Single-Nucleotide Polymorphisms Related to Glioblastoma Risk and Worldwide Epidemiology: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Search Strategy
2.3. Study Selection
2.4. Data Extraction
2.5. Quality Assessment
2.6. Data Analysis
2.7. Epidemiological Data Correlation
3. Results
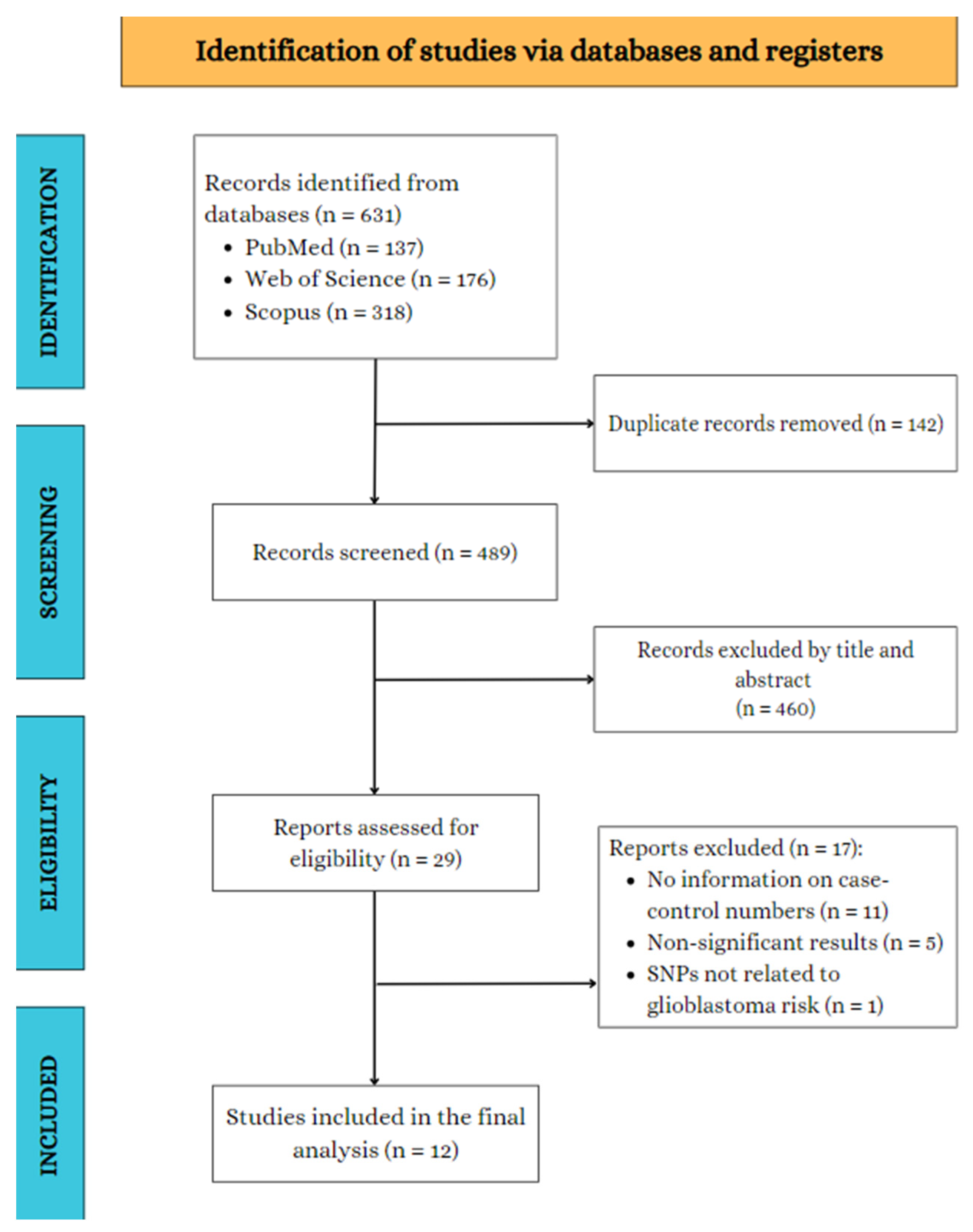
3.1. Search Results
3.2. Characteristics of Studies, Genes/SNPs, and Participants
3.3. Quality Assessment
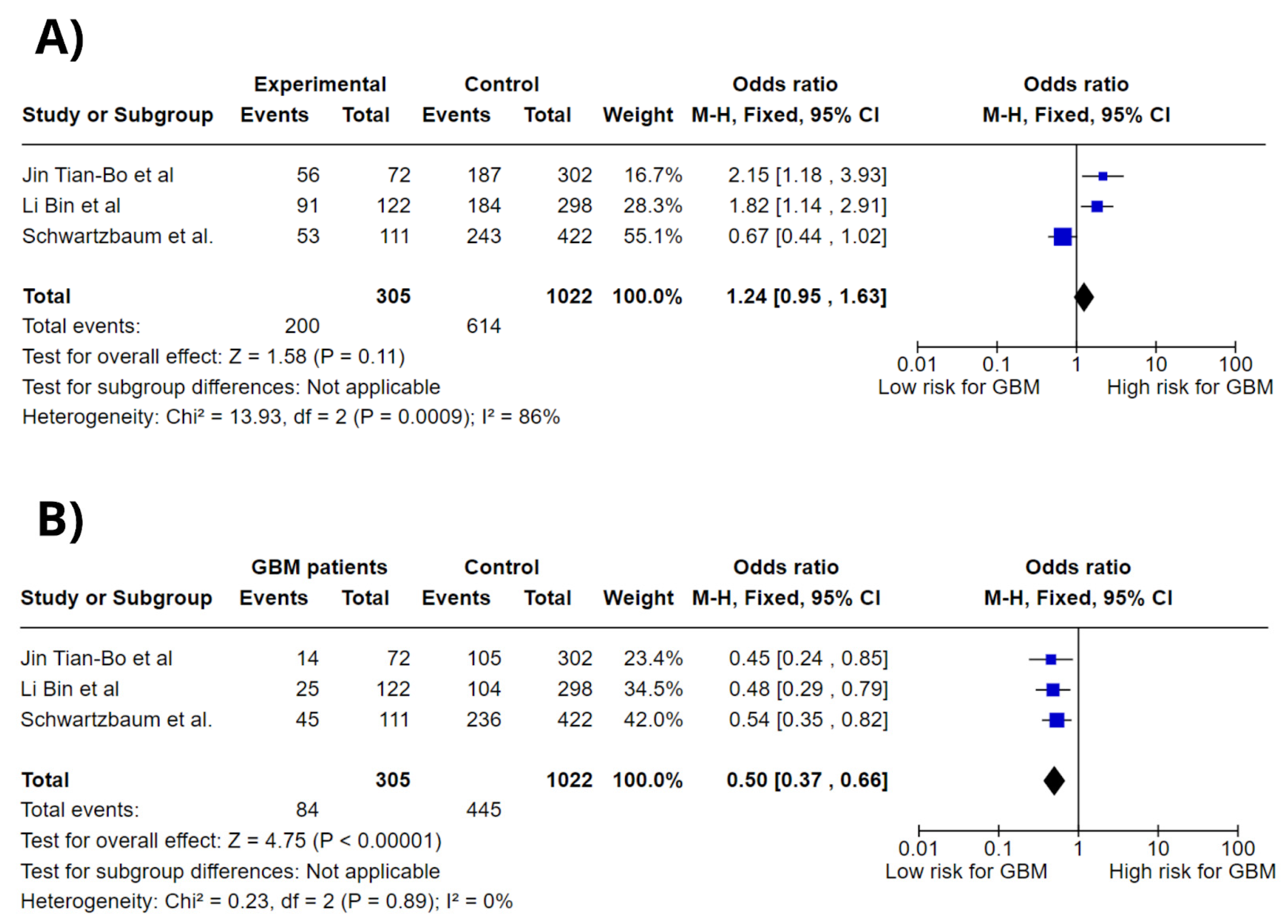
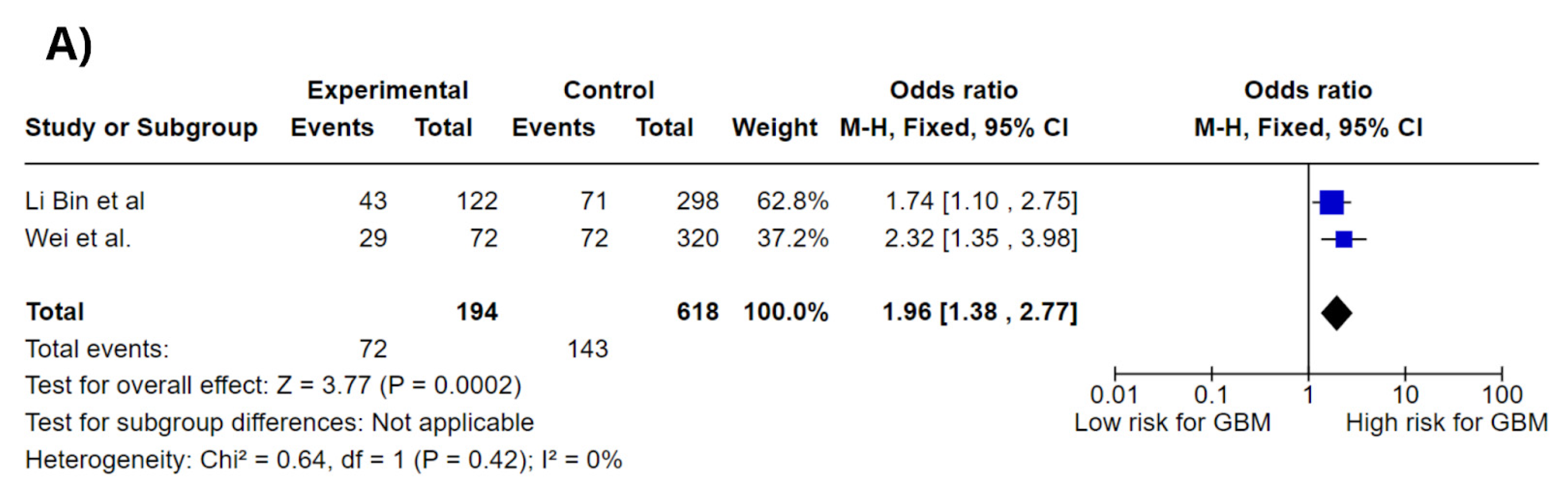
