Abstract
Background: COVID-19, caused by SARS-CoV-2, has posed significant challenge to global healthcare systems, necessitating reliable biomarkers to predict disease severity and mortality. This systematic review and meta-analysis evaluated the prognostic value of novel biomarkers in COVID-19 patients. The aim of this study was to identify and prioritize the most prognostically relevant novel biomarkers associated with COVID-19 outcomes. Methods: We conducted a systematic review and meta-analysis of the available evidence. A systematic search of PubMed and Web of Science was performed to identify studies on the COVID-19 biomarkers. Observational studies that compared poor (severe disease/mortality) and good outcomes were included. For continuous measures, standard mean differences (SMDs) with 95% confidence intervals (CIs) were calculated. Pooled sensitivity, specificity, diagnostic odds ratio (DOR), and summary receiver operating characteristic (SROC) curve analyses for the biomarkers were used. The risk of bias was assessed using the Newcastle–Ottawa scale. Results: Of the 2907 screened studies, 38 were included (21 in the meta-analysis). MR-proADM showed higher levels of prediction for poor outcomes (SMD = 1.40, 95% CI: 1.11–1.69; AUC 0.74–0.96; sensitivity, 85%; specificity, 71%). The neutrophil-to-lymphocyte ratio (NLR) showed a high correlation with disease severity (SMD = 1.07, 95% CI: 0.79–1.35; AUC 0.73–0.98; sensitivity, 86%; specificity, 78%). Increased KL-6 levels were associated with lung injury (SMD = 1.22, 95% CI: 0.24–2.19; AUC 0.85–0.95). Other biomarkers (suPAR, miR-155, Galectin-3) showed promise but lacked sufficient data for pooled analysis. Heterogeneity was observed among the included studies in terms of diagnostic accuracy. These findings indicate that elevated levels of MR-proADM, NLR, and KL-6 are significantly associated with COVID-19 prognostic accuracy to guide patient management. Conclusions: MR-proADM, NLR, and KL-6 levels demonstrated strong prognostic value for COVID-19 severity and mortality. These biomarkers can enhance clinical decision-making.
1. Introduction
In 2019, SARS-CoV-2 emerged in Wuhan, China, and spread worldwide, causing a pandemic responsible for more than 768 million cases and almost 7 million deaths globally [1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent of the current global coronavirus disease 2019 (COVID-19) pandemic [2]. COVID-19 presents as an asymptomatic or mild-to-severe-to-lethal form. Mild COVID-19 cases are characterized by flu-like symptoms, such as fever, monoarthralgia, cough, and nasal congestion [3]. Severe COVID-19 cases are characterized by acute respiratory distress syndrome (ARDS) triggered by a hyper-inflammatory response known as a cytokine storm, with respiratory failure, multiorgan disease, and death [4]. Patients with ARDS have a persistent elevation of inflammatory cytokines (IL-6, IL-8, and TNF-α) and are probably associated with disease severity and prognosis [5,6].
In this setting, clinical findings and analytical parameters are crucial for identifying high-risk patients, providing better treatment, and improving clinical outcomes [7,8]. During the COVID-19 pandemic, extensive evidence has been produced, and new biomarkers have been described, linking the roles of cytokines, traps, and nucleosomes [9]. Despite progress in treatment and prevention, immune mechanisms and host responses remain unclear [10]. Several biomarkers have been associated with the severity of COVID-19, including D-dimer, C-reactive protein, ferritin, and lactate dehydrogenase. However, their use has not yet been tested in a clinical setting [11].
New biomarkers such as Mid-Regional pro-Adrenomedullin (MR-proADM), neutrophil-to-lymphocyte ratio (NLR), and Krebs von den Lungen (KL-6) have been tested in different settings with better specificity than conventional markers [12]. MR-proADM is a vasoactive peptide linked to endothelial dysfunction and organ failure. Elevated levels are correlated with severe COVID-19, ARDS, and mortality [13]. The NLR is a simple and cost-effective marker of systemic inflammation. This reflects immune dysregulation, where high neutrophils and low lymphocytes predict worse outcomes [14]. KL-6 is a glycoprotein produced by damaged lung epithelial cells and is strongly associated with pulmonary fibrosis and severe respiratory failure in COVID-19 [15]. Other emerging biomarkers, such as the soluble urokinase Plasminogen Activator Receptor (suPAR), MicroRNA-155 (miR-155), and Galectin-3, indicate immune activation and hyperinflammation. miR-155 is a regulator of inflammatory signaling pathways that are potentially linked to cytokine storms [16]. Galectin-3 is involved in fibrosis and chronic inflammation, which may predict long-term lung damage [17].
Given the numerous studies published on novel biomarkers for COVID-19, there is a critical need to synthesize and evaluate the clinical relevance of these findings. Although these biomarkers show promise, their clinical utility remains understudied in large-scale meta-analyses. This systematic review and meta-analysis aimed to identify and prioritize the most prognostically relevant novel biomarkers associated with COVID-19 outcomes, quantify their strength of correlation with clinical endpoints, and assess their potential utility in guiding risk stratification and patient management strategies.
2. Material and Methods
This study was conducted in accordance with the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) statement [18] and was registered in the PROSPERO International Prospective Register of Systematic Reviews (No. CRD42024594709).
2.1. Inclusion and Exclusion Criteria
We chose the following studies for this review: observational studies regarding biomarkers in COVID-19 adult patients, comparing poor and good outcomes. There were no restrictions on the type of study (RCT, quasi-randomized trials, or case reports). Reviews, case reports, guidelines, systematic reviews, meta-analyses, and animal studies were also excluded.
