Predicting Response to [177Lu]Lu-PSMA Therapy in mCRPC Using Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Protocol
2.1.1. Patient Cohorts
2.1.2. PET Imaging Protocols
2.1.3. [177Lu]Lu-PSMA-617 Protocol
2.2. PSMA-RLT Response Evaluation
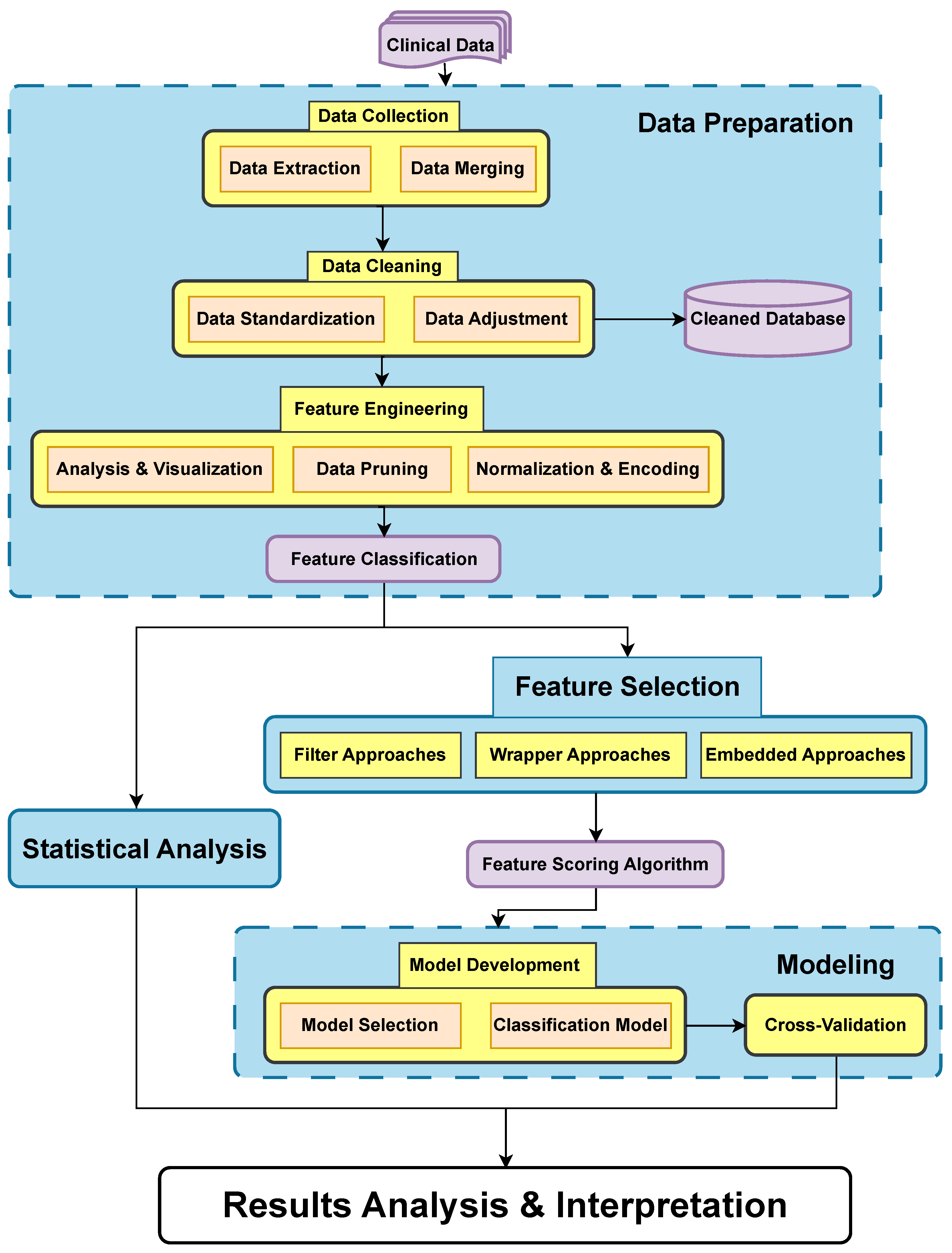
2.3. Data Analysis and Model Development
2.3.1. Data Preparation
Data Collection and Integration
Data Cleaning
Feature Engineering
2.3.2. Statistical Analysis
Mann–Whitney U Test
- : The distributions of the two groups are equal.
