Linking a Population Biobank with National Health Registries—The Estonian Experience
Abstract
:1. From a Biobank to Personalized Medicine
1.1. Objectives
1.2. Brief History
2. Biobanking and Legislation
HGRA and Consent Form
- -
- While the funding for research projects conducted by the EGCUT is competitively based, the specimen collection stored in the biobank and the database itself is maintained by the state as stated in the HGRA. This might contribute to the reasons that the biobank is strongly supported by the public as shown by annual telephone polls over the past decade [1].
- -
- Tissue samples, genetic information and the description of health (phenotypic data) all have a unique code and are anonymous. However, decoding is permitted for specific purposes described by the HGRA. These include renewal of the description of health or in order to contact participants. Decoding the data allows health descriptions to be linked with the genetic data of a participant, which is essential for genome-based health research.
- -
- The HGRA permits the participants to be re-contacted in order to renew and supplement the data available at the EGCUT. Re-contacting also makes it possible to obtain new tissue samples either when the first sample has been destroyed or does not contain sufficient DNA or when other types of tissue samples are necessary for specific projects (epigenetics, expression studies).
- -
- The act permits additional information to be received from other databases to supplement the description of the state of health of the participants.
- -
- The Gene Donor Consent Form signed by all participants is a “broad consent”. This type of consent requests permission to use the samples collected for future research purposes in general (with the objectives to study the links between genes, environmental factors and lifestyle with physical characteristics, health and disease), without specifically identifying each of the projects. This makes it possible to conduct a wide range of research, which would have been difficult if not impossible to foresee at the time of participant recruitment. Such research projects are only conducted after approval from the Research Ethics Committee of the University of Tartu.
- -
- The broad consent form also asks for permission to receive additional information from other databases. Linking with other databases and registries makes it possible to renew and enhance the database of the EGCUT on a regular basis without having to re-contact the participants.
