Population Heterogeneity and Selection of Coronary Artery Disease Polygenic Scores
Abstract
1. Introduction
2. Materials and Methods
2.1. Population Characteristics
2.2. PGS Selection
2.3. Statistical Analysis
3. Results
3.1. Population Characteristics and Quality Control
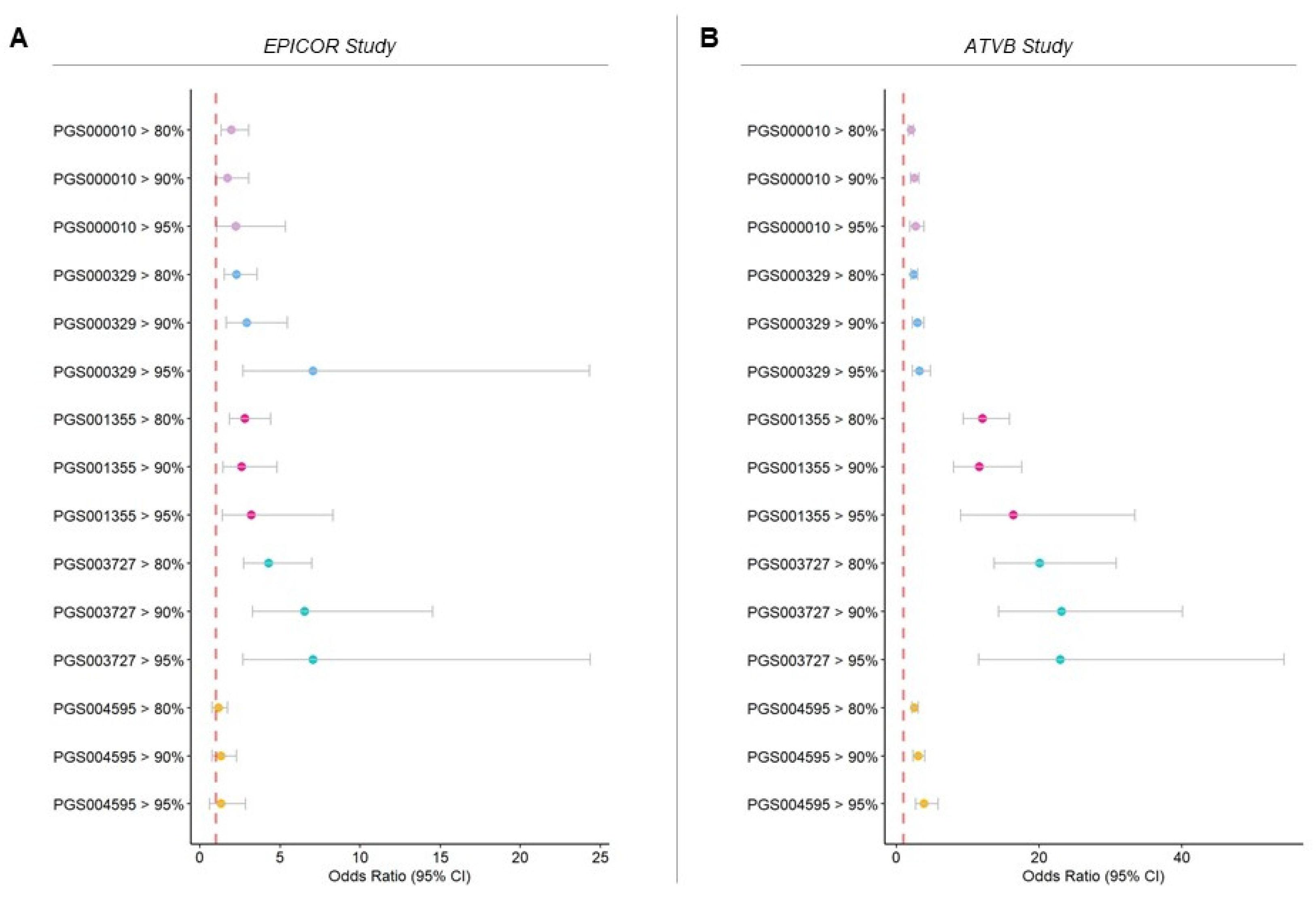
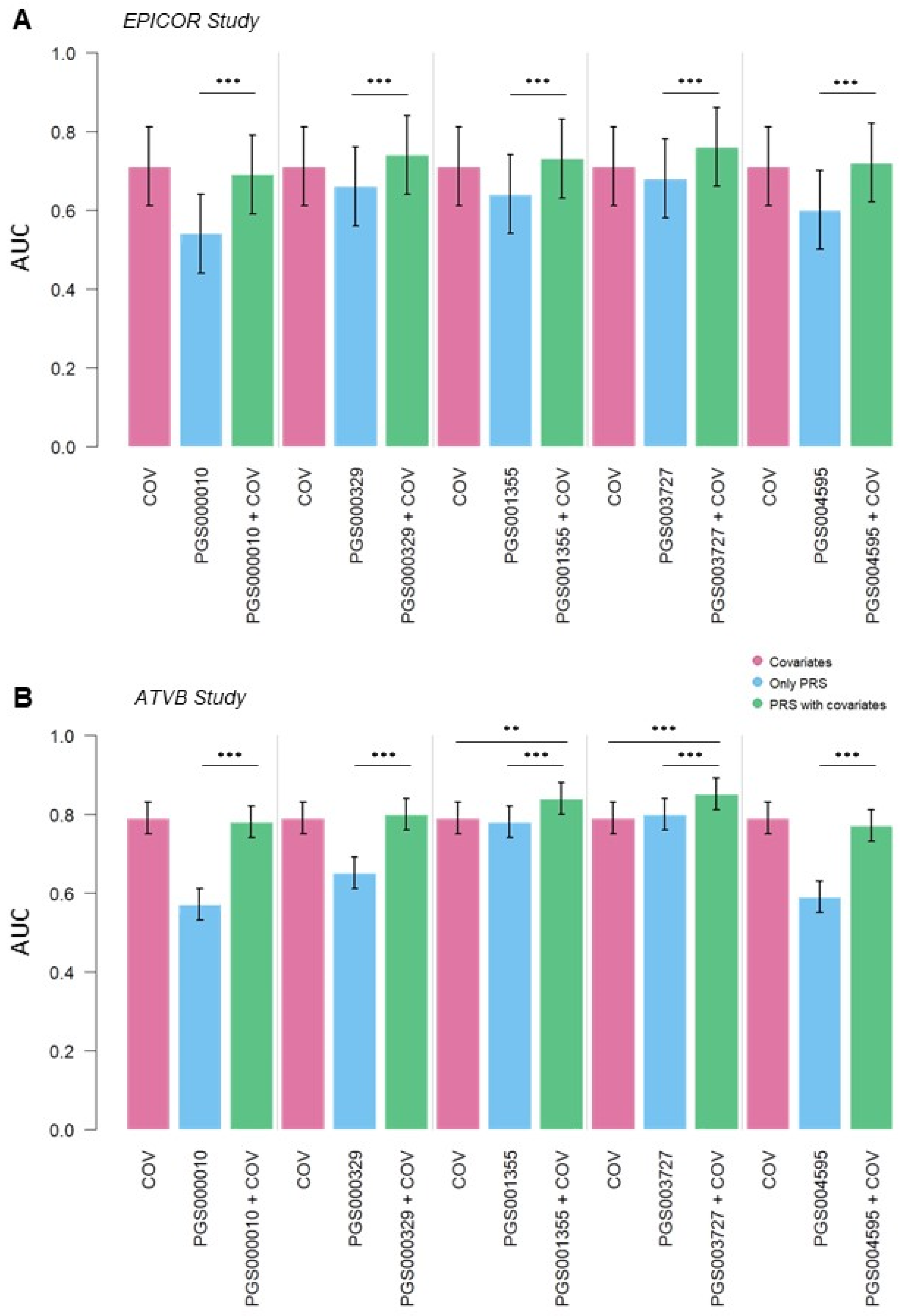
3.2. CAD Risk Predictive Power of PGS
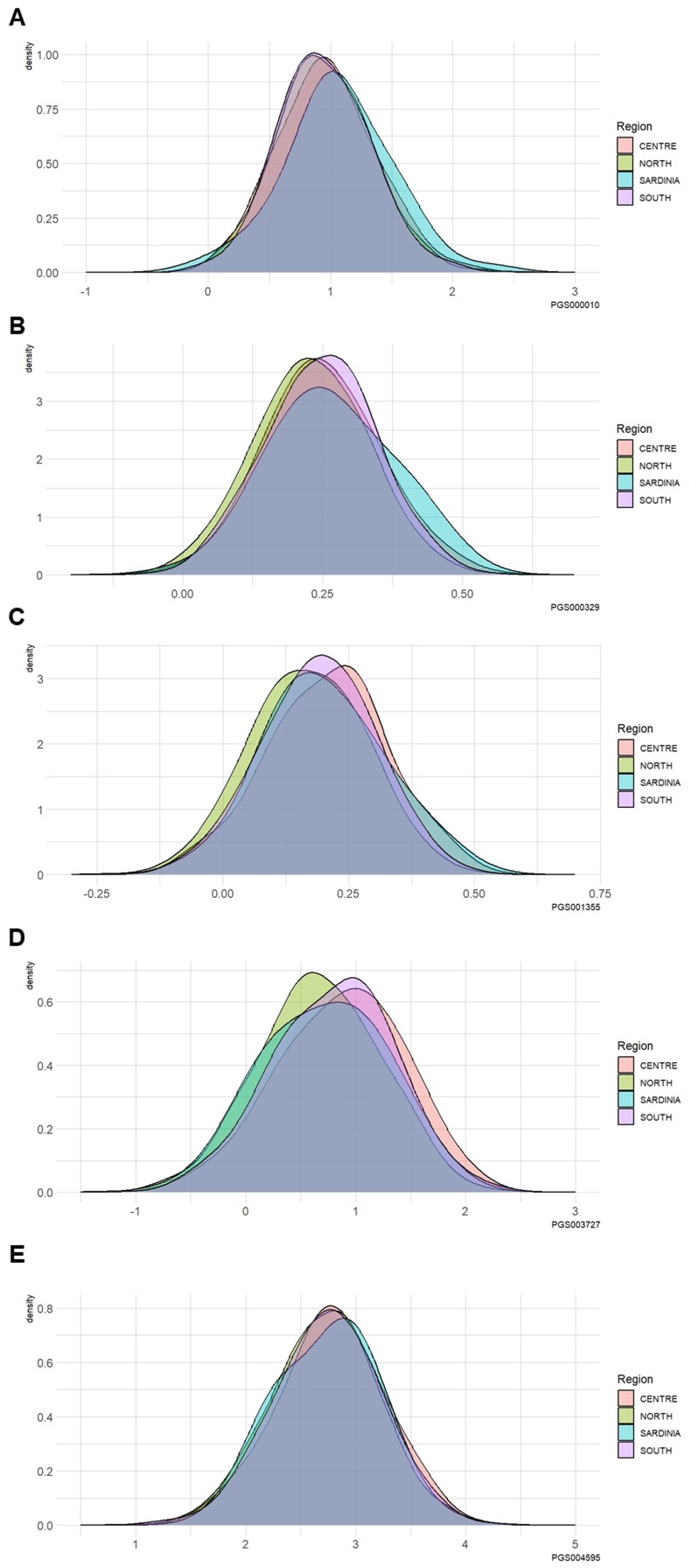
3.3. Differences in the PRS among Different Geographical Italian Macro-Areas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ralapanawa, U.; Sivakanesan, R. Epidemiology and the Magnitude of Coronary Artery Disease and Acute Coronary Syndrome: A Narrative Review. J. Epidemiology Glob. Health 2021, 11, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Drobni, Z.D.; Kolossvary, M.; Karady, J.; Jermendy, A.L.; Tarnoki, A.D.; Tarnoki, D.L.; Simon, J.; Szilveszter, B.; Littvay, L.; Voros, S.; et al. Heritability of Coronary Artery Disease: Insights From a Classical Twin Study. Circ. Cardiovasc. Imaging 2022, 15, e013348. [Google Scholar] [CrossRef] [PubMed]
- Latimer, J.; Batty, J.A.; Neely, R.D.G.; Kunadian, V. PCSK9 inhibitors in the prevention of cardiovascular disease. J. Thromb. Thrombolysis 2016, 42, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Schunkert, H. Genetics of coronary artery disease in the post-GWAS era. J. Intern. Med. 2021, 290, 980–992. [Google Scholar] [CrossRef]
- Hua, B.; Springer, M. Widespread Cumulative Influence of Small Effect Size Mutations on Yeast Quantitative Traits. Cell Syst. 2018, 7, 590–600.e6. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef]
- Lewis, C.M.; Vassos, E. Polygenic risk scores: From research tools to clinical instruments. Genome Med. 2020, 12, 44. [Google Scholar] [CrossRef]
- Lello, L.; Raben, T.G.; Hsu, S.D.H. Sibling validation of polygenic risk scores and complex trait prediction. Sci. Rep. 2020, 10, 13190. [Google Scholar] [CrossRef]