- : The distributions of the two groups are not equal.
- and represent the sample sizes of the two groups.
- is the sum of the ranks for the first group (class NFBTP).
Chi-Square Test
- : The two categorical variables are independent.
- : The two categorical variables are not independent.
- represents the observed frequency for category i.
- is the expected frequency for category i, calculated under the assumption that the two variables are independent.
2.3.3. Feature Selection
| Algorithm 1 Comprehensive Feature Scoring Algorithm |
| Initialize as empty dictionary |
| Retrieve feature ranking lists |
| for all in do |
| for all in do |
| total features in |
| if not in then |
| end if |
| end for |
| end for |
| by values descending |
| return |
Filter Approaches
Wrapper Approaches
Embedded Approaches
2.3.4. Modeling
Model Development
- Logistic Regression (LR);
- Random Forest Classifier (RF);
- XGBoost Classifier;
- Decision Tree Classifier (DT);
- K-Nearest Neighbor (KNN);
- Support Vector Machines (SVM);
- Naive Bayes [20] (NB).
Cross-Validation
3. Results
3.1. Statistical Analysis
3.2. Model Development and Evaluation
4. Discussion
4.1. Advancements in Predictive Factors and Imaging Techniques for RLT Response in Prostate Cancer
4.2. Study Limitations and Potential Insights from [18F]Choline PET/CT
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PC | Prostate cancer |
| PSMA | Prostate specific membrane antigen |
| FDG | Fluorodeoxyglucose |
| RLT | Radioligand therapy |
| ML | Machine learning |
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Kasperzyk, J.L.; Finn, S.P.; Flavin, R.; Fiorentino, M.; Lis, R.; Hendrickson, W.K.; Clinton, S.K.; Sesso, H.D.; Giovannucci, E.L.; Stampfer, M.J.; et al. Prostate-Specific Membrane Antigen Protein Expression in Tumor Tissue and Risk of Lethal Prostate Cancer. Cancer Epidemiol. Biomarkers Prev. 2013, 22, 2354–2363. [Google Scholar] [CrossRef] [PubMed]
- Perner, S.; Hofer, M.D.; Kim, R.; Shah, R.B.; Li, H.; Möller, P.; Hautmann, R.E.; Gschwend, J.E.; Kuefer, R.; Rubin, M.A. Prostate-specific membrane antigen expression as a predictor of prostate cancer progression. Hum. Pathol. 2007, 38, 696–701. [Google Scholar] [CrossRef] [PubMed]
- Sweat, S.D.; Pacelli, A.; Murphy, G.P.; Bostwick, D.G. Prostate-specific membrane antigen expression is greatest in prostate adenocarcinoma and lymph node metastases. Urology 1998, 52, 637–640. [Google Scholar] [CrossRef]
- Sartor, O.; de Bono, J.; Chi, K.N.; Fizazi, K.; Herrmann, K.; Rahbar, K.; Tagawa, S.T.; Nordquist, L.T.; Vaishampayan, N.; El-Haddad, G.; et al. Lutetium-177–PSMA-617 for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2021, 385, 1091–1103. [Google Scholar] [CrossRef]
- Hofman, M.S.; Emmett, L.; Sandhu, S.; Iravani, A.; Joshua, A.M.; Goh, J.C.; Pattison, D.A.; Tan, T.H.; Kirkwood, I.D.; Ng, S.; et al. [177Lu]Lu-PSMA-617 versus cabazitaxel in patients with metastatic castration-resistant prostate cancer (TheraP): A randomised, open-label, phase 2 trial. Lancet 2021, 397, 797–804. [Google Scholar] [CrossRef]
- FDA Approves Pluvicto/Locametz for Metastatic Castration-Resistant Prostate Cancer. J. Nucl. Med. 2022, 63, 13N.