2.2. Search Strategy
We searched two electronic databases, PubMed and Web of Science, through July 2024, using relevant keywords. WOS: (TS = (biomarkers) AND ((TS = (“COVID-19” OR “SARS-CoV-2”)) AND TS = (prognosis OR diagnosis))) AND DT = (Article). PubMed: (((biomarkers [MeSH Terms]) OR (biomarkers [Title/Abstract])) AND (“COVID-19” [Title/Abstract] OR “SARS-CoV-2” [Title/Abstract])) AND ((prognosis OR diagnosis [MeSH Terms]) OR (prognosis [Title/Abstract] OR diagnosis [Title/Abstract])).
2.3. Selection of Studies
Eligibility screening was conducted based on our inclusion criteria in two steps. First, we screened titles and abstracts for eligibility and then retrieved and screened the full-text articles of the selected abstracts for inclusion in this review. Two reviewers (F.N. and B.R.) independently conducted the literature search, screening, and retrieved trials. They screened the titles and abstracts. Following this preliminary screening, S.S. and D.M. retrieved full reports of potentially relevant trials and applied the inclusion criteria to the full reports using an eligibility form. If the eligibility was not clear, the trial authors were contacted for clarification. We scrutinized eligible trials to ensure that each trial was included only once. We listed the trials that were not eligible for inclusion and explained the reasons for their exclusion.
2.4. Data Extraction and Quality Assessment
We collected data from the included studies using a standard data extraction template by a pair of distinct reviewers (S.S. and D.M.), and the CHC arbitrated any conflicts. The domains of the extracted data included the number of patients, year of publication, country of study, biomarker level, and outcomes. Two reviewers (S.S. and D.M.) independently assessed the risk of bias for each study. The quality of each study was categorized using the Newcastle–Ottawa scale for cohort and case-control studies by group selection (0–4 points), comparability (0–2 points), and exposure/outcome reliability (0–3 points) [19]. Studies were categorized as good quality if they scored ≥7 points, fair quality if they scored 5–6 points, and poor quality if they scored <5 points.
2.5. Statistical Analysis
Statistical analysis was performed using the RevMan 5.4 software (Nordic Cochrane Center, Cochrane Collaboration, Copenhagen, Sweden) and STATA 12. For continuous measures, standard mean differences (SMDs) with 95% confidence intervals (CIs) were calculated. When the continuous outcome was reported in a study as median, range, and interquartile range, the means and variances were calculated using the formula described by Wan et al. [20]. We used the χ2 test to assess the heterogeneity between studies. Statistical significance was set at p < 0.05, and a random-effects model was used. I2 tests were used to assess the degree of heterogeneity and were interpreted as follows: 0–25% might be important, 26–50% indicates moderate heterogeneity, 51–75% suggests substantial heterogeneity, and 76–100% reflects considerable heterogeneity [21]. Otherwise, a fixed-effects model was used to pool outcomes. Outcomes reported in these studies were included in the meta-analysis. Sensitivity analysis, excluding one study at a time, was used to explore the source of heterogeneity in the analyses. Publication bias among the studies was investigated by visual inspection of the funnel plot and quantitative analysis using the Egger’s test. A random effects model was used to calculate the pooled sensitivity, specificity, and diagnostic odds ratio (DOR) as a biomarker to predict poor clinical outcome in COVID-19 patients. We constructed a summary receiver operating characteristic (SROC) curve analysis by plotting the summary points of sensitivity and specificity.
We summarized the values of the area under the receiver operating characteristic curve as well as the sensitivity and specificity of the included studies.
3. Results
Of the 2907 papers initially retrieved for screening, 96 were eligible for full-text screening, and 58 were excluded. Thus, 38 studies were included (Figure 1).
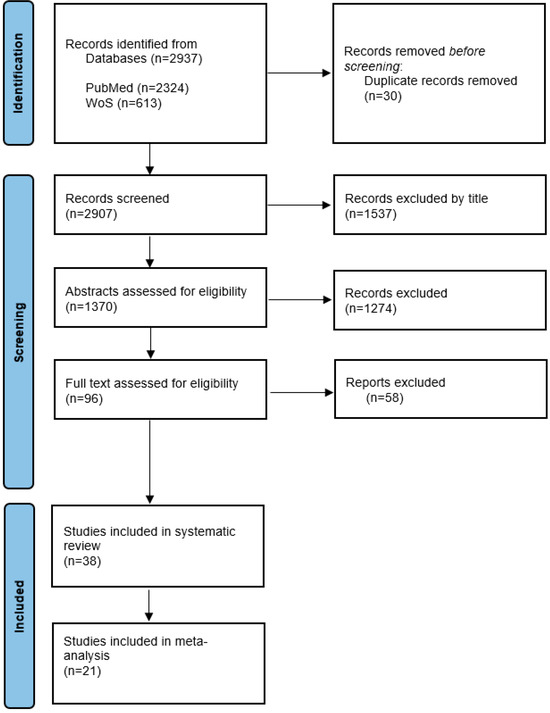
Figure 1.
Systematic reviews and meta-analysis flow diagram.
3.1. Characteristics of Included Studies
A total of 2937 studies were retrieved through the initial search (Figure 1), of which 30 duplicates were removed. After screening the titles of the remaining articles, 1537 articles were excluded. The abstracts of the remaining 1370 studies were analyzed based on the inclusion and exclusion criteria, and after excluding 1274 studies, the full texts of 96 studies were assessed. Finally, 38 studies were selected for systematic review, and 21 studies were included in the meta-analysis. Seventeen studies were excluded from the meta-analysis because of a lack of available data.
The baseline characteristics of the included studies are summarized in Table 1.

Table 1.
Characteristics of included studies.