- Elliott, J.; Bodinier, B.; Bond, T.A.; Chadeau-Hyam, M.; Evangelou, E.; Moons, K.G.M.; Dehghan, A.; Muller, D.C.; Elliott, P.; Tzoulaki, I. Predictive Accuracy of a Polygenic Risk Score–Enhanced Prediction Model vs. a Clinical Risk Score for Coronary Artery Disease. JAMA 2020, 323, 636–645. [Google Scholar] [CrossRef]
- Schnitzer, F.; Forer, L.; Schönherr, S.; Gieger, C.; Grallert, H.; Kronenberg, F.; Peters, A.; Lamina, C. Association between a polygenic and family risk score on the prevalence and incidence of myocardial infarction in the KORA-F3 study. Atherosclerosis 2022, 352, 10–17. [Google Scholar] [CrossRef]
- Lambert, S.A.; Gil, L.; Jupp, S.; Ritchie, S.C.; Xu, Y.; Buniello, A.; McMahon, A.; Abraham, G.; Chapman, M.; Parkinson, H.; et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 2021, 53, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Duncan, L.; Shen, H.; Gelaye, B.; Meijsen, J.; Ressler, K.; Feldman, M.; Peterson, R.; Domingue, B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019, 10, 3328. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.Q.; Sallah, N.; Dunca, D.; Trivedi, B.; Hunt, K.A.; Hodgson, S.; Lambert, S.A.; Arciero, E.; Wright, J.; Griffiths, C.; et al. Transferability of genetic loci and polygenic scores for cardiometabolic traits in British Pakistani and Bangladeshi individuals. Nat. Commun. 2022, 13, 4664. [Google Scholar] [CrossRef] [PubMed]
- Amariuta, T.; Ishigaki, K.; Sugishita, H.; Ohta, T.; Koido, M.; Dey, K.K.; Matsuda, K.; Murakami, Y.; Price, A.L.; Kawakami, E.; et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat. Genet. 2020, 52, 1346–1354. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, J.; Ni, G.; Yang, J.; Visscher, P.M.; Yengo, L. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat. Commun. 2020, 11, 3865. [Google Scholar] [CrossRef]
- Durvasula, A.; Lohmueller, K.E. Negative selection on complex traits limits phenotype prediction accuracy between populations. Am. J. Hum. Genet. 2021, 108, 620–631. [Google Scholar] [CrossRef]