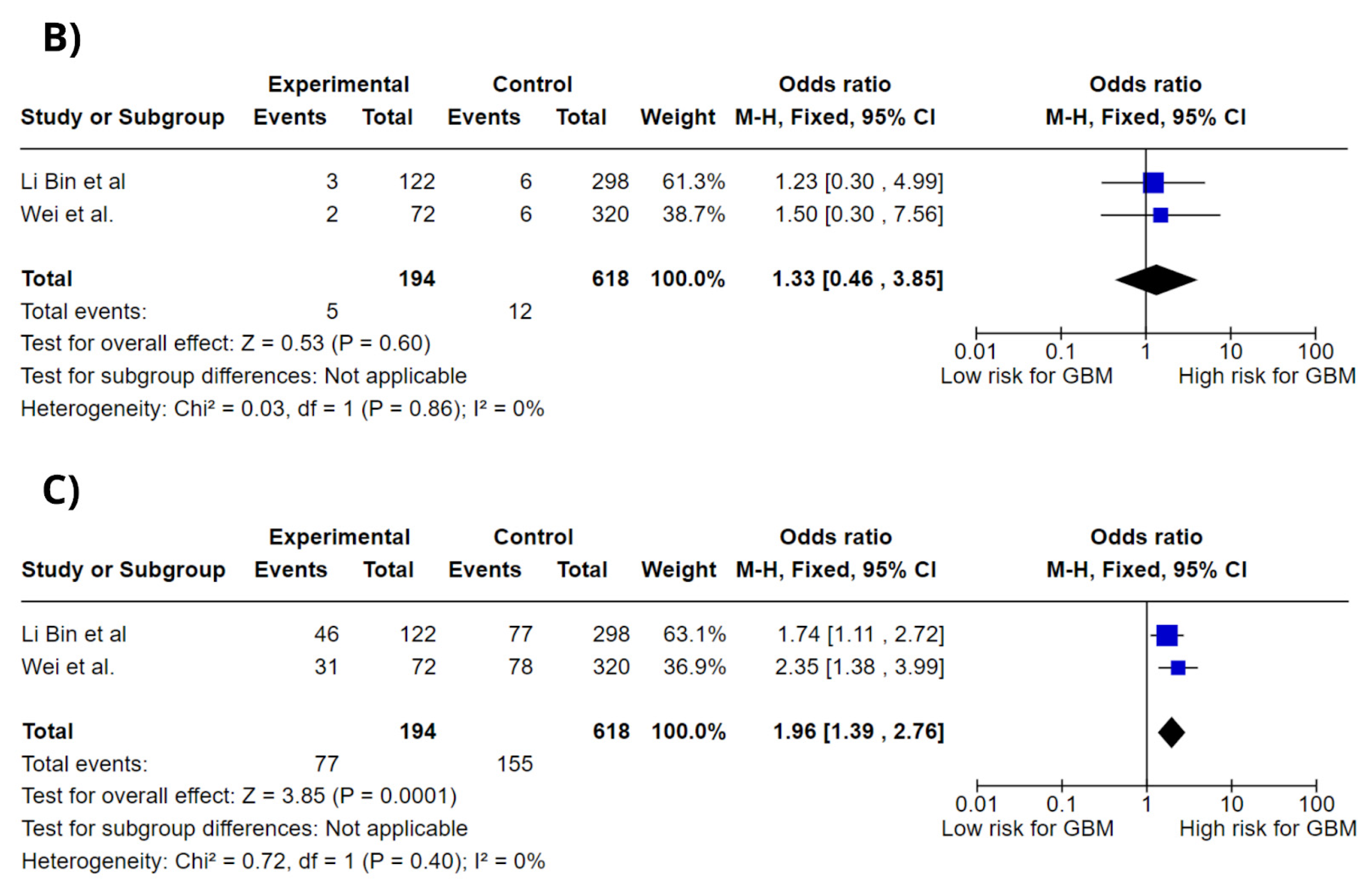
3.4. Meta-Analysis
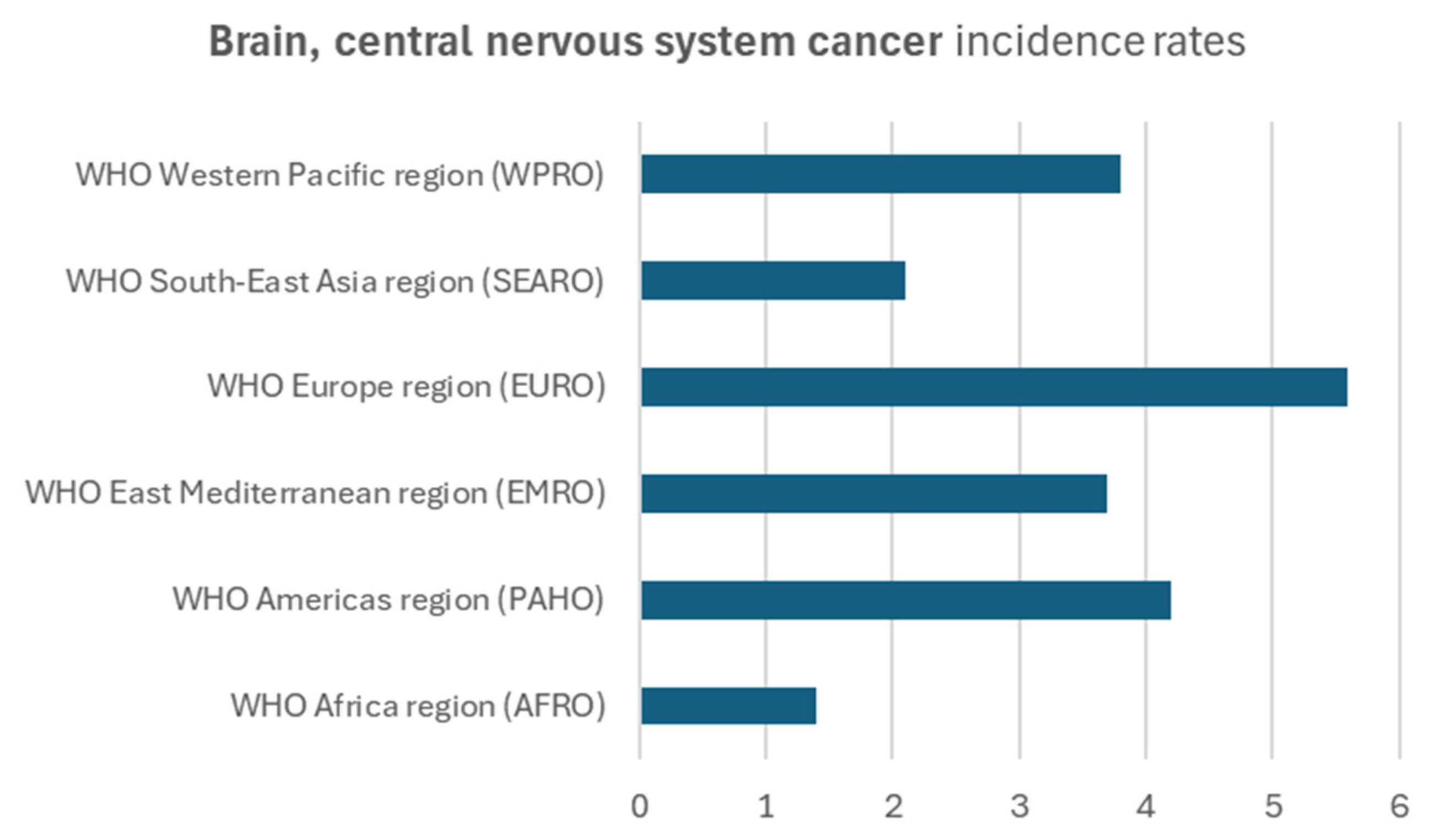
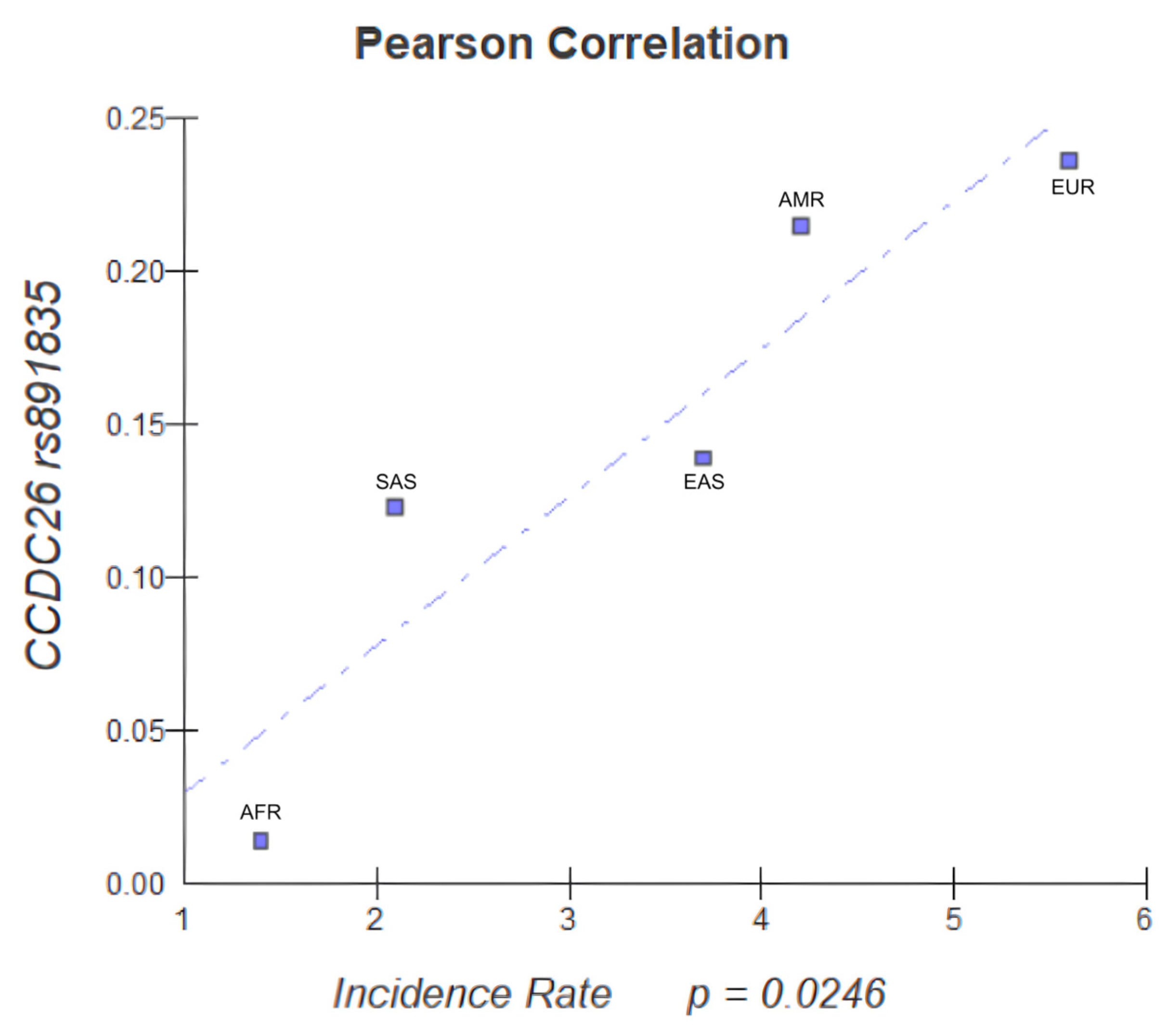
3.5. Epidemiological Correlation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weller, M.; Lim, M.; Ashby, L.S.; Reardon, D.A.; Bendszus, M. Glioblastoma Nature Reviews Disease Primers. Neuro Oncol. 2021, 23, 1617–1637. [Google Scholar]
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; A Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2016-2020. Neuro-Oncol. 2023, 25, iv1–iv99. [Google Scholar] [CrossRef]