3.2. Biomarkers
3.2.1. MR-proADM
Nine studies [38,39,40,41,42,43,44,45,46] involving 905 patients (211 with poor outcomes vs. 694 with good outcomes) were investigated. One study was excluded because of a lack of statistical data [37]. The pooled standard mean difference between the two groups was 1.40 (95% CI 1.11, 1.69), showing a strong association between higher levels of MR-proADM and poor outcome in COVID-19 patients (Figure 2a). The AUC was 0.74–0.96, showing a high diagnostic accuracy. The heterogeneity was substantial among the studies (I2 = 63%).
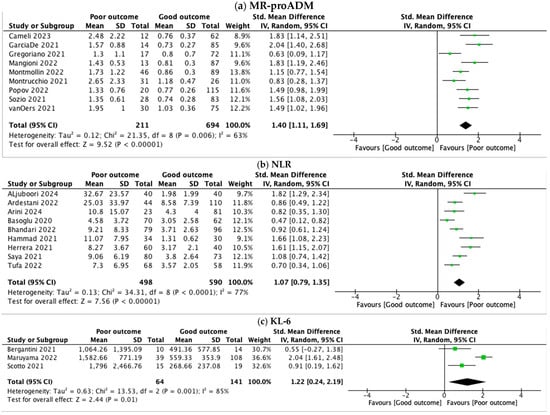
Figure 2.
Forest plots of the meta-analysis of MR-proADM, NLR, and KL-6, pooled by standard mean difference (SMD) and confidence interval (CI). (a) MR-proADM levels were 1.4 SMD higher in severe COVID-19 (95% CI: 1.11–1.69), with substantial heterogeneity (I2 = 63%). (b) NLR was 1.07 SMD higher in patients with severe COVID-19 (95% CI: 0.79–1.35), with considerable heterogeneity (I2 = 77%). (c) KL-6 levels were 1.22 SMD higher in severe COVID-19 (95% CI: 0.24–2.19), with considerable heterogeneity (I2 = 85%).
3.2.2. NLR
Regarding NLR, we reviewed nine studies [22,23,24,25,27,28,29,30,31] with a total of 1088 patients (498 with poor outcome vs. 590 with good outcome). One study was excluded because of a lack of statistical data [26]. NLR was higher in patients with poor outcomes (SMD 1.07, 95% CI 0.79, 1.35) (Figure 2b), showing a significant correlation with severity. The AUC was 0.73–0.98, which shows a robust predictive performance. The heterogeneity between studies was substantial (I2 = 77%).
3.2.3. KL-6
We analyzed KL-6 levels in 205 patients (64 with poor outcomes vs. 141 with good outcomes) from three studies [34,35,36]. Two studies lacked statistical data [32,33]. As shown in Figure 2c, the pooled standard mean difference between the two groups indicated a significant increase in KL-6 levels among patients with poor outcome (SMD 1.22, 95% CI 0.24, 2.19). As for the other biomarkers evaluated, the heterogeneity was substantial (I2 = 85%).
3.2.4. Other Biomarkers: Amyloid A, miR-155, Galectin-3, and SuPAR
We were unable to assess the standard mean difference for the other four biomarkers evaluated due to a lack of data: Amyloid A [47,48,49], miR-155 [50,51,52], Galectin-3 [53,54], and suPAR [55,56,57,58,59].
3.2.5. Receiver Operating Curve Analysis for New Biomarkers as Predictors of Poor Outcome in COVID-19 Patients
The values of the area under the ROC curve, sensitivity, and specificity are summarized in Table 2. The ROC curves for the biomarkers MR-proADM and NLR are shown in Figure 3a and Figure 3b, respectively. The pooled sensitivity and specificity of MR-proADM were 0.85 (95% CI, 0.79–0.90) and 0.71 (95%CI, 0.64–0.76), respectively, showing they are effective for risk stratification. The DOR was 22.71 (95%CI, 8.97–57.48). The pooled sensitivity and specificity of NLR were 0.86 (0.79–0.91) and 0.78 (0.64–0.87), respectively, showing that they are useful for the early detection of high-risk patients.

Table 2.
ROC analysis for outcome prediction according to baseline biomarker values.

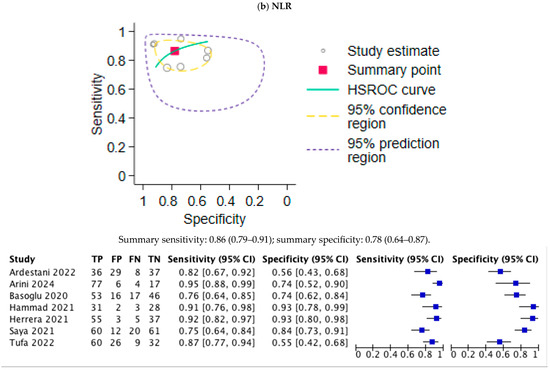
Figure 3.
Summary receiver operating characteristic (SROC) curves of the meta-analysis of MR-proADM and NLR. TP, true positive; FP, false positive; FN, false negative; TN, true negative. (a) Summary AUC for MR-proADM was 0.85 (95%CI: 0.74–0.96) with optimal cut-off at 1.01 nmol/L (sensitivity 85%, specificity 71%). (b) Summary AUC for NLR was 0.86 (95%CI: 0.79–0.91) with optimal cut-off at 6.4 (sensitivity 86%, specificity 78%).
3.3. Quality Assessment
The included studies were evaluated using the Newcastle–Ottawa scale. Most studies (26 of 38, 68.4%) scored >7, indicating good quality and supporting reliability. Seven studies (18.4%) scored 5–6 (fair quality), often because of inadequate control for confounders, and five studies (13.2%) scored <5 (poor quality), primarily because of unclear outcome ascertainment or small sample sizes. A low risk of bias (>7 points) dominated the pooled analysis (MR-proADM and NLR). Heterogeneity was noted (I2 = 63–85%), but sensitivity analyses confirmed robustness.