- Fiorito, G.; Di Gaetano, C.; Guarrera, S.; Rosa, F.; Feldman, M.W.; Piazza, A.; Matullo, G. The Italian genome reflects the history of Europe and the Mediterranean basin. Eur. J. Hum. Genet. 2016, 24, 1056–1062. [Google Scholar] [CrossRef]
- Di Gaetano, C.; Voglino, F.; Guarrera, S.; Fiorito, G.; Rosa, F.; Di Blasio, A.M.; Manzini, P.; Dianzani, I.; Betti, M.; Cusi, D.; et al. An overview of the genetic structure within the Italian population from genome-wide data. PLoS ONE 2012, 7, e43759. [Google Scholar] [CrossRef]
- Sarno, S.; Boattini, A.; Carta, M.; Ferri, G.; Alù, M.; Yao, D.Y.; Ciani, G.; Pettener, D.; Luiselli, D. An ancient Mediterranean melting pot: Investigating the uniparental genetic structure and population history of sicily and southern Italy. PLoS ONE 2014, 9, e96074. [Google Scholar] [CrossRef]
- Sazzini, M.; Abondio, P.; Sarno, S.; Gnecchi-Ruscone, G.A.; Ragno, M.; Giuliani, C.; De Fanti, S.; Ojeda-Granados, C.; Boattini, A.; Marquis, J.; et al. Genomic history of the Italian population recapitulates key evolutionary dynamics of both Continental and Southern Europeans. BMC Biol. 2020, 18, 51. [Google Scholar] [CrossRef]
- Raveane, A.; Aneli, S.; Montinaro, F.; Athanasiadis, G.; Barlera, S.; Birolo, G.; Boncoraglio, G.; Di Blasio, A.M.; Di Gaetano, C.; Pagani, L.; et al. Population structure of modern-day Italians reveals patterns of ancient and archaic ancestries in Southern Europe. Sci. Adv. 2019, 5, eaaw3492. [Google Scholar] [CrossRef] [PubMed]
- Guarrera, S.; Fiorito, G.; Onland-Moret, N.C.; Russo, A.; Agnoli, C.; Allione, A.; Di Gaetano, C.; Mattiello, A.; Ricceri, F.; Chiodini, P.; et al. Gene-specific DNA methylation profiles and LINE-1 hypomethylation are associated with myocardial infarction risk. Clin. Epigenetics 2015, 7, 133. [Google Scholar] [CrossRef] [PubMed]
