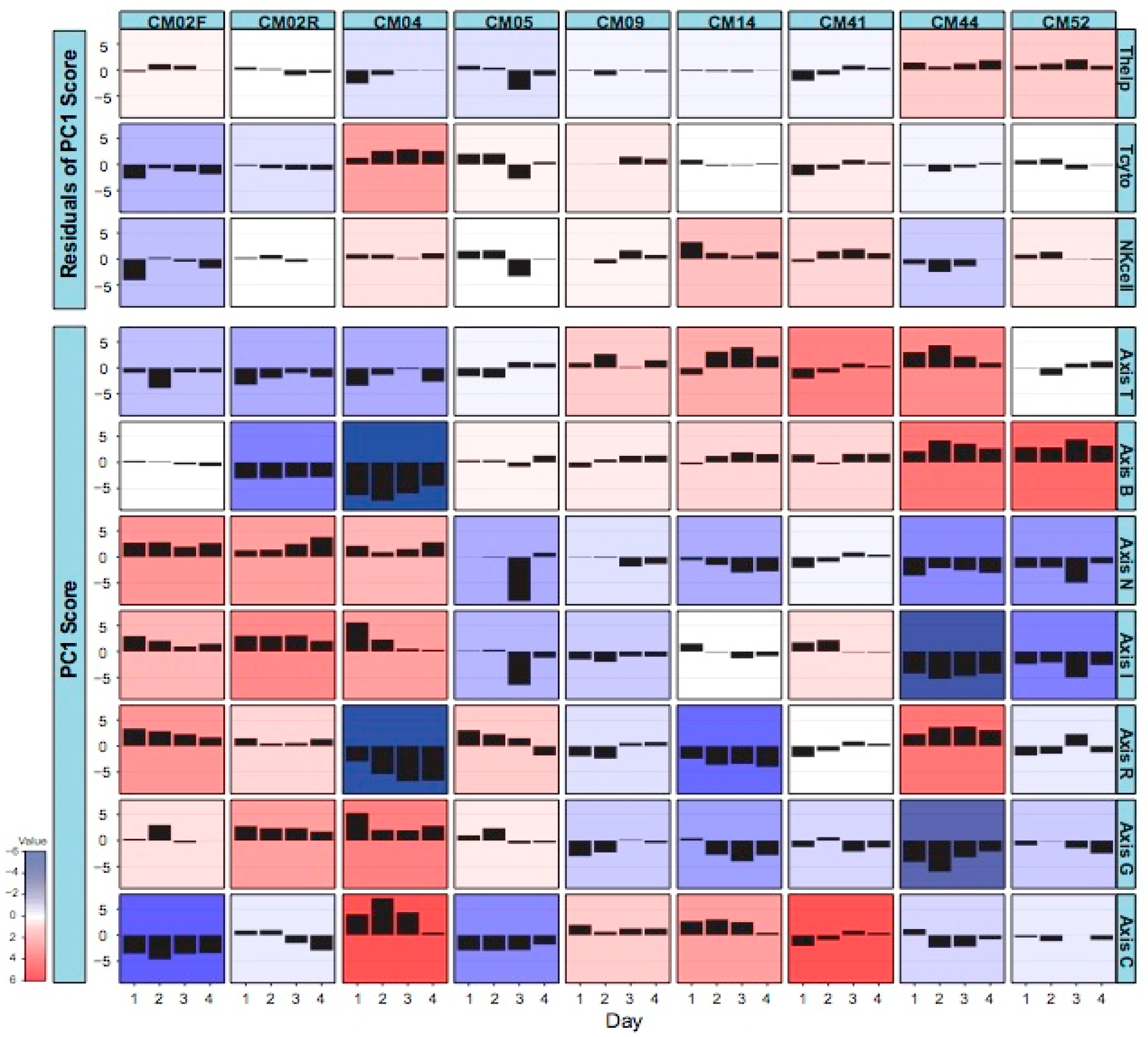
3.2. Modular Analysis Resolves the Major Immune Activities Engaged in Each Individual
- CM02, who presented with severe anemia clinical manifestations and relatively low parasitemia, had generally low cytotoxic T-lymphocyte and natural killer cell activity, which may explain her failure to resolve the infection. She also had particularly low mitotic activity during the initial infection and low B cell function during the relapse. All this was compensated by elevated inflammatory and interferon responses.