4. Discussion
We investigated the diagnostic accuracy of new biomarkers for predicting poor outcomes in patients with severe COVID-19. This systematic review and meta-analysis identified MR-proADM, NLR, and KL-6 as robust predictors of severe COVID-19 outcomes, with AUC values exceeding 0.80 for mortality and ICU admission. Our findings extend beyond conventional biomarkers (e.g., CRP, D-dimer, and ferritin) by highlighting the pathophysiology-specific markers of endothelial dysfunction (MR-proADM), immune dysregulation (NLR), and lung injury (KL-6). One of the key aspects of COVID-19 is the body’s inflammatory response, which plays a crucial role in determining the severity of the illness. In this setting, identifying inflammatory biomarkers is fundamental for predicting the disease severity and patient prognosis.
Many studies have demonstrated the association between disease severity and the most common inflammation biomarkers, such as C-reactive protein (CRP), ferritin, interleukin-6 (IL6), and D-dimer. However, several novel biomarkers have been proposed and analyzed during the pandemic. In this article, we review the literature on seven new biomarkers studied in COVID-19 and their association with patient outcomes.
MR-pro-Adrenomedullin (MR-proADM) is a biomarker that has emerged as a potential indicator of organ failure and poor prognosis in COVID-19 patients. It is produced by endothelial cells and reflects overall vascular health. Elevated levels of MR-proADM have been associated with severe disease outcomes, making it a valuable tool for risk stratification in hospitalized COVID-19 patients. Rapid assessment can aid clinicians in identifying individuals at a higher risk of complications, thereby optimizing patient management. Cameli et al. [39] identified MR-proADM as a significant prognostic biomarker for COVID-19 patients. Their study found that patients with MR-proADM levels ≥1.02 nmol/L at admission had a markedly higher mortality rate (39.1%) than those with lower levels of proADM (2.2%). This demonstrates the effectiveness of this biomarker in predicting in-hospital mortality and the need for respiratory support, making it a crucial tool for clinical decision-making in COVID-19 management. García et al. [40] conducted a study in Spain, where they highlighted the prognostic value of MR-proADM in COVID-19 patients. Their findings showed that MR-proADM levels above 1.01 nmol/L had the highest performance in predicting 28-day mortality, with an area under the curve (AUC) of 0.905. Additionally, MR-proADM was independently associated with a 10.47 times higher risk of mortality (hazard ratio [HR]: 10.470) and 6.803 times higher risk of progression to severe disease (HR: 6.803). These results underscore the role of biomarkers in early prognostic assessment and clinical decision-making.
In accordance with the previously mentioned studies, our meta-analysis showed a significant association between higher levels of MR-proADM and poor outcome in COVID-19 patients. The biomarker has good sensitivity (85%) and moderate specificity (71%) for predicting poor outcomes, with an AUC of 0.74–0.96 across studies. Our meta-analysis (SMD = 1.40, AUC 0.74–0.96) confirmed its role in endothelial injury and multiorgan failure. Unlike CRP, MR-proADM reflects microvascular leakage, which is a hallmark of severe COVID-19. Thus, MR-proADM may be useful for risk stratification in hospital settings owing to its strong correlation with disease severity and mortality.
The neutrophil-to-lymphocyte ratio (NLR) has been identified as an indicator of inflammation and disease severity in COVID-19 patients. An increased NLR reflects an exaggerated immune response, characterized by an increase in neutrophils (inflammatory markers) and decrease in lymphocytes (key for adaptive immunity). This biomarker has proven useful in distinguishing between mild and severe cases, suggesting that elevated levels may predict a poor disease prognosis. Saya et al. [30] identified NLR to be able to predict mortality with high accuracy, with a 7.4 cut-off value, while Tufa et al. [31] stated that NLR is a biomarker with only modest accuracy for predicting disease severity and mortality. Our data are in line with those of previous studies, highlighting a positive correlation between NLR and poor outcomes. This method showed high sensitivity (86%) and specificity (78%), with AUC values ranging from 0.73 to 0.98. A cut-off value >6.4 supports its use even in resource-limited settings. Its rise precedes clinical deterioration by 48–72 h, offering a time window for intervention. The clinical utility of this biomarker is easy, and cost-effective, and can aid in the early identification of high-risk patients.
KL-6 (Krebs von den Lungen-6) is a glycoprotein biomarker associated with lung epithelial damage. In COVID-19 patients, elevated KL-6 levels indicate severe lung involvement, particularly in those requiring mechanical ventilation. This biomarker helps predict disease severity and potential development of pulmonary fibrosis, making it useful for the early identification of high-risk patients and monitoring lung recovery over time. Alessandro et al. conducted two studies, showing that increased KL-6 serum concentrations are associated with disease severity [33] and with fibrotic lung alterations [32]. Similarly, Maruyama et al. [35] found that the peak KL-6 value had precise accuracy in the discrimination of patients with poor prognosis when using a cut-off of 966 U/mL cut-off. According to our meta-analysis, higher KL-6 levels were a predictive factor of poor outcomes in hospitalized COVID-19 patients, with AUC values ranging from 0.85 to 0.95. This biomarker may be particularly valuable for assessing lung injury and progression to ARDS in COVID-19 patients.
Limited data precluded meta-analysis for other biomarkers (Amyloid A, miR-155, Galectin-3, and suPAR), but individual studies suggested diagnostic potential, particularly for suPAR (AUC: 0.71–0.81) and miR-155 (AUC: 0.91–0.93).