- Palli, D.; Berrino, F.; Vineis, P.; Tumino, R.; Panico, S.; Masala, G.; Saieva, C.; Salvini, S.; Cerati, M.; Pala, V.; et al. A molecular epidemiology project on diet and cancer: The epic-Italy prospective study. Design and baseline characteristics of participants. Tumori J. 2003, 89, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Atherosclerosis; Thrombosis; Vascular Biology Italian Study Group. No Evidence of Association Between Prothrombotic Gene Polymorphisms and the Development of Acute Myocardial Infarction at a Young Age. Circulation 2003, 107, 1117–1122. [Google Scholar] [CrossRef]
- Project, N.E.S.; Do, R.; Stitziel, N.O.; Won, H.-H.; Jørgensen, A.B.; Duga, S.; Merlini, P.A.; Kiezun, A.; Farrall, M.; Goel, A.; et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015, 518, 102–106. [Google Scholar] [CrossRef]
- Birolo, G.; Aneli, S.; Di Gaetano, C.; Cugliari, G.; Russo, A.; Allione, A.; Casalone, E.; Giorgio, E.; Paraboschi, E.M.; Ardissino, D.; et al. Functional and clinical implications of genetic structure in 1686 Italian exomes. Hum. Mutat. 2021, 42, 272–289. [Google Scholar] [CrossRef]
- Mega, J.L.; Stitziel, N.O.; Smith, J.G.; Chasman, D.I.; Caulfield, M.J.; Devlin, J.J.; Nordio, F.; Hyde, C.L.; Cannon, C.P.; Sacks, F.M.; et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: An analysis of primary and secondary prevention trials. Lancet 2015, 385, 2264–2271. [Google Scholar] [CrossRef]
- Mars, N.; Koskela, J.T.; Ripatti, P.; Kiiskinen, T.T.J.; Havulinna, A.S.; Lindbohm, J.V.; Ahola-Olli, A.; Kurki, M.; Karjalainen, J.; Palta, P.; et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med. 2020, 26, 549–557. [Google Scholar] [CrossRef]
- Ye, Y.; Chen, X.; Han, J.; Jiang, W.; Natarajan, P.; Zhao, H. Interactions Between Enhanced Polygenic Risk Scores and Lifestyle for Cardiovascular Disease, Diabetes, and Lipid Levels. Circ. Genom. Precis. Med. 2021, 14, e003128. [Google Scholar] [CrossRef]