- Orasanu, C.I.; Aschie, M.; Deacu, M.; Bosoteanu, M.; Vamesu, S.; Enciu, M.; Bălţătescu, G.I.; Cozaru, G.C.; Mitroi, A.F.; Voda, R.I. Implications of Cellular Immaturity in Necrosis and Microvascularization in Glioblastomas IDH-Wild-Type. Clin. Pract. 2022, 12, 1054–1068. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.; Ostrom, Q.; Byun, J.; Wimberly, C.; Armstrong, G.; Amos, C.; GICC Collaborators; Bondy, M. Epid-18. the genomic architecture of glioblastoma predisposition is highly polygenic in patients with a positive family history of glioma. Neuro-Oncol. 2023, 25, v119. [Google Scholar] [CrossRef]
- Nurminen, R.; Afyounian, E.; Paunu, N.; Katainen, R.; Isomäki, M.; Nurminen, A.; Scaravilli, M.; Tolppanen, J.; Fey, V.; Kivinen, A.; et al. Previously reported CCDC26 risk variant and novel germline variants in GALNT13, AR, and MYO10 associated with familial glioma in Finland. Sci. Rep. 2024, 14, 11562. [Google Scholar] [CrossRef]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for Glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef]
- Sundar, R.; Cho, B.-C.; Brahmer, J.R.; Soo, R.A. Nivolumab in NSCLC: Latest evidence and clinical potential. Ther. Adv. Med Oncol. 2015, 7, 85–96. [Google Scholar] [CrossRef]
- Wells, G.A.; Shea, B.; O'Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomized Studies in Meta-Analyses. Ottawa Hospital Research Institute. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed on 25 November 2024).