There are some limitations of our systematic review: There were differences in the type of studies (prospective or retrospective), inclusion and exclusion criteria, patients used, and cut-off values of included studies; also, pooled data were only used for analysis. In addition, there was significant heterogeneity (I2: 63–85%) between the included studies for diagnostic accuracy, which might be caused by the characteristics of the included patients, study designs, different control groups, and outcome definitions.
5. Conclusions
Inflammation is a hallmark of COVID-19 and contributes to disease severity and poor outcomes. Along with classical biomarkers, many novel inflammatory biomarkers have been investigated during the pandemic. Higher MR-proADM, NLR, and KL-6 levels were associated with an increased risk of ICU admission and death. These are validated biomarkers for predicting COVID-19 severity and mortality. These biomarkers may complement traditional markers (CRP, D-dimer, and ferritin), offering superior prognostic precision by targeting distinct disease mechanisms and improving clinical decision making. This study provides novel evidence for new biomarkers with a strong prognostic value for COVID-19. Their integration into clinical practice could improve risk stratification, therapeutic decisions, and patient outcomes by enabling early intervention and personalized care. These findings highlight the importance and need for large-scale multicenter studies to validate novel biomarker performance and cut-off values, which can help better diagnose and assess standardized protocols and prognosis in inflammatory diseases.
Author Contributions
Conceptualization, C.H.-C., S.W.S.L. and D.M.; methodology, C.H.-C. and S.W.S.L.; investigation, S.W.S.L. and D.M.; writing—original draft preparation, S.W.S.L. and D.M.; documentation F.N. and B.R.-D.; writing—review and editing, C.H.-C. and V.M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created.
Acknowledgments
We are thankful to Blanca San Jose for providing the documentation support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Marks, K.M.; Gulick, R.M. COVID-19. Ann. Intern. Med. 2023, 176, ITC145–ITC160. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, R.T.; Lynch, J.B.; Del Rio, C. Mild or Moderate COVID-19. N. Engl. J. Med. 2020, 383, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Berlin, D.A.; Gulick, R.M.; Martinez, F.J. Severe COVID-19. N. Engl. J. Med. 2020, 383, 2451–2460. [Google Scholar] [CrossRef]
- Salton, F.; Confalonieri, P.; Campisciano, G.; Cifaldi, R.; Rizzardi, C.; Generali, D.; Pozzan, R.; Tavano, S.; Bozzi, C.; Lapadula, G.; et al. Cytokine Profiles as Potential Prognostic and Therapeutic Markers in SARS-CoV-2-Induced ARDS. J. Clin. Med. 2022, 11, 2951. [Google Scholar] [CrossRef]
- Li, J.; Wei, J.; Xu, Z.; Jiang, C.; Li, M.; Chen, J.; Li, Y.; Yang, M.; Gu, Y.; Wang, F.; et al. Cytokine/Chemokine Expression Is Closely Associated Disease Severity of Human Adenovirus Infections in Immunocompetent Adults and Predicts Disease Progression. Front. Immunol. 2021, 12, 691879. [Google Scholar] [CrossRef]
- Rasool, G.; Riaz, M.; Abbas, M.; Fatima, H.; Qamar, M.M.; Zafar, F.; Mahmood, Z. COVID-19: Clinical laboratory diagnosis and monitoring of novel coronavirus infected patients using molecular, serological and biochemical markers: A review. Int. J. Immunopathol. Pharmacol. 2022, 36, 03946320221115316. [Google Scholar] [CrossRef]
- Alnor, A.; Sandberg, M.B.; Gils, C.; Vinholt, P.J. Laboratory Tests and Outcome for Patients with Coronavirus Disease 2019: A Systematic Review and Meta-Analysis. J. Appl. Lab. Med. 2020, 5, 1038–1049. [Google Scholar] [CrossRef]
- Plebani, M. COVID-19 and Biomarkers: The Contribution of the Journal. J. Clin. Med. 2023, 12, 1853. [Google Scholar] [CrossRef]
- Ranjbar, M.; Cusack, R.P.; Whetstone, C.E.; Brister, D.L.; Wattie, J.; Wiltshire, L.; Alsaji, N.; Le Roux, J.; Cheng, E.; Srinathan, T.; et al. Immune Response Dynamics and Biomarkers in COVID-19 Patients. Int. J. Mol. Sci. 2024, 25, 6427. [Google Scholar] [CrossRef]
- Park, J.O.; Cho, H.K.; Jeon, C.H.; Kim, S.-H.; Park, I.H.; Kim, K.M.; Lee, J.; Wi, Y.M. Characteristics and biomarkers associated with mortality in COVID-19 patients presenting to the emergency department. Epidemiol. Infect. 2024, 152, e76. [Google Scholar] [CrossRef] [PubMed]