- Ahmadzadehfar, H.; Essler, M. Predictive Factors of Response and Overall Survival in Patients with Castration-Resistant Metastatic Prostate Cancer Undergoing 177Lu-PSMA Therapy. J. Nucl. Med. 2018, 59, 1033–1034. [Google Scholar] [CrossRef]
- Manafi-Farid, R.; Harsini, S.; Saidi, B.; Ahmadzadehfar, H.; Herrmann, K.; Briganti, A.; Walz, J.; Beheshti, M. Factors predicting biochemical response and survival benefits following radioligand therapy with [177Lu]Lu-PSMA in metastatic castrate-resistant prostate cancer: A review. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4028–4041. [Google Scholar] [CrossRef]
- Alongi, P.; Laudicella, R.; Lanzafame, H.; Farolfi, A.; Mapelli, P.; Picchio, M.; Burger, I.A.; Iagaru, A.; Minutoli, F.; Evangelista, L. PSMA and Choline PET for the Assessment of Response to Therapy and Survival Outcomes in Prostate Cancer Patients: A Systematic Review from the Literature. Cancers 2022, 14, 1770. [Google Scholar] [CrossRef]
- Moazemi, S.; Erle, A.; Khurshid, Z.; Lütje, S.; Muders, M.; Essler, M.; Schultz, T.; Bundschuh, R.A. Decision-support for treatment with 177Lu-PSMA: Machine learning predicts response with high accuracy based on PSMA-PET/CT and clinical parameters. Ann. Transl. Med. 2021, 9, 818. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Miu, X.; Wang, H.; Li, X. A Bayesian optimization tunning integrated multi-stacking classifier framework for the prediction of radiodermatitis from 4D-CT of patients underwent breast cancer radiotherapy. Front. Oncol. 2023, 13, 1152020. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-W.; Tsai, Y.-H.; Chang, F.-R.; Lin, W.-C. Ensemble feature selection in medical datasets: Combining filter, wrapper, and embedded feature selection results. Expert Syst. 2020, 37, e12553. [Google Scholar] [CrossRef]
- Bashir, S.; Khattak, I.U.; Khan, A.; Khan, F.H.; Gani, A.; Shiraz, M. A Novel Feature Selection Method for Classification of Medical Data Using Filters, Wrappers, and Embedded Approaches. Complexity 2022, 2022, 8190814. [Google Scholar] [CrossRef]
- Boellaard, R.; Delgado-Bolton, R.; Oyen, W.J.; Giammarile, F.; Tatsch, K.; Eschner, W.; Verzijlbergen, F.J.; Barrington, S.F.; Pike, L.C.; Weber, W.A.; et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: Version 2.0. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 328–354. [Google Scholar] [CrossRef]
- Fendler, W.P.; Eiber, M.; Beheshti, M.; Bomanji, J.; Calais, J.; Ceci, F.; Cho, S.Y.; Fanti, S.; Giesel, F.L.; Goffin, K.; et al. PSMA PET/CT: Joint EANM procedure guideline/SNMMI procedure standard for prostate cancer imaging 2.0. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 1466–1486. [Google Scholar] [CrossRef]