- Patel, A.P.; Wang, M.; Ruan, Y.; Koyama, S.; Clarke, S.L.; Yang, X.; Tcheandjieu, C.; Agrawal, S.; Fahed, A.C.; Ellinor, P.T.; et al. A multi-ancestry polygenic risk score improves risk prediction for coronary artery disease. Nat. Med. 2023, 29, 1793–1803. [Google Scholar] [CrossRef]
- Oni-Orisan, A.; Haldar, T.; Cayabyab, M.A.; Ranatunga, D.K.; Hoffmann, T.J.; Iribarren, C.; Krauss, R.M.; Risch, N. Polygenic Risk Score and Statin Relative Risk Reduction for Primary Prevention of Myocardial Infarction in a Real-World Population. Clin. Pharmacol. Ther. 2022, 112, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Modi, A.; Vizzari, M.T.; Catalano, G.; Agostini, R.B.; Vai, S.; Lari, M.; Vergata, C.; Zaro, V.; Liccioli, L.; Fedi, M.; et al. Genetic structure and differentiation from early bronze age in the mediterranean island of sicily: Insights from ancient mitochondrial genomes. Front. Genet. 2022, 13, 945227. [Google Scholar] [CrossRef] [PubMed]
- Ghiani, M.; Calò, M.; Autuori, L.; Mameli, G.; Succa, V.; Vacca, L.; Cerutti, N.; Massa, E.R.; Vona, G. New data on the genetic structure of the population of Sicily: Analysis of the Alia population (Palermo, Italy). Am. J. Hum. Biol. 2002, 14, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, A.; Sidore, C.; Achilli, A.; Angius, A.; Posth, C.; Furtwängler, A.; Brandini, S.; Capodiferro, M.R.; Gandini, F.; Zoledziewska, M.; et al. Mitogenome Diversity in Sardinians: A Genetic Window onto an Island’s Past. Mol. Biol. Evol. 2017, 34, 1230–1239. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kanai, M.; Tan, T.; Kamariza, M.; Tsuo, K.; Yuan, K.; Zhou, W.; Okada, Y.; Huang, H.; Turley, P.; et al. Polygenic prediction across populations is influenced by ancestry, genetic architecture, and methodology. Cell Genom. 2023, 3, 100408. [Google Scholar] [CrossRef]
- Scutari, M.; Mackay, I.; Balding, D. Using Genetic Distance to Infer the Accuracy of Genomic Prediction. PLOS Genet. 2016, 12, e1006288. [Google Scholar] [CrossRef]