- Snopkowska Lesniak, S.W.; Maschio, D.; Henriquez-Camacho, C.; Moreno Cuerda, V. Biomarkers for SARS-CoV-2 infection. A narrative review. Front. Med. 2025, 12, 1563998. [Google Scholar] [CrossRef] [PubMed]
- Oblitas, C.-M.; Galeano-Valle, F.; Ramírez-Navarro, J.; López-Cano, J.; Monterrubio-Manrique, Á.; García-Gámiz, M.; Sancho-González, M.; Arenal-López, S.; Álvarez-Sala Walther, L.-A.; Demelo-Rodríguez, P. Mid-Regional Pro-Adrenomedullin, Methemoglobin and Carboxyhemoglobin as Prognosis Biomarkers in Critically Ill Patients with COVID-19: An Observational Prospective Study. Viruses 2021, 13, 2445. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, H.; Li, C.; Yu, J.; Zhu, M. Characteristics of cytokines/chemokines associated with disease severity and adverse prognosis in COVID-19 patients. Front. Immunol. 2024, 15, 1464545. [Google Scholar] [CrossRef]
- Sanduzzi Zamparelli, S.; Fucci, V.; Rea, G.; Perna, F.; Bocchino, M.; Sanduzzi Zamparelli, A. The Role of SARS-CoV-2 Nucleocapsidic Antigen and Krebs von den Lungen 6 Serum Levels in Predicting COVID-19 Pneumonia Outcome. Diagnostics 2024, 14, 642. [Google Scholar] [CrossRef]
- Haroun, R.A.-H.; Osman, W.H.; Amin, R.E.; Hassan, A.K.; Abo-Shanab, W.S.; Eessa, A.M. Circulating plasma miR-155 is a potential biomarker for the detection of SARS-CoV-2 infection. Pathology 2022, 54, 104–110. [Google Scholar] [CrossRef]
- Gajovic, N.; Markovic, S.S.; Jurisevic, M.; Jovanovic, M.; Arsenijevic, N.; Mijailovic, Z.; Jovanovic, M.; Jovanovic, I. Galectin-3 as an important prognostic marker for COVID-19 severity. Sci. Rep. 2023, 13, 1460. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Lo, C.K.-L.; Mertz, D.; Loeb, M. Newcastle-Ottawa Scale: Comparing reviewers’ to authors’ assessments. BMC Med. Res. Methodol. 2014, 14, 45. [Google Scholar] [CrossRef]
- Wan, X.; Wang, W.; Liu, J.; Tong, T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med. Res. Methodol. 2014, 14, 135. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Al-Juboori, R.S.F.; Al-Bayaa, Y.J. The role of C-reactive protein, procalcitonin, interleukin-6 and neutrophil/lymphocyte ratio as a laboratory biomarker in COVID-19. Egypt. J. Immunol. 2024, 31, 93–101. [Google Scholar] [PubMed]
- Ardestani, S.K.; Salehi, M.R.; Attaran, B.; Hashemi, S.M.; Sadeghi, S.; Ghaffarpour, S.; Tuserkani, F.; Ghazanfari, T. Neutrophil to Lymphocyte Ratio (NLR) and Derived NLR Combination: A Cost-effective Predictor of Moderate to Severe COVID-19 Progression. Iran. J. Allergy Asthma Immunol. 2022, 21, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Arini, I.A.; Masyeni, S.; Widhidewi, N.W. Relationship between neutrophil-lymphocyte ratio and platelet-lymphocyte ratio with the severity of COVID-19. Narra J. 2024, 4, e262. [Google Scholar] [CrossRef]
- Bhandari, S.; Rankawat, G.; Mathur, S.; Kumar, A.; Sahlot, R.; Jain, A. Circulatory Cytokine Levels as a Predictor of Disease Severity in COVID-19: A Study from Western India. J. Assoc. Physicians India 2022, 70, 11–12. [Google Scholar]
- Bohra, G.K.; Bhatia, P.K.; Khichar, S.; Garg, M.K.; Sharma, P. Association of Inflammatory markers with COVID-19 Outcome among Hospitalized adult Patients. J. Assoc. Physicians India 2022, 70, 11–12. [Google Scholar]
- Hammad, R.; Eldosoky, M.A.E.R.; Fouad, S.H.; Elgendy, A.; Tawfeik, A.M.; Alboraie, M.; Abdelmaksoud, M.F. Circulating cell-free DNA, peripheral lymphocyte subsets alterations and neutrophil lymphocyte ratio in assessment of COVID-19 severity. Innate Immun. 2021, 27, 240–250. [Google Scholar] [CrossRef]
- Basoglu, O.K.; Ozhan, M.H.; Ekren, P.K.; Ak, G.; Tasbakan, M.S.; Sayiner, A. Communication: The Follow-Up of Biomarkers Better Predicts the Poor Outcome in COVID-19 Patients. Ann. Clin. Lab. Sci. 2020, 50, 848–851. [Google Scholar]
- Herrera-Van Oostdam, A.S.; Castañeda-Delgado, J.E.; Oropeza-Valdez, J.J.; Borrego, J.C.; Monárrez-Espino, J.; Zheng, J.; Mandal, R.; Zhang, L.; Soto-Guzmán, E.; Fernández-Ruiz, J.C.; et al. Immunometabolic signatures predict risk of progression to sepsis in COVID-19. PLoS ONE 2021, 16, e0256784. [Google Scholar] [CrossRef]
- Sayah, W.; Berkane, I.; Guermache, I.; Sabri, M.; Lakhal, F.Z.; Yasmine Rahali, S.; Djidjeli, A.; Lamara Mahammed, L.; Merah, F.; Belaid, B.; et al. Interleukin-6, procalcitonin and neutrophil-to-lymphocyte ratio: Potential immune-inflammatory parameters to identify severe and fatal forms of COVID-19. Cytokine 2021, 141, 155428. [Google Scholar] [CrossRef]