- Little, J.; Higgins, J.P.; Ioannidis, J.P.; Moher, D.; Gagnon, F.; von Elm, E.; Khoury, M.J.; Cohen, B.; Davey-Smith, G.; Grimshaw, J.; et al. Strengthening the reporting of genetic association studies (STREGA): An extension of the STROBE statement. PLOS Med. 2009, 6, e1000022. [Google Scholar] [CrossRef]
- Global Cancer Observatory. Cancer Today: Global Cancer Observatory Statistics on Brain and CNS Cancers; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Jin, T.; Li, X.; Zhang, J.; Wang, H.; Geng, T.; Li, G.; Gao, G.; Chen, C. Genetic association between selected cytokine genes and Glioblastoma in the Han Chinese population. BMC Cancer 2013, 13, 236. [Google Scholar] [CrossRef]
- Wei, X.-B.; Jin, T.-B.; Li, G.; Geng, T.-T.; Zhang, J.-Y.; Chen, C.-P.; Gao, G.-D.; Chen, C.; Gong, Y.-K. CCDC26 gene polymorphism and Glioblastoma risk in the Han Chinese population. Asian Pac. J. Cancer Prev. 2014, 15, 3629–3633. [Google Scholar] [CrossRef]
- Schwartzbaum, J.; Ahlbom, A.; Malmer, B.; Lönn, S.; Brookes, A.J.; Doss, H.; Debinski, W.; Henriksson, R.; Feychting, M. Polymorphisms associated with asthma are inversely related to Glioblastoma multiforme. Cancer Res. 2005, 65, 6459–6465. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, Y.; Yan, M.; Li, J.; Li, J.; Du, S.; Jin, T.; Gong, Y. Association of NFIL3, XRCC5 polymorphisms with Glioblastoma in Han Chinese population. Int. J. Clin. Exp. Pathol. 2017, 10, 4838–4846. [Google Scholar]
- Mesic, A.; Rogar, M.; Hudler, P.; Bilalovic, N.; Eminovic, I.; Komel, R. Genetic variations in AURORA cell cycle kinases are associated with Glioblastoma multiforme. Sci. Rep. 2021, 11, 17444. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Liu, P.F.; Liu, H.E.; Ma, L.X.; Li, G. Association between STAT5 polymorphisms and Glioblastoma risk in Han Chinese population. Pathol. Res. Pract. 2014, 210, 582–585. [Google Scholar] [CrossRef]
- Custodio, A.C.; Almeida, L.O.D.; Pinto, G.R.; Santos, M.J.D.; Almeida, J.R.; Clara, C.A.; Rey, J.A.; Casartelli, C. GSTP1 Ile105Val polymorphism in astrocytomas and Glioblastomas. Genet. Mol. Res. 2010, 9, 2328–2334. [Google Scholar] [CrossRef]
- Dong, Y.-S.; Hou, W.-G.; Li, X.-L.; Jin, T.-B.; Li, Y.; Feng, D.-Y.; Liu, D.-B.; Gao, G.-D.; Yin, Z.-M.; Qin, H.-Z. Genetic association of CHEK2, GSTP1, and ERCC1 with Glioblastoma in the Han Chinese population. Tumour Biol. 2014, 35, 4937–4941. [Google Scholar] [CrossRef]
- McKean-Cowdin, R.; Barnholtz-Sloan, J.; Inskip, P.D.; Ruder, A.M.; Butler, M.; Rajaraman, P.; Razavi, P.; Patoka, J.; Wiencke, J.K.; Bondy, M.L.; et al. Associations between polymorphisms in DNA repair genes and Glioblastoma. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1118–1126. [Google Scholar] [CrossRef]
- Rodriguez-Hernandez, I.; Perdomo, S.; Santos-Briz, A.; Garcia, J.L.; Gomez-Moreta, J.A.; Cruz, J.J.; Gonzalez-Sarmiento, R. Analysis of DNA repair gene polymorphisms in Glioblastoma. Gene 2014, 536, 79–83. [Google Scholar] [CrossRef]
- Jin, T.-B.; Li, X.-L.; Yang, H.; Jiri, M.; Shi, X.-G.; Yuan, D.-Y.; Kang, L.-L.; Li, S.-Q. Association of polymorphisms in FLT3, EGFR, ALOX5, and NEIL3 with Glioblastoma in the Han Chinese population. Med. Oncol. 2013, 30, 718. [Google Scholar] [CrossRef]
- Al-Khatib, S.M.; Abdo, N.; Al-Eitan, L.N.; Al-Mistarehi, A.-H.W.; Zahran, D.J.; Al Ajlouni, M.; Kewan, T.Z. The Impact of the Genetic Polymorphism in DNA Repair Pathways on Increased Risk of Glioblastoma Multiforme in the Arab Jordanian Population: A Case-Control Study. Appl. Clin. Genet. 2020, 13, 115–126. [Google Scholar] [CrossRef]
- Kohanbash, G.; McKaveney, K.; Sakaki, M.; Ueda, R.; Mintz, A.H.; Amankulor, N.; Fujita, M.; Ohlfest, J.R.; Okada, H. GM-CSF Promotes the Immunosuppressive Activity of Glioma-Infiltrating Myeloid Cells through Interleukin-4 Receptor-α. Cancer Res. 2013, 73, 6413–6423. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, P.; Mo, W.; Zhang, Y.; Zhou, W.; Deng, T.; Zhou, M.; Chen, X.; Wang, S.; Wang, C. lncRNA-CCDC26, as a novel biomarker, predicts prognosis in acute myeloid leukemia. Oncol. Lett. 2019, 18, 2203–2211. [Google Scholar] [CrossRef] [PubMed]