- Reisberg, S.; Iljasenko, T.; Läll, K.; Fischer, K.; Vilo, J. Comparing distributions of polygenic risk scores of type 2 diabetes and coronary heart disease within different populations. PLoS ONE 2017, 12, e0179238. [Google Scholar] [CrossRef]
- Cavazos, T.B.; Witte, J.S. Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. Hum. Genet. Genom. Adv. 2020, 2, 100017. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jørsboe, E.; Andersen, M.K.; Skotte, L.; Stæger, F.F.; Færgeman, N.J.; Hanghøj, K.; Santander, C.G.; Senftleber, N.K.; Diaz, L.J.; Overvad, M.; et al. An LDLR missense variant poses high risk of familial hypercholesterolemia in 30% of Greenlanders and offers potential of early cardiovascular disease intervention. Hum. Genet. Genom. Adv. 2022, 3, 100118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hajar, R. Risk Factors for Coronary Artery Disease: Historical Perspectives. Hear. Views 2017, 18, 109–114. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.-H.; O’rEilly, P.F. Tutorial: A guide to performing polygenic risk score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Viigimaa, M.; Jürisson, M.; Pisarev, H.; Kalda, R.; Alavere, H.; Irs, A.; Saar, A.; Fischer, K.; Läll, K.; Kruuv-Käo, K.; et al. Effectiveness and feasibility of cardiovascular disease personalized prevention on high polygenic risk score subjects: A randomized controlled pilot study. Eur. Heart J. Open 2022, 15, oeac079. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | EPICOR Study (576 Individuals) | ATVB Study (3359 Individuals) | |||||
|---|---|---|---|---|---|---|---|
| Unit | Pre-clinical CAD individuals (286) | Disease-free individuals (290) | p-value | CAD Patients (1691) | Disease-free individuals (1668) | p-value | |
| Age | years mean (SD) | 53 (±7.3) | 53 (±7.5) | 0.94 | 40 (±4.9) | 40 (±4.9) | 0.752 |
| Sex | |||||||
| Male | 184 | 186 | 1.0 | 1498 | 1474 | 0.871 | |
| Female | 102 | 104 | 193 | 194 | |||
| BMI | kg/m2 mean (SD) | 27 (±3.7) | 26 (±3.9) | 0.0002 *** | 27 (±4.2) | 25 (±3.3) | <0.0001 *** |