- Tufa, A.; Gebremariam, T.H.; Manyazewal, T.; Asrat, Y.; Getinet, T.; Hundie, T.G.; Webb, D.-L.; Hellström, P.M.; Genet, S. Limited value of neutrophil-to-lymphocyte ratio and serum creatinine as point-of-care biomarkers of disease severity and infection mortality in patients hospitalized with COVID-19. PLoS ONE 2022, 17, e0275391. [Google Scholar] [CrossRef] [PubMed]
- d’Alessandro, M.; Bergantini, L.; Cameli, P.; Curatola, G.; Remediani, L.; Bennett, D.; Bianchi, F.; Perillo, F.; Volterrani, L.; Mazzei, M.A.; et al. Serial KL-6 measurements in COVID-19 patients. Intern. Emerg. Med. 2021, 16, 1541–1545. [Google Scholar] [CrossRef] [PubMed]
- d’Alessandro, M.; Cameli, P.; Refini, R.M.; Bergantini, L.; Alonzi, V.; Lanzarone, N.; Bennett, D.; Rana, G.D.; Montagnani, F.; Scolletta, S.; et al. Serum KL-6 concentrations as a novel biomarker of severe COVID-19. J. Med. Virol. 2020, 92, 2216–2220. [Google Scholar] [CrossRef]
- Bergantini, L.; Bargagli, E.; d’Alessandro, M.; Refini, R.M.; Cameli, P.; Galasso, L.; Scapellato, C.; Montagnani, F.; Scolletta, S.; Franchi, F.; et al. Prognostic bioindicators in severe COVID-19 patients. Cytokine 2021, 141, 155455. [Google Scholar] [CrossRef]
- Maruyama, S.; Nakamori, Y.; Nakano, H.; Tsuyumu, K.; Kanayama, S.; Iwamura, H.; Wada, D.; Yoshihara, T.; Saito, F.; Yoshiya, K.; et al. Peak value of serum KL-6 may be useful for predicting poor prognosis of severe COVID-19 patients. Eur. J. Med. Res. 2022, 27, 69. [Google Scholar] [CrossRef]
- Scotto, R.; Pinchera, B.; Perna, F.; Atripaldi, L.; Giaccone, A.; Sequino, D.; Zappulo, E.; Sardanelli, A.; Schiano Moriello, N.; Stanziola, A.; et al. Serum KL-6 Could Represent a Reliable Indicator of Unfavourable Outcome in Patients with COVID-19 Pneumonia. Int. J. Environ. Res. Public Health 2021, 18, 2078. [Google Scholar] [CrossRef]
- García de Guadiana-Romualdo, L.; Martínez Martínez, M.; Rodríguez Mulero, M.D.; Esteban-Torrella, P.; Hernández Olivo, M.; Alcaraz García, M.J.; Campos-Rodríguez, V.; Sancho-Rodríguez, N.; Galindo Martínez, M.; Alcaraz, A.; et al. Circulating MR-proADM levels, as an indicator of endothelial dysfunction, for early risk stratification of mid-term mortality in COVID-19 patients. Int. J. Infect. Dis. 2021, 111, 211–218. [Google Scholar] [CrossRef]
- Gregoriano, C.; Koch, D.; Kutz, A.; Haubitz, S.; Conen, A.; Bernasconi, L.; Hammerer-Lercher, A.; Saeed, K.; Mueller, B.; Schuetz, P. The vasoactive peptide MR-pro-adrenomedullin in COVID-19 patients: An observational study. Clin. Chem. Lab. Med. 2021, 59, 995–1004. [Google Scholar] [CrossRef]
- Cameli, P.; Pordon, E.; d’Alessandro, M.; Marzi, M.L.; Galasso, L.; Biuzzi, C.; Bergantini, L.; Bargagli, E.; Scolletta, S.; Franchi, F. MR-proADM as Prognostic Factor of Outcome in COVID-19 Patients. Biomedicines 2023, 11, 1680. [Google Scholar] [CrossRef]
- García de Guadiana-Romualdo, L.; Calvo Nieves, M.D.; Rodríguez Mulero, M.D.; Calcerrada Alises, I.; Hernández Olivo, M.; Trapiello Fernández, W.; González Morales, M.; Bolado Jiménez, C.; Albaladejo-Otón, M.D.; Fernández Ovalle, H.; et al. MR-proADM as marker of endotheliitis predicts COVID-19 severity. Eur. J. Clin. Investig. 2021, 51, e13511. [Google Scholar] [CrossRef]
- de Montmollin, E.; Peoc’h, K.; Marzouk, M.; Ruckly, S.; Wicky, P.-H.; Patrier, J.; Jaquet, P.; Sonneville, R.; Bouadma, L.; Timsit, J.-F. Mid-Regional Pro-Adrenomedullin as a Prognostic Factor for Severe COVID-19 ARDS. Antibiotics 2022, 11, 1166. [Google Scholar] [CrossRef] [PubMed]
- Mangioni, D.; Oggioni, M.; Chatenoud, L.; Liparoti, A.; Uceda Renteria, S.; Alagna, L.; Biscarini, S.; Bolis, M.; Di Modugno, A.; Mussa, M.; et al. Prognostic Value of Mid-Region Proadrenomedullin and In Vitro Interferon Gamma Production for In-Hospital Mortality in Patients with COVID-19 Pneumonia and Respiratory Failure: An Observational Prospective Study. Viruses 2022, 14, 1683. [Google Scholar] [CrossRef] [PubMed]
- Montrucchio, G.; Sales, G.; Rumbolo, F.; Palmesino, F.; Fanelli, V.; Urbino, R.; Filippini, C.; Mengozzi, G.; Brazzi, L. Effectiveness of mid-regional pro-adrenomedullin (MR-proADM) as prognostic marker in COVID-19 critically ill patients: An observational prospective study. PLoS ONE 2021, 16, e0246771. [Google Scholar] [CrossRef]
- Popov, D.; Borovkova, U.; Rybka, M.; Ramnyonok, T.; Golukhova, E. Mid-regional pro-adrenomedullin as a predictor of in-hospital mortality in adult patients with COVID-19: A single-centre prospective study. Anaesthesiol. Intensive Ther. 2022, 54, 242–246. [Google Scholar] [CrossRef]