- Zou, L. Recurrent structural variations of lncRNA gene CCDC26 in diffuse intrinsic pontine glioma. bioRxiv 2021. [Google Scholar] [CrossRef]
- Li, S.; Jin, T.; Zhang, J.; Lou, H.; Yang, B.; Li, Y.; Chen, C.; Zhang, Y. Polymorphisms of TREH, IL-4R and CCDC26 genes associated with risk of glioma. Cancer Epidemiol. 2012, 36, 283–287. [Google Scholar] [CrossRef]
| Authors, Year | Country | Participants (Male/Female) | Mean Age | Mean Age at Diagnosis (Years) | Genotyping Method | GBM Histologically Confirmed | Ethnicity | Main Results |
|---|---|---|---|---|---|---|---|---|
| Mesic et al., 2021 [15] | Bosnia and Herzegovina | 129 (68/61) | 50 (19–81) | 58 | castPCR | Yes | European | rs2289590 in AURKB and rs11084490 in AURKC were associated with a reduced GBM risk |
| Liu et al., 2014 [16] | China | 115 (58.5%/41.5%) | 45.3 | 42.9 | PCR | Yes | Chinese | STAT5b rs2293157 G/T was associated with increased GBM risk |
| Custódio et al., 2010 [17] | Brazil | 80 (52/28) | 45 | NR | PCR-RFLP | NR | Brazilian | GSTP1 rs947894 polymorphism was associated with increased GBM risk |
| Dong et al., 2014 [18] | China | 72 (28/44) | 44 ± 15 | NR | locus-specific PCR | Yes | Chinese | Gene polymorphisms in CHEK2, GSTP1, and ERCC1 may be involved in GBM in the Han Chinese population |
| Jin Tianbo et al., 2013 [11] | China | 72 (28/44) | NR | 44 ± 15 | locus-specific PCR | Yes | Chinese | Polymorphisms within FLT3, EGFR, NEIL3, and ALOX5 may contribute to the occurrence of GBM in the Han Chinese population |
| Wei et al., 2014 [12] | China | 72 (28/44) | 44 ± 15 | 41 ± 18 | locus-specific PCR | Yes | Chinese | Genetic contribution of CCDC26 to GBM progression among Han Chinese |
| McKean-Cowdin et al., 2009 [19] | United States of America | 1015 (619/396) | 56.3 ± 12.6 | 56 | castPCR | Yes | North American | The C allele of the PARP1 rs1136410 variant was associated with decreased GBM risk, G allele of the PRKDC rs7003908 was associated with increased GBM risk |
| Schwartzbaum et al., 2005 [13] | United States of America | 111 (59.18% male) | 56 | 65 | DASH PCR | NR | North American | IL-4RA rs1805015 TC, CC and IL-4RA rs1801275 AG, AA were positively associated with GBM while IL-13 rs1800925 CT, TT was negatively associated with GBM |
| Rodriguez-Hernandez et al., 2013 [20] | Spain | 115 (59.1% male) | 63.2 ± 10.1 | NR | castPCR | NR | European | MLH1 rs1800734 and ERCC2 rs13181 polymorphisms might constitute glioblastoma susceptibility factors |
| Jin Tian-Bo et al., 2013 [21] | China | 72 (28/44) | NR | 44 ± 15 | locus-specific PCR | Yes | Chinese | rs12645561 in NEIL3 and rs2291427 in ALOX5 were associated with increased GBM risk |
| Li Bin et al., 2017 [14] | China | 122 (70/52) | 46.89 | NR | locus-specific PCR | NR | Chinese | NFIL3, XRCC5, CCDC26, TP53 and IL-4R genes associated with the risk of glioblastoma |
| Al-Khatib et al., 2020 [22] | Jordan | 84 (60% male) | 45.4 | NR | locus-specific PCR | Yes | Arabic | BRCA1 SNPs rs1799966 and rs799917 are associated with the risk of developing GBM in the Arab Jordanian population |
| Gene | SNPs | Study 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL-4R | rs1801275 | X | X | X | 3 | |||||||||
| IL-4R | rs1805015 | X | 1 | |||||||||||
| CCDC26 | rs891835 | X | X | 2 | ||||||||||
| CCDC26 | rs6470745 | X | 1 | |||||||||||
| GSTP1 | rs1695 | X | 1 | |||||||||||
| GSTP1 | rs947894 | X | 1 | |||||||||||
| AURKB | rs2289590 | X | 1 | |||||||||||
| AURKC | rs11084490 | X | 1 | |||||||||||
| STAT5b | rs2293157 | X | 1 | |||||||||||
| CHEK2 | rs2267130 | X | 1 | |||||||||||
| IL-3 | rs20541 | X | 1 | |||||||||||
| IL-10 | rs1800871 | X | 1 | |||||||||||
| PARP1 | rs1136410 | X | 1 | |||||||||||
| PRKDC | rs7003908 | X | 1 | |||||||||||
| MLH1 | rs1800734 | X | 1 | |||||||||||
| ERCC2 | rs13181 | X | 1 | |||||||||||
| FLT3 | rs3829382 | X | 1 | |||||||||||
| XRCC5 | rs9288516 | X | 1 | |||||||||||
| NFIL3 | rs7021746 | X | 1 | |||||||||||
| TP53 | rs1042522 | X | 1 | |||||||||||
| BRCA1 | rs799917 | X | 1 | |||||||||||
| EGFR | rs9642393 | X | 1 |
| Studies | Selection | Item 2 | Item 3 | Item 4 | Comparability | Item 1b | Exposure | Item 2 | Item 3 | Total Score |