- Kratochwil, C.; Fendler, W.P.; Eiber, M.; Hofman, M.S.; Emmett, L.; Calais, J.; Osborne, J.R.; Iravani, A.; Koo, P.; Lindenberg, L.; et al. Joint EANM/SNMMI procedure guideline for the use of 177Lu-labeled PSMA-targeted radioligand-therapy (177Lu-PSMA-RLT). Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 2830–2845. [Google Scholar] [CrossRef]
- Scher, H.I.; Morris, M.J.; Stadler, W.M.; Higano, C.; Basch, E.; Fizazi, K.; Antonarakis, E.S.; Beer, T.M.; Carducci, M.A.; Chi, K.N.; et al. Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J. Clin. Oncol. 2016, 34, 1402–1418. [Google Scholar] [CrossRef]
- Hartrampf, P.E.; Serfling, S.E.; Michalski, K.; Buck, A.K.; Werner, R.A. PSMA PET/CT for Response Assessment of 177Lu-PSMA Therapy. Semin. Nucl. Med. 2024, 54, 69–76. [Google Scholar] [CrossRef]
- Bokhare, A.; Jha, P. Machine learning models applied in analyzing breast cancer classification accuracy. Int. J. Artif. Intell. 2023, 2252, 1371. [Google Scholar] [CrossRef]
- Powers, D. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness & Correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar]
- Larracy, R.; Phinyomark, A.; Scheme, E. Machine Learning Model Validation for Early Stage Studies with Small Sample Sizes. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2021, 2021, 2314–2319. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhang, T.; Chen, S.; Bu, T.; Zhao, J.; Ni, X.; Shi, B.; Gan, H.; Wei, Y.; Wang, Q.; et al. Nomogram to predict the presence of PSMA-negative but FDG-positive lesion in castration-resistant prostate cancer: A multicenter cohort study. Ther. Adv. Med. Oncol. 2024, 16, 17588359231220506. [Google Scholar] [CrossRef] [PubMed]
- Groener, D.; Schneider, S.; Baumgarten, J.; Happel, C.; Klimek, K.; Mader, N.; Nguyen Ngoc, C.; Wichert, J.; Mandel, P.; Tselis, N.; et al. Baseline [68Ga]Ga-PSMA-11 PET/CT before [177Lu]Lu-PSMA-617 Radioligand Therapy: Value of PSMA-Uptake Thresholds in Predicting Targetable Lesions. Cancers 2023, 15, 473. [Google Scholar] [CrossRef] [PubMed]
- Giovanella, L.; Garo, M.L.; Cuzzocrea, M.; Paone, G.; Herrmann, K. Prognostic role of early prostate specific antigen changes after [177 Lu]Lu-PSMA radioligand therapy of metastasized prostate cancer: A meta-analysis. Eur. J. Clin. Investig. 2023, 53, e14014. [Google Scholar] [CrossRef]
- Eisazadeh, R.; Mirshahvalad, S.A.; Schwieghofer-Zwink, G.; Hehenwarter, L.; Rendl, G.; Gampenrieder, S.; Greil, R.; Pirich, C.; Beheshti, M. Pre-treatment 68 Ga-PSMA-11 PET/CT Prognostic Value in Predicting Response to 177Lu-PSMA-I&T Therapy and Patient Survival. Mol. Imaging Biol. 2024, 26, 360–369. [Google Scholar] [CrossRef]