- -
- The participants have the right to be aware of their data available in the database and they have the right to receive genetic counseling upon accessing that data. In other words, the participants have the right to have the research results returned. The participants can also grant their physician access to that information. This makes it possible to conduct research projects where personal genome information is introduced into medical practice.
3. Building, Maintaining, and Enhancing the Database
3.1. Study Design
3.2. Data Collected
3.3. Re-Contacting Participants
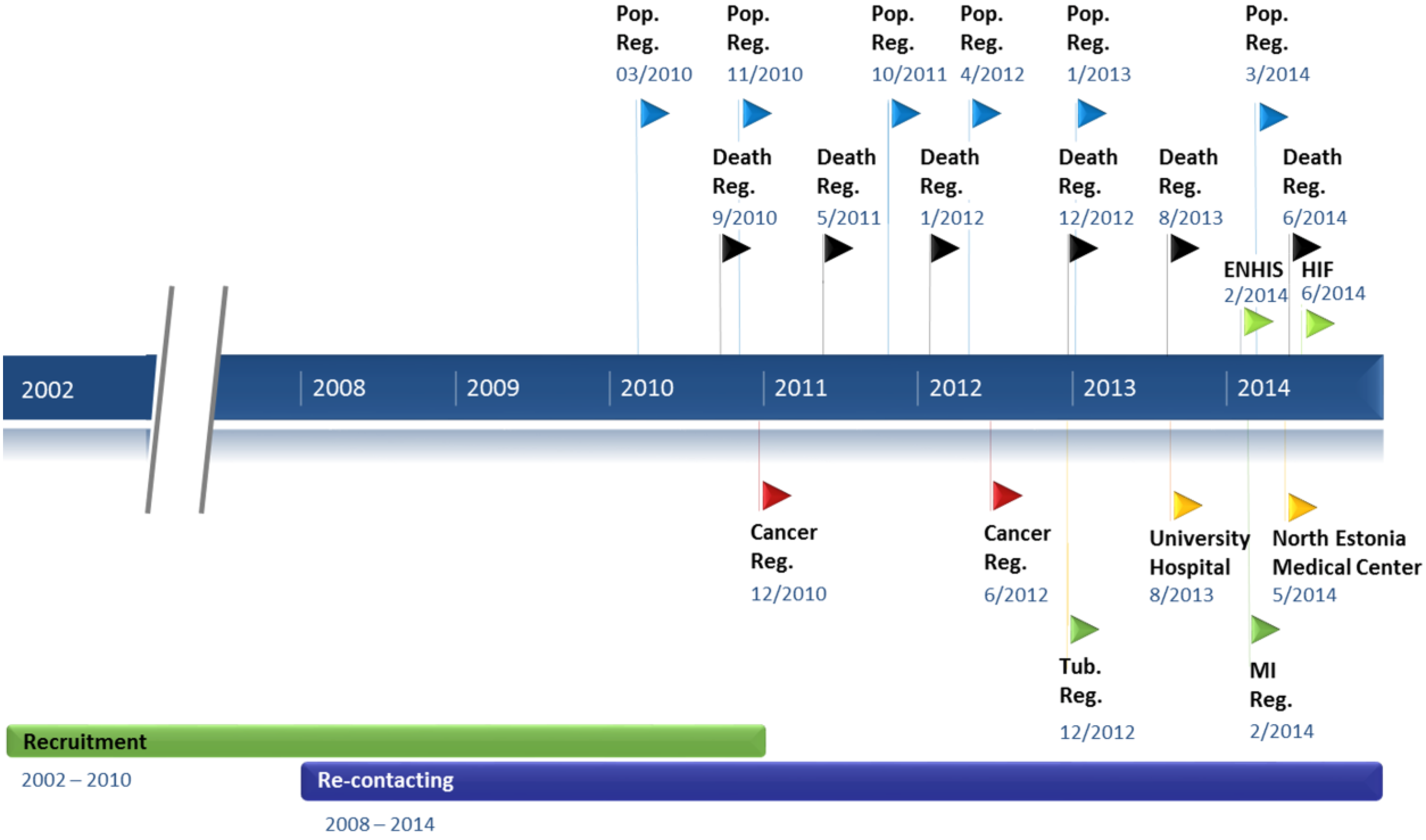
3.4. Data Updates from Registries and Hospital Databases
| Data Source | No. of Participants Found * | Percentage of EGCUT Participants Represented |
|---|---|---|
| Population Register | 51,800 | 99.8 |
| Estonian Health Insurance Fund | 51,607 | 99.5 |
| ENHIS | 39,880 | 76.9 |
| Tartu University Hospital | 22,492 | 43.4 |
| North Estonia Medical Centre | 21,202 | 40.9 |
| Estonian Cancer Registry | 2644 | 5.1 |
| Estonian Causes of Death Registry | 2349 | 4.5 |
| Myocardial Infarction Registry | 945 | 1.8 |
| Estonian Tuberculosis Registry | 260 | 0.5 |
3.5. Research Potential
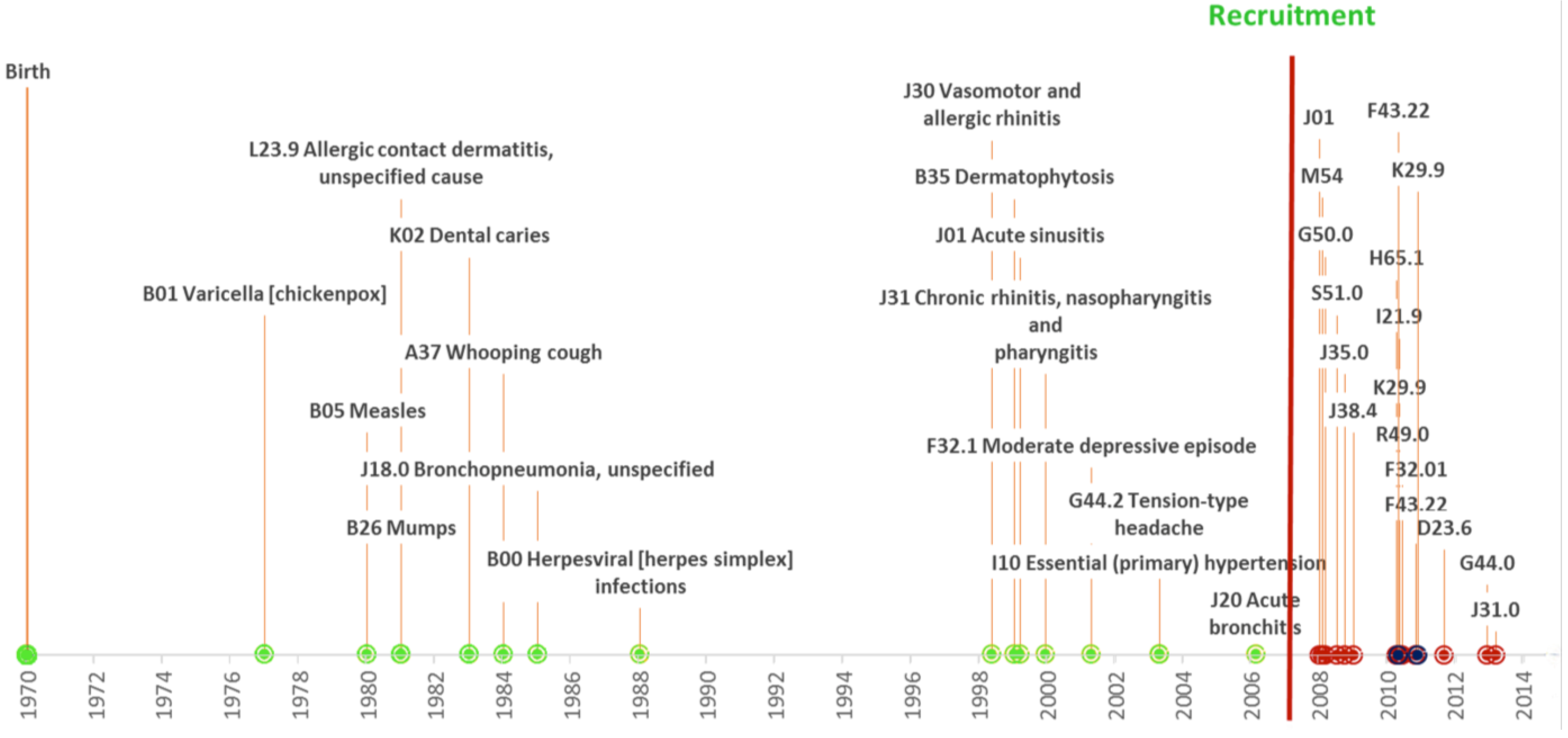
4. Info-Technology, E-Solutions and Related Projects
4.1. National Infrastructure
4.2. Management of the Data Collection
4.3. Future of the EGCUT Information System
5. Translating Research Results to Medicine
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Leitsalu, L.; Haller, T.; Esko, T.; Tammesoo, M.L.; Alavere, H.; Snieder, H.; Perola, M.; Ng, P.C.; Mägi, R.; Milani, L.; et al. Cohort profile: Estonian biobank of the Estonian Genome center, University of Tartu. Int. J. Epidemiol. 2014. [Google Scholar] [CrossRef]
- Riigikogu. Human Genes Research Act. 2000. Available online: https://www.riigiteataja.ee/en/eli/531102013003/consolide (accessed on 27 June 2014).
- Gene Donor Consent Form. Available online: http://www.geenivaramu.ee/en/access-biobank (accessed on 14 October 2014).
- Estonian Biobank. Available online: http://www.geenivaramu.ee/en/access-biobank (accessed on 2 January 2015).
- Fischer, K.; Kettunen, J.; Würtz, P.; Haller, T.; Havulinna, A.S.; Kangas, A.J.; Soininen, P.; Esko, T.; Tammesoo, M.L.; Mägi, R.; et al. Biomarker profiling by nuclear magnetic resonance spectroscopy for the prediction of all-cause mortality: An observational study of 17,345 persons. PLOS Med. 2014, 11. [Google Scholar] [CrossRef]