|---|---|---|---|---|---|---|---|---|---|---|
| Item 1 | Item 1a | Item 1 | ||||||||
| Mesic et al., 2021 [15] | * | * | * | * | * | * | * | * | 8 | |
| Liu et al., 2014 [16] | * | * | * | * | * | * | * | * | * | 9 |
| Custódio et al., 2010 [17] | * | * | * | * | * | * | * | * | * | 9 |
| Dong et al., 2014 [18] | * | * | * | * | * | * | * | * | 8 | |
| Jin Tianbo et al., 2013 [11] | * | * | * | * | * | * | * | * | * | 9 |
| Wei et al., 2014 [12] | * | * | * | * | * | * | * | * | 8 | |
| McKean-Cowdin et al., 2009 [19] | * | * | * | * | * | * | * | * | * | 9 |
| Schwartzbaum et al., 2005 [13] | * | * | * | * | * | * | * | * | 8 | |
| Rodriguez-Hernandez et al., 2013 [20] | * | * | * | * | * | * | * | * | 8 | |
| Jin Tian-Bo et al., 2013 [21] | * | * | * | * | * | * | * | 7 | ||
| Li Bin et al., 2017 [14] | * | * | * | * | * | * | * | 7 | ||
| Al-Khatib et al., 2020 [22] | * | * | * | * | * | * | * | 7 |
| Studies | Description of Genotyping Methods and Errors | Description of Modeling Population Stratification | Description of Modeling Haplotype Variation | Hardy-Weinberg Equilibrium Was Considered | Statement of Whether the Study is the First Report of a Genetic Association a Replication Effort, or Both | Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotyping Methods and Platforms | Error Rates and Call Rates | Genotyping in Batches | Laboratory/Center Where the Genotyping Was Done | The Numbers of Individuals Was Successful Genotyping | ||||||
| Mesic et al., 2021 [15] | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | 8 |
| Liu et al., 2014 [16] | Yes | Yes | No | Yes | Yes | Yes | No | Yes | Yes | 7 |
| Custódio et al., 2010 [17] | Yes | No | No | Yes | Yes | Yes | No | Yes | Yes | 6 |
| Dong et al., 2014 [18] | Yes | No | No | Yes | Yes | Yes | No | Yes | Yes | 6 |
| Jin Tianbo et al., 2013 [11] | Yes | Yes | No | Yes | Yes | Yes | No | Yes | Yes | 7 |
| Wei et al., 2014 [12] | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| McKean-Cowdin et al., 2009 [19] | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | 8 |
| Schwartzbaum et al., 2005 [13] | Yes | Yes | No | Yes | Yes | No | No | No | No | 4 |
| Rodriguez-Hernandez et al., 2013 [20] | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | 7 |
| Jin Tian-Bo et al., 2013 [21] | Yes | Yes | No | Yes | Yes | No | No | Yes | Yes | 6 |
| Li Bin et al., 2017 [14] | Yes | No | No | Yes | Yes | Yes | No | Yes | Yes | 6 |
| Al-Khatib et al., 2020 [22] | Yes | No | No | Yes | No | No | No | Yes | Yes | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilioli da Costa Nunes, G.; Cezar Aquino de Moraes, F.; de Cássia Calderaro Coelho, R.; Rodrigues Fernandes, M.; Emanuel Batista dos Santos, S.; Pereira Carneiro dos Santos, N. Single-Nucleotide Polymorphisms Related to Glioblastoma Risk and Worldwide Epidemiology: A Systematic Review and Meta-Analysis. J. Pers. Med. 2025, 15, 401. https://doi.org/10.3390/jpm15090401
Gilioli da Costa Nunes G, Cezar Aquino de Moraes F, de Cássia Calderaro Coelho R, Rodrigues Fernandes M, Emanuel Batista dos Santos S, Pereira Carneiro dos Santos N. Single-Nucleotide Polymorphisms Related to Glioblastoma Risk and Worldwide Epidemiology: A Systematic Review and Meta-Analysis. Journal of Personalized Medicine. 2025; 15(9):401. https://doi.org/10.3390/jpm15090401
Chicago/Turabian StyleGilioli da Costa Nunes, Giovanna, Francisco Cezar Aquino de Moraes, Rita de Cássia Calderaro Coelho, Marianne Rodrigues Fernandes, Sidney Emanuel Batista dos Santos, and Ney Pereira Carneiro dos Santos. 2025. "Single-Nucleotide Polymorphisms Related to Glioblastoma Risk and Worldwide Epidemiology: A Systematic Review and Meta-Analysis" Journal of Personalized Medicine 15, no. 9: 401. https://doi.org/10.3390/jpm15090401
APA StyleGilioli da Costa Nunes, G., Cezar Aquino de Moraes, F., de Cássia Calderaro Coelho, R., Rodrigues Fernandes, M., Emanuel Batista dos Santos, S., & Pereira Carneiro dos Santos, N. (2025). Single-Nucleotide Polymorphisms Related to Glioblastoma Risk and Worldwide Epidemiology: A Systematic Review and Meta-Analysis. Journal of Personalized Medicine, 15(9), 401. https://doi.org/10.3390/jpm15090401