| Total Cholesterol | mmol/L mean (SD) | 6.2 (±1.2) | 6.0 (±1.2) | 0.20 | 5.7 (±1.4) | 5.2 (±1.0) | <0.0001 *** |
| Hypercholesterolemia | 236 | 192 | 0.015 * | 933 | 690 | <0.0001 *** | |
| HDL | mmol/L mean (SD) | 1.4 (±0.4) | 1.6 (±0.4) | <0.0001 *** | 1.1 (±0.3) | 1.3 (±0.3) | <0.0001 *** |
| LDL | mmol/L mean (SD) | 4.0 (±1.0) | 3.7 (±1.0) | 0.012 * | 3.7 (±1.4) | 3.2 (±0.9) | <0.0001 *** |
| Triglycerides | mmol/L mean (SD) | 1.8 (±1.2) | 1.6 (±0.9) | 0.016 * | 2.0 (±1.5) | 1.3 (±0.8) | <0.0001 *** |
| Glycaemia | mmol/L mean (SD) | 5.8 (±1.8) | 5.5 (±1.0) | 0.166 | 6.2 (±2.2) | 5.0 (±0.8) | <0.0001 *** |
| Diabetes | 7 | 2 | 0.105 | 131 | 14 | <0.0001 *** | |
| Hypertension | 122 | 96 | 0.03 * | 459 | 148 | <0.0001 *** | |
| PAS | mmHg mean (SD) | 140 (±19) | 136 (±19) | 0.008 ** | 132 (±21) | 124 (±14) | <0.0001 *** |
| PAD | mmHg mean (SD) | 86 (±9) | 85 (±11) | 0.427 | 83 (±14) | 82 (±41) | <0.0001 *** |
| Smoke | |||||||
| Yes | 121 | 68 | <0.0001 *** | 709 | 294 | <0.0001 *** | |
| No | 86 | 123 | (reference) | 220 | 527 | (reference) | |
| Former | 79 | 99 | 0.54 | 758 | 845 | <0.0001 *** | |
| Kolmogorov–Smirnov Test Macro-Area Comparison | PGS000010 (p-value) | PGS000329 (p-value) | PGS001355 (p-value) | PGS003727 (p-value) | PGS004595 (p-value) |
| North vs. Center | 0.27 | 0.01 | <0.0001 | <0.0001 | 0.05 |
| North vs. Sardinia | 0.03 | 0.06 | 0.14 | 0.73 | 0.69 |
| North vs. South | 0.98 | 0.0004 | <0.0001 | <0.0001 | 0.49 |
| Center vs. Sardinia | 0.15 | 0.44 | 0.51 | 0.18 | 0.43 |
| Center vs. South | 0.29 | 0.85 | 0.02 | 0.13 | 0.44 |
| Sardinia vs. South | 0.03 | 0.26 | 0.90 | 0.50 | 0.64 |
| Kolmogorov–Smirnov Test Sex comparison (Female vs. Male) | PGS000010 (p-value) | PGS000329 (p-value) | PGS001355 (p-value) | PGS003727 (p-value) | PGS004595 (p-value) |
| North (230 vs. 1800) | 0.98 | 0.18 | 0.35 | 0.13 | 0.43 |
| Center (46 vs. 374) | 0.67 | 0.73 | 0.43 | 0.21 | 0.37 |