- Kohane, I.S. Using electronic health records to drive discovery in disease genomics. Nat. Rev. Genet. 2011, 12, 417–428. [Google Scholar] [CrossRef] [PubMed]
- E-Health. Overview of Estonian Electronic Health Record (EHR) System. Available online: http://www.e-tervis.ee/index.php/en/news-and-arcticles/432-overview-of-estonian-electronic-health-record-ehr-system (accessed on 3 July 2014).
- Sepper, R.; Ross, P.; Tiik, M. Nationwide Health Data Management System: A novel approach for integrating biomarker measurements with comprehensive health records in large populations studies. J. Proteome Res. 2011, 10, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Coalition Agreement. 2014. Available online: https://valitsus.ee/sites/default/files/content-editors/failid/2014_coalition_agreement_0.pdf (accessed on 3 July 2014).
- Government. The Government Commissioned a Personalised Medicine Implementation Pilot Project 2015–2018 from the Ministries. Available online: https://valitsus.ee/en/news/government-commissioned-personalised-medicine-implementation-pilot-project-2015–2018-ministries (accessed on 3 July 2014).
- Ginsburg, G. Medical genomics: Gather and use genetic data in health care. Nature 2014, 508, 451–453. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leitsalu, L.; Alavere, H.; Tammesoo, M.-L.; Leego, E.; Metspalu, A. Linking a Population Biobank with National Health Registries—The Estonian Experience. J. Pers. Med. 2015, 5, 96-106. https://doi.org/10.3390/jpm5020096
Leitsalu L, Alavere H, Tammesoo M-L, Leego E, Metspalu A. Linking a Population Biobank with National Health Registries—The Estonian Experience. Journal of Personalized Medicine. 2015; 5(2):96-106. https://doi.org/10.3390/jpm5020096
Chicago/Turabian StyleLeitsalu, Liis, Helene Alavere, Mari-Liis Tammesoo, Erkki Leego, and Andres Metspalu. 2015. "Linking a Population Biobank with National Health Registries—The Estonian Experience" Journal of Personalized Medicine 5, no. 2: 96-106. https://doi.org/10.3390/jpm5020096
APA StyleLeitsalu, L., Alavere, H., Tammesoo, M.-L., Leego, E., & Metspalu, A. (2015). Linking a Population Biobank with National Health Registries—The Estonian Experience. Journal of Personalized Medicine, 5(2), 96-106. https://doi.org/10.3390/jpm5020096