- CM04, who experienced renal and liver failure, also had a notable inflammatory response and low B cell activity, but this was offset by high cytotoxic T cell and mitotic activity. His reticulocyte numbers were relatively low, consistent with the absence of anemia, and his interferon response settled down upon hospitalization.
- CM05, with severe thrombocytopenia, was notable for a dramatic drop in inflammation and interferon signaling specifically on day 3 and for a relatively high cytotoxic T cell function on days 1 and 2.
- CM09, with severe thrombocytopenia and high parasitemia, had a rather stable profile without any dominant characteristics, perhaps correlating with his less complicated disease or with his age (69), which suggests that he may have built up more tolerance to malaria than the younger patients.
- CM14, with hepatic failure and severe thrombocytopenia, was notable for very high NK cell signatures and relatively low reticulocytes.
- CM41, who experienced severe thrombocytopenia and jaundice, presumably due to liver dysfunction, had relatively high NK cell and cytotoxic T cell activity, as well as elevated cell cycle gene expression, similar to CM14.
- CM44 and CM52, both infected with P. vivax, had similar transcriptome profiles. CM52 was a 3-year-old boy at the time of this episode of malaria and was mainly hospitalized for respiratory distress. CM44 presented a more extreme condition (severe thrombocytopenia, jaundice, and renal injury) with respect to her combination of low inflammation (perhaps related to a relatively low parasitemia) and particularly high B cell and helper T cell activity.
3.3. Patient-Specific Gene Expression Reinforces the Conclusions of the Modular Analysis
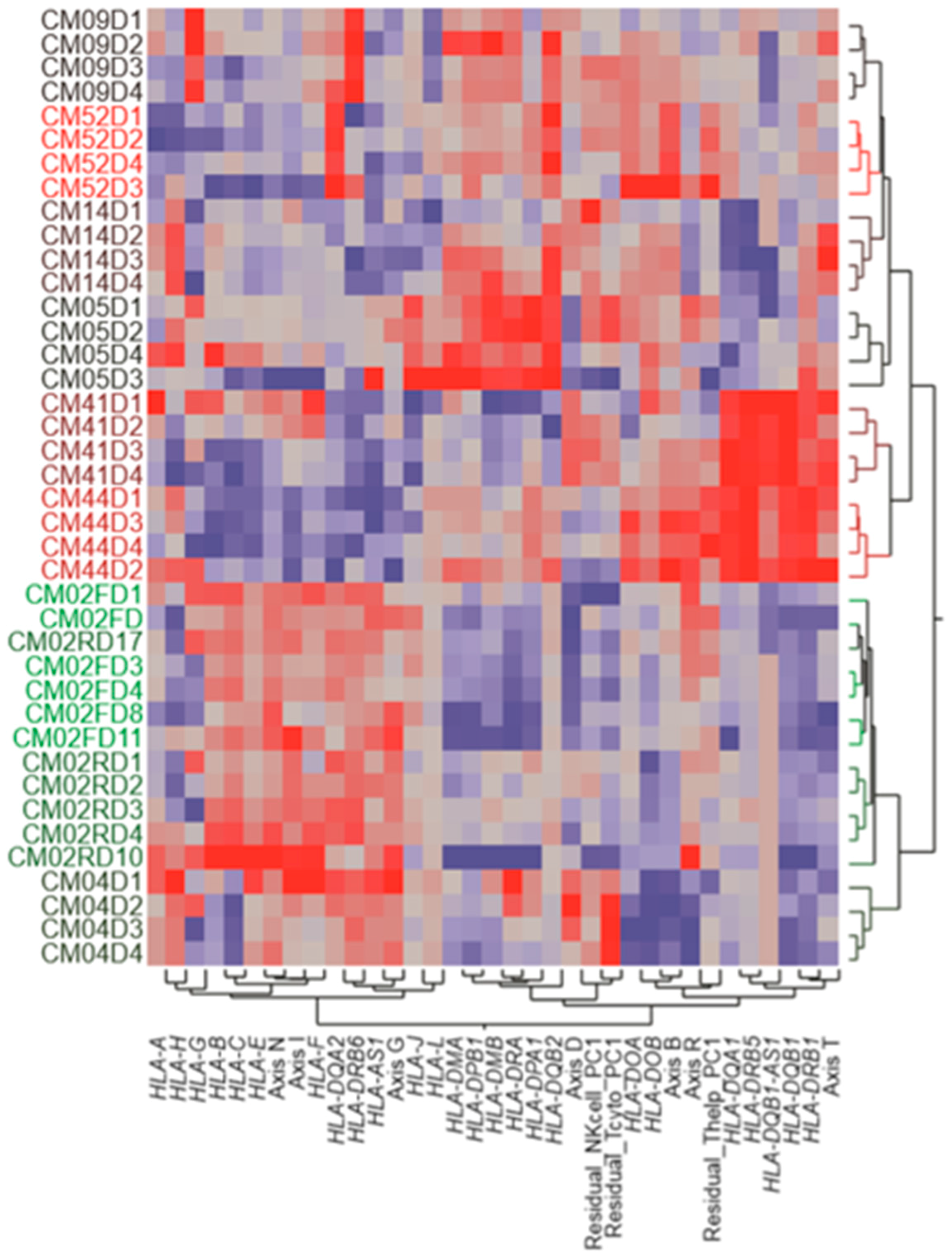
3.4. Variable Expression of HLA Complex Genes among Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). World Malaria Report; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Chaparro-Narváez, P.E.; Lopez-Perez, M.; Rengifo, L.M.; Padilla, J.; Herrera, S.; Arévalo-Herrera, M. Clinical and epidemiological aspects of complicated malaria in Colombia, 2007–2013. Malar. J. 2016, 15, 269. [Google Scholar] [CrossRef] [PubMed]
- Sypniewska, P.; Duda, J.F.; Locatelli, I.; Althaus, C.R.; Althaus, F.; Genton, B. Clinical and laboratory predictors of death in African children with features of severe malaria: A systematic review and meta-analysis. BMC Med. 2017, 15, 147. [Google Scholar] [CrossRef] [PubMed]