- Laudicella, R.; Minutoli, F.; Russo, S.; Siracusa, M.; Bambaci, M.; Pagano, B.; Baldari, S. mCRPC progression of disease after [177Lu]Lu-PSMA-617 detected on [18F]Choline: A case of PCa heterogeneity. Urol. Case Rep. 2024, 54, 102750. [Google Scholar] [CrossRef]
- Gafita, A.; Calais, J.; Grogan, T.R.; Hadaschik, B.; Wang, H.; Weber, M.; Sandhu, S.; Kratochwil, C.; Esfandiari, R.; Tauber, R.; et al. Nomograms to predict outcomes after 177Lu-PSMA therapy in men with metastatic castration-resistant prostate cancer: An international, multicentre, retrospective study. Lancet Oncol. 2021, 22, 1115–1125. [Google Scholar] [CrossRef]
- Gafita, A.; Martin, A.J.; Emmett, L.; Eiber, M.; Iravani, A.; Fendler, W.P.; Buteau, J.; Sandhu, S.; Azad, A.A.; Herrmann, K.; et al. Validation of Prognostic and Predictive Models for Therapeutic Response in Patients Treated with [177Lu]Lu-PSMA-617 Versus Cabazitaxel for Metastatic Castration-resistant Prostate Cancer (TheraP): A Post Hoc Analysis from a Randomised, Open-label, Phase 2 Trial. Eur. Urol. Oncol. 2024; in press. [Google Scholar] [CrossRef]

| Patient | Age | WHO a | Gleason Score | PSA Level before RLT (ng/mL) | PSA Level after RLT (ng/mL) | Number of Cures |
|---|---|---|---|---|---|---|
| 1 | 73 | 2 | 9 | 560 | >1200 | 1 |
| 2 | 61 | 1 | 7 | 385 | 28.1 | 6 |
| 3 | 64 | 0 | 8 | 68 | 915.1 | 3 |
| 4 | 50 | 1 | 8 | 68.4 | 1636 | 4 |
| 5 | 78 | 0 | 7 | 14.2 | 1.52 | 6 |
| 6 | 83 | 1 | 6 | 46.79 | 5.88 | 6 |
| 7 | 79 | 1 | 6 | 47 | 52 | 4 |
| 8 | 75 | 1 | 10 | 22.4 | 376 | 5 |
| 10 | 85 | 1 | 7 | 92.77 | 15.86 | 2 |
| 11 | 68 | 1 | 7 | 37 | 13.87 | 6 |
| 12 | 73 | 1 | 7 | 0.8 | 0.12 | 5 |
| 15 | 75 | 1 | 8 | 198.4 | 152 | 2 |
| 16 | 77 | 1 | 7 | 87.31 | 7 | 6 |
| 19 | 79 | 1 | 7 | 51.7 | 158.1 | 5 |
| 20 | 63 | 1 | 9 | 21.99 | 3.8 | 6 |
| 21 | 83 | 1 | 8 | 65.263 | 0.4 | 6 |
| 22 | 73 | 2 | 8 | 27.11 | <0.006 | 6 |
| 23 | 80 | 2 | 8 | 98.42 | 3.82 | 6 |
| 24 | 75 | 1 | 9 | 134.59 | 243.82 | 4 |
| 28 | 60 | 1 | 9 | 172.68 | 243 | 1 |
| 30 | 64 | 1 | 7 | 19.3 | 35.42 | 6 |
| 31 | 74 | 0 | 7 | 77.28 | 2 | 6 |
| 32 | 70 | 0 | NA b | 42.55 | 1.49 | 6 |
| 33 | 78 | 1 | 8 | 93.3 | 179 | 6 |
| 36 | 63 | 1 | 9 | 2009 | >5000 | 3 |
| Parameters | Description | |
|---|---|---|
| Radiomics Features | Imaging Parameters | Statistical features such as mean, minimum, maximum, peak values of ROI |
| Invasion | Bone, pelvic tissue, liver, lung, and lymph node invasion | |
| Tracer Status | Consistency and fixed states of PSMA, Choline, and FDG tracers | |
| Clinical Parameters | Age | Patient’s age (range from 50 to 85) |
| Gleason Score | Describes abnormality degree of cancer cells in prostate (range from 6 to 10) | |
| TNM Staging | Tumor size, lymph node involvement, and metastasis staging | |
| WHO Classification | World Health Organization classification of the disease | |