| Sardinia (14 vs. 54) | 0.42 | 0.85 | 0.57 | 0.97 | 0.51 |
| South (84 vs. 660) | 0.91 | 0.54 | 0.61 | 0.16 | 0.35 |
| Kolmogorov–Smirnov Test Cases vs. Controls comparison | PGS000010 (p-value) | PGS000329 (p-value) | PGS001355 (p-value) | PGS003727 (p-value) | PGS004595 (p-value) |
| North (903 vs. 1127) | <0.0001 | 0 | <0.0001 | <0.0001 | <0.0001 |
| Center (269 vs. 151) | 0.13 | <0.0001 | <0.0001 | <0.0001 | 0.002 |
| Sardinia (42 vs. 26) | 0.12 | 0.02 | 0.007 | 0.0001 | 0.53 |
| South (419 vs. 325) | 0.0002 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Debernardi, C.; Savoca, A.; De Gregorio, A.; Casalone, E.; Rosselli, M.; Herman, E.J.; Di Primio, C.; Tumino, R.; Sieri, S.; Vineis, P.; et al. Population Heterogeneity and Selection of Coronary Artery Disease Polygenic Scores. J. Pers. Med. 2024, 14, 1025. https://doi.org/10.3390/jpm14101025
Debernardi C, Savoca A, De Gregorio A, Casalone E, Rosselli M, Herman EJ, Di Primio C, Tumino R, Sieri S, Vineis P, et al. Population Heterogeneity and Selection of Coronary Artery Disease Polygenic Scores. Journal of Personalized Medicine. 2024; 14(10):1025. https://doi.org/10.3390/jpm14101025
Chicago/Turabian StyleDebernardi, Carla, Angelo Savoca, Alessandro De Gregorio, Elisabetta Casalone, Miriam Rosselli, Elton Jalis Herman, Cecilia Di Primio, Rosario Tumino, Sabina Sieri, Paolo Vineis, and et al. 2024. "Population Heterogeneity and Selection of Coronary Artery Disease Polygenic Scores" Journal of Personalized Medicine 14, no. 10: 1025. https://doi.org/10.3390/jpm14101025
APA StyleDebernardi, C., Savoca, A., De Gregorio, A., Casalone, E., Rosselli, M., Herman, E. J., Di Primio, C., Tumino, R., Sieri, S., Vineis, P., Panico, S., Sacerdote, C., Ardissino, D., Asselta, R., & Matullo, G. (2024). Population Heterogeneity and Selection of Coronary Artery Disease Polygenic Scores. Journal of Personalized Medicine, 14(10), 1025. https://doi.org/10.3390/jpm14101025