- Arévalo-Herrera, M.; Lopez-Perez, M.; Medina, L.; Moreno, A.; Gutierrez, J.B.; Herrera, S. Clinical profile of Plasmodium falciparum and Plasmodium vivax infections in low and unstable malaria transmission settings of Colombia. Malar. J. 2015, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Arévalo-Herrera, M.; Rengifo, L.; Lopez-Perez, M.; Arce-Plata, M.I.; Garcia, J.; Herrera, S. Complicated malaria in children and adults from three settings of the Colombian Pacific Coast: A prospective study. PLoS ONE 2017, 12, e0185435. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Severe malaria. Trop. Med. Int. Health. 2014, 19 (Suppl. 1), 7–131. [Google Scholar] [CrossRef] [PubMed]
- Tobon-Castano, A.; Giraldo-Castro, C.; Blair, S. Prognostic value of clinical and parasitological signs for severe malaria in patients from Colombia. Biomedica 2012, 32 (Suppl. 1), 79–94. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, B.A.; Thakkinstian, A.; White, N.J.; Sirivichayakul, C.; Dondorp, A.M.; Chokejindachai, W. Severe vivax malaria: A systematic review and meta-analysis of clinical studies since 1900. Malar. J. 2014, 13, 481. [Google Scholar] [CrossRef] [PubMed]
- Quispe, A.M.; Pozo, E.; Guerrero, E.; Durand, S.; Baldeviano, G.C.; Edgel, K.A.; Graf, P.C.; Lescano, A.G. Plasmodium vivax hospitalizations in a monoendemic malaria region: Severe vivax malaria? Am. J. Trop. Med. Hyg. 2014, 91, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, L.M.; Guzman, M.; Carmona, J.; Lopera, T.; Blair, S. Clinical and epidemiologic characteristics of 291 hospitalized patients for malaria in Medellin (Colombia). Acta Med. Colomb. 2000, 25, 163–170. [Google Scholar]
- Langhorne, J.; Ndungu, F.M.; Sponaas, A.M.; Marsh, K. Immunity to malaria: More questions than answers. Nat. Immunol. 2008, 9, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Gazzinelli, R.T.; Kalantari, P.; Fitzgerald, K.A.; Golenbock, D.T. Innate sensing of malaria parasites. Nat. Rev. Immunol. 2014, 14, 744–757. [Google Scholar] [CrossRef] [PubMed]
- McCall, M.B.; Sauerwein, R.W. Interferon-gamma-central mediator of protective immune responses against the pre-erythrocytic and blood stage of malaria. J. Leukoc. Biol. 2010, 88, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Peña, M.L.; Vallejo, A.; Herrera, S.; Gibson, G.; Arévalo-Herrera, M. Transcription profiling of malaria-naïve and semi-immune Colombian volunteers in a Plasmodium vivax sporozoite challenge. PLoS Negl. Trop. Dis. 2015, 9, e0003978. [Google Scholar]