- Sozio, E.; Tascini, C.; Fabris, M.; D’Aurizio, F.; De Carlo, C.; Graziano, E.; Bassi, F.; Sbrana, F.; Ripoli, A.; Pagotto, A.; et al. MR-proADM as prognostic factor of outcome in COVID-19 patients. Sci. Rep. 2021, 11, 5121. [Google Scholar] [CrossRef]
- van Oers, J.A.H.; Kluiters, Y.; Bons, J.A.P.; de Jongh, M.; Pouwels, S.; Ramnarain, D.; de Lange, D.W.; de Grooth, H.-J.; Girbes, A.R.J. Endothelium-associated biomarkers mid-regional proadrenomedullin and C-terminal proendothelin-1 have good ability to predict 28-day mortality in critically ill patients with SARS-CoV-2 pneumonia: A prospective cohort study. J. Crit. Care 2021, 66, 173–180. [Google Scholar] [CrossRef]
- Cheng, L.; Yang, J.-Z.; Bai, W.-H.; Li, Z.-Y.; Sun, L.-F.; Yan, J.-J.; Zhou, C.-L.; Tang, B.-P. Prognostic value of serum amyloid A in patients with COVID-19. Infection 2020, 48, 715–722. [Google Scholar] [CrossRef]
- Tufa, A.; Gebremariam, T.H.; Manyazewal, T.; Getinet, T.; Webb, D.-L.; Hellström, P.M.; Genet, S. Inflammatory mediators profile in patients hospitalized with COVID-19: A comparative study. Front. Immunol. 2022, 13, 964179. [Google Scholar] [CrossRef]
- Haroun, R.A.-H.; Osman, W.H.; Eessa, A.M. Interferon-γ-induced protein 10 (IP-10) and serum amyloid A (SAA) are excellent biomarkers for the prediction of COVID-19 progression and severity. Life Sci. 2021, 269, 119019. [Google Scholar] [CrossRef]
- Soltane, R.; Almulla, N.; Alasiri, A.; Elashmawy, N.F.; Qumsani, A.T.; Alshehrei, F.M.; Keshek, D.E.-G.; Alqadi, T.; Al-Ghamdi, S.B.; Allayeh, A.K. A Comparative Analysis of MicroRNA Expression in Mild, Moderate, and Severe COVID-19: Insights from Urine, Serum, and Nasopharyngeal Samples. Biomolecules 2023, 13, 1681. [Google Scholar] [CrossRef]
- Gaytán-Pacheco, N.; Ibáñez-Salazar, A.; Herrera-Van Oostdam, A.S.; Oropeza-Valdez, J.J.; Magaña-Aquino, M.; Adrián López, J.; Monárrez-Espino, J.; López-Hernández, Y. miR-146a, miR-221, and miR-155 are Involved in Inflammatory Immune Response in Severe COVID-19 Patients. Diagnostics 2022, 13, 133. [Google Scholar] [CrossRef] [PubMed]
- Kadhim, H.; Ghareeb, A.; Alhilal, M. Expression level of non-coding (MiR-155) gene as biomarker for severity of coronaviruses infection among vaccinated and non-vaccinated Iraqi patients. Hum. Antibodies 2024, 32, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Karsli, E.; Anabarli Metin, D.; Canacik, O.; Sabirli, R.; Kaymaz, B.; Kurt, O.; Koseler, A. Galectin-3 as a Potential Prognostic Biomarker for COVID-19 Disease: A Case-Control Study. Cureus 2022, 14, e28805. [Google Scholar] [CrossRef]
- Kartal Baykan, E.; Şebin, E.; Karaşahin, Ö.; Baykan, A.R.; Cerrah, S.; Göğebakan, H.; Sevinç, C.; Kahraman, M.; Yavuz, Y.C. Galectin-3: Can it be a diagnostic tool for pneumonia in COVID-19 patients? Turk. J. Med. Sci. 2021, 51, 2256–2262. [Google Scholar] [CrossRef]
- Vasbinder, A.; Padalia, K.; Pizzo, I.; Machado, K.; Catalan, T.; Presswalla, F.; Anderson, E.; Ismail, A.; Hutten, C.; Huang, Y.; et al. SuPAR, biomarkers of inflammation, and severe outcomes in patients hospitalized for COVID-19: The International Study of Inflammation in COVID-19. J. Med. Virol. 2024, 96, e29389. [Google Scholar] [CrossRef]
- Vassiliou, A.G.; Zacharis, A.; Vrettou, C.S.; Keskinidou, C.; Jahaj, E.; Mastora, Z.; Orfanos, S.E.; Dimopoulou, I.; Kotanidou, A. Comparison of the Mortality Prediction Value of Soluble Urokinase Plasminogen Activator Receptor (suPAR) in COVID-19 and Sepsis. Diagnostics 2022, 12, 1261. [Google Scholar] [CrossRef]
- Arnold, D.T.; Attwood, M.; Barratt, S.; Morley, A.; Elvers, K.T.; McKernon, J.; Donald, C.; Oates, A.; Noel, A.; MacGowan, A.; et al. Predicting outcomes of COVID-19 from admission biomarkers: A prospective UK cohort study. Emerg. Med. J. 2021, 38, 543–548. [Google Scholar] [CrossRef]
- Chalkias, A.; Skoulakis, A.; Papagiannakis, N.; Laou, E.; Tourlakopoulos, K.; Pagonis, A.; Michou, A.; Ntalarizou, N.; Mermiri, M.; Ragias, D.; et al. Circulating suPAR associates with severity and in-hospital progression of COVID-19. Eur. J. Clin. Investig. 2022, 52, e13794. [Google Scholar] [CrossRef]
- Molfino, A.; Anastasi, E.; Assanto, E.; Toccini, L.; Imbimbo, G.; Gigante, A.; Viggiani, V.; Farina, A.; Picconi, O.; Angeloni, A.; et al. Association between serum levels of GDF-15, suPAR, PIVKA-II, sdLDL and clinical outcomes in hospitalized COVID-19 patients. Intern. Emerg. Med. 2024, 19, 1557–1566. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).