| Biological Parameters | PSA Level | Baseline serum prostate-specific antigen level |
| Complete Blood Count | Hemoglobin, Leukocytes, Neutrophils, Lymphocytes, Platelets | |
| Liver Function Tests | ASAT, ALAT, Total Bilirubin, Albumin, ALP | |
| Kidney Function Tests | GFR, Creatinine | |
| Variable | FBTP | NFBTP a | p-Value |
|---|---|---|---|
| Max: g/mL_Choline_Kidney | 14.97 (3.33) | 18.34 (3.25) | 0.013 |
| Min: g/mL_Choline_Kidney | 6.30 (1.41) | 7.72 (1.37) | 0.013 |
| Mean: g/mL_Choline_Kidney | 9.64 (2.27) | 11.91 (2.35) | 0.023 |
| Peak: g/mL_Choline_Kidney | 11.79 (2.54) | 14.84 (2.82) | 0.013 |
| Std. dev: g/mL_Choline_Kidney | 1.85 (0.50) | 2.36 (0.51) | 0.021 |
| Max: g/mL_Choline_Bone+ b | 6.91 (6.57) | 11.81 (5.98) | 0.040 |
| Min: g/mL_Choline_Bone+ | 2.92 (2.79) | 4.98 (2.53) | 0.035 |
| Mean: g/mL_Choline_Bone+ | 4.17 (4.29) | 7.36 (3.92) | 0.035 |
| Peak: g/mL_Choline_Bone+ | 4.86 (4.75) | 7.94 (3.52) | 0.035 |
| Std. dev: g/mL_Choline_Bone+ | 0.98 (1.08) | 1.71 (1.03) | 0.027 |
| Leukocytes (G/L) | 5.23 [4.65, 5.96] | 6.66 [5.60, 8.43] | 0.013 |
| Neutrophils (G/L) | 3.29 [2.77, 3.64] | 4.05 [3.13, 5.79] | 0.040 |
| ALP (Alkaline Phosphatase) | 91.00 [62.00, 112.00] | 188.50 [95.50, 351.50] | 0.035 |
| Invasion score of Pelvis c | 1.00 [0.00, 3.00] | 5.50 [3.25, 7.88] | 0.033 |
| Difference_PSMA-FDG d | (0.5/0/0.5) | (0/0.67/0.33) | 0.025 |
| Difference_PSMA-choline | (0.64/0.27/0.09) | (0.5/0.29/0.21) | 0.0505 e |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, K.; Magnier, B.; L’hostis, S.; Borrely, F.; Le Bon, S.; Houede, N.; Mamou, A.; Maimoun, L.; Kotzki, P.O.; Boudousq, V. Predicting Response to [177Lu]Lu-PSMA Therapy in mCRPC Using Machine Learning. J. Pers. Med. 2024, 14, 1068. https://doi.org/10.3390/jpm14111068
Gong K, Magnier B, L’hostis S, Borrely F, Le Bon S, Houede N, Mamou A, Maimoun L, Kotzki PO, Boudousq V. Predicting Response to [177Lu]Lu-PSMA Therapy in mCRPC Using Machine Learning. Journal of Personalized Medicine. 2024; 14(11):1068. https://doi.org/10.3390/jpm14111068
Chicago/Turabian StyleGong, Kaiyuan, Baptiste Magnier, Salomé L’hostis, Fanny Borrely, Sébastien Le Bon, Nadine Houede, Adel Mamou, Laurent Maimoun, Pierre Olivier Kotzki, and Vincent Boudousq. 2024. "Predicting Response to [177Lu]Lu-PSMA Therapy in mCRPC Using Machine Learning" Journal of Personalized Medicine 14, no. 11: 1068. https://doi.org/10.3390/jpm14111068
APA StyleGong, K., Magnier, B., L’hostis, S., Borrely, F., Le Bon, S., Houede, N., Mamou, A., Maimoun, L., Kotzki, P. O., & Boudousq, V. (2024). Predicting Response to [177Lu]Lu-PSMA Therapy in mCRPC Using Machine Learning. Journal of Personalized Medicine, 14(11), 1068. https://doi.org/10.3390/jpm14111068