- Idaghdour, Y.; Quinlan, J.; Goulet, J.; Berghout, J.; Gbeha, E.; Bruat, V.; de Malliard, T.; Grenier, J.C.; Gomez, S.; Gros, P.; et al. Evidence for additive and interaction effects of host genotype and infection in malaria. Proc. Natl. Acad. Sci. USA 2012, 109, 16786–16793. [Google Scholar] [CrossRef] [PubMed]
- Berg, A.; Otterdal, K.; Patel, S.; Gonca, M.; David, C.; Dalen, I.; Nymo, S.; Nilsson, M.; Nordling, S.; Magnusson, P.U. Complement activation correlates with disease severity and contributes to cytokine responses in Plasmodium falciparum malaria. J. Infect. Dis. 2015, 212, 1835–1840. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Biryukov, S.; Angov, E.; Landmesser, M.E.; Spring, M.D.; Ockenhouse, C.F.; Stoute, J.A. Complement and antibody-mediated enhancement of red blood cell invasion and growth of malaria parasites. EBioMedicine 2016, 9, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNAseq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Rouphael, N.; Duraisingham, S.; Romero-Steiner, S.; Presnell, S.; Davis, C.; Schmidt, D.S.; Johnson, S.C.; Milton, A.; Rajam, G.; et al. Molecular signatures of antibody responses derived from a systems biological study of 5 human vaccines. Nat. Immunol. 2014, 15, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Chaussabel, D.; Quinn, C.; Shen, J.; Patel, P.; Glaser, C.; Baldwin, N.; Stichweh, D.; Blankenship, D.; Li, L.; Munagala, I.; et al. A modular analysis framework for blood genomics studies: Application to systemic lupus erythematosus. Immunity 2008, 29, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Preininger, M.; Arafat, D.; Kim, J.; Nath, A.P.; Idaghdour, Y.; Brigham, K.L.; Gibson, G. Blood-informative transcripts define nine common axes of peripheral blood gene expression. PLoS Genet. 2013, 9, e1003362. [Google Scholar] [CrossRef] [PubMed]
- WHO. Severe falciparum malaria. World Health Organization, communicable diseases cluster. Trans. R. Soc. Trop. Med. Hyg. 2000, 94, 1–90. [Google Scholar]
- MinSalud, Ministerio de la Protección Social. Colombia. Guía de Atención Clínica de Malaria Ministerio de la Protección Social, Bogotá; Ministerio de la Protección Social: Bogota, Colombia, 2010; p. 132.
- Matheson, L.A.; Duong, T.T.; Rosenberg, A.M.; Yeung, R.S. Assessment of sample collection and storage methods for multicenter immunologic research in children. J. Immunol. Methods 2008, 339, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Sigurgeirsson, B.; Emanuelsson, O.; Lundeberg, J. Analysis of stranded information using an automated procedure for strand specific RNA sequencing. BMC Genom. 2014, 15, 631. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Whitney, A.R.; Diehn, M.; Popper, S.J.; Alizadeh, A.A.; Boldrick, J.C.; Relman, D.A.; Brown, P.O. Individuality and variation in gene expression patterns in human blood. Proc. Natl. Acad. Sci. USA 2003, 100, 1896–1901. [Google Scholar] [CrossRef] [PubMed]
- Mo, A.; Marigorta, U.M.; Arafat, D.; Chan, L.H.K.; Ponder, L.A.; Jang, S.R.; Prince, R.; Kugathasan, S.; Prahalas, S.; Gibson, G. Disease-specific regulation of gene expression in a comparative analysis of juvenile idiopathic arthritis and inflammatory bowel disease. Genome Med. 2018, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, R.; Sivadas, A.; Agrawal, V.; Tian, H.; Arafat, D.; Gibson, G. Omic personality: Implications of stable transcript and methylation profiles for personalized medicine. Genome Med. 2015, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, R.; Hong, S.; Cantarel, B.; Baldwin, N.; Baisch, J.; Edens, M.; Cepika, A.M.; Acs, P.; Turner, J.; Anguiano, E.; et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell 2016, 165, 1548–1550. [Google Scholar] [CrossRef] [PubMed]
- Brodin, P.; Jojic, V.; Gao, T.; Bhattacharya, S.; Angel, C.J.; Furman, D.; Shen-Orr, S.; Dekker, C.L.; Swan, G.E.; Butte, A.J.; et al. Variation in the human immune system is largely driven by non-heritable influences. Cell 2015, 160, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Carr, E.J.; Dooley, J.; Garcia-Perez, J.E.; Lagou, V.; Lee, J.C.; Wouters, C.; Meyts, I.; Goris, A.; Boeckxstaens, G.; Linterman, M.A.; et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016, 17, 461–468. [Google Scholar] [CrossRef] [PubMed]
| Subject | Site | Sex | Parasite Spp/Counts | Clinical Criteria § | Laboratory Criteria | Age | Hospital Days |
|---|---|---|---|---|---|---|---|
| CM09 | Valle del Cauca | M | P.f./25.1 | OI, RD | ST | 69 | 5 |
| CM14 | Chocó | M | P.f./19.6 | OI | Hep, ST, MH | 37 | 5 |
| CM41 | Chocó | F | P.f./142 | OI, Jau | ST | 49 | 4 |
| CM44 | Chocó | F | P.v./5.7 | OI, Jau | MH, RI, ST | 25 | 5 |
| CM52 | Chocó | M | P.v./5.7 | RD | None | 3 | 4 |
| CM05 | Chocó | F | P.f./11.6 | RD | ST | 38 | 4 |
| CM02 * | Chocó | F | P.f./5.2, 4.3 | RD | SA | 28 | 11, 18 |
| CM04 | Nariño | M | P.f./20.8 | OI | RI, Hep | 22 | 4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas-Peña, M.L.; Duan, M.; Arafat, D.; Rengifo, L.; Herrera, S.; Arévalo-Herrera, M.; Gibson, G. Individualized Transcriptional Resolution of Complicated Malaria in a Colombian Study. J. Pers. Med. 2018, 8, 29. https://doi.org/10.3390/jpm8030029
Rojas-Peña ML, Duan M, Arafat D, Rengifo L, Herrera S, Arévalo-Herrera M, Gibson G. Individualized Transcriptional Resolution of Complicated Malaria in a Colombian Study. Journal of Personalized Medicine. 2018; 8(3):29. https://doi.org/10.3390/jpm8030029
Chicago/Turabian StyleRojas-Peña, Mónica L., Meixue Duan, Dalia Arafat, Lina Rengifo, Socrates Herrera, Myriam Arévalo-Herrera, and Greg Gibson. 2018. "Individualized Transcriptional Resolution of Complicated Malaria in a Colombian Study" Journal of Personalized Medicine 8, no. 3: 29. https://doi.org/10.3390/jpm8030029
APA StyleRojas-Peña, M. L., Duan, M., Arafat, D., Rengifo, L., Herrera, S., Arévalo-Herrera, M., & Gibson, G. (2018). Individualized Transcriptional Resolution of Complicated Malaria in a Colombian Study. Journal of Personalized Medicine, 8(3), 29. https://doi.org/10.3390/jpm8030029






