Low CDKN1B Expression Associated with Reduced CD8+ T Lymphocytes Predicts Poor Outcome in Breast Cancer in a Machine Learning Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Tissue Microarray Construction and Immunohistochemistry
2.3. Immunohistochemistry for CD8+ T Cells
2.4. GSEA, In Silico Cytometry, and Pathway Network Analyses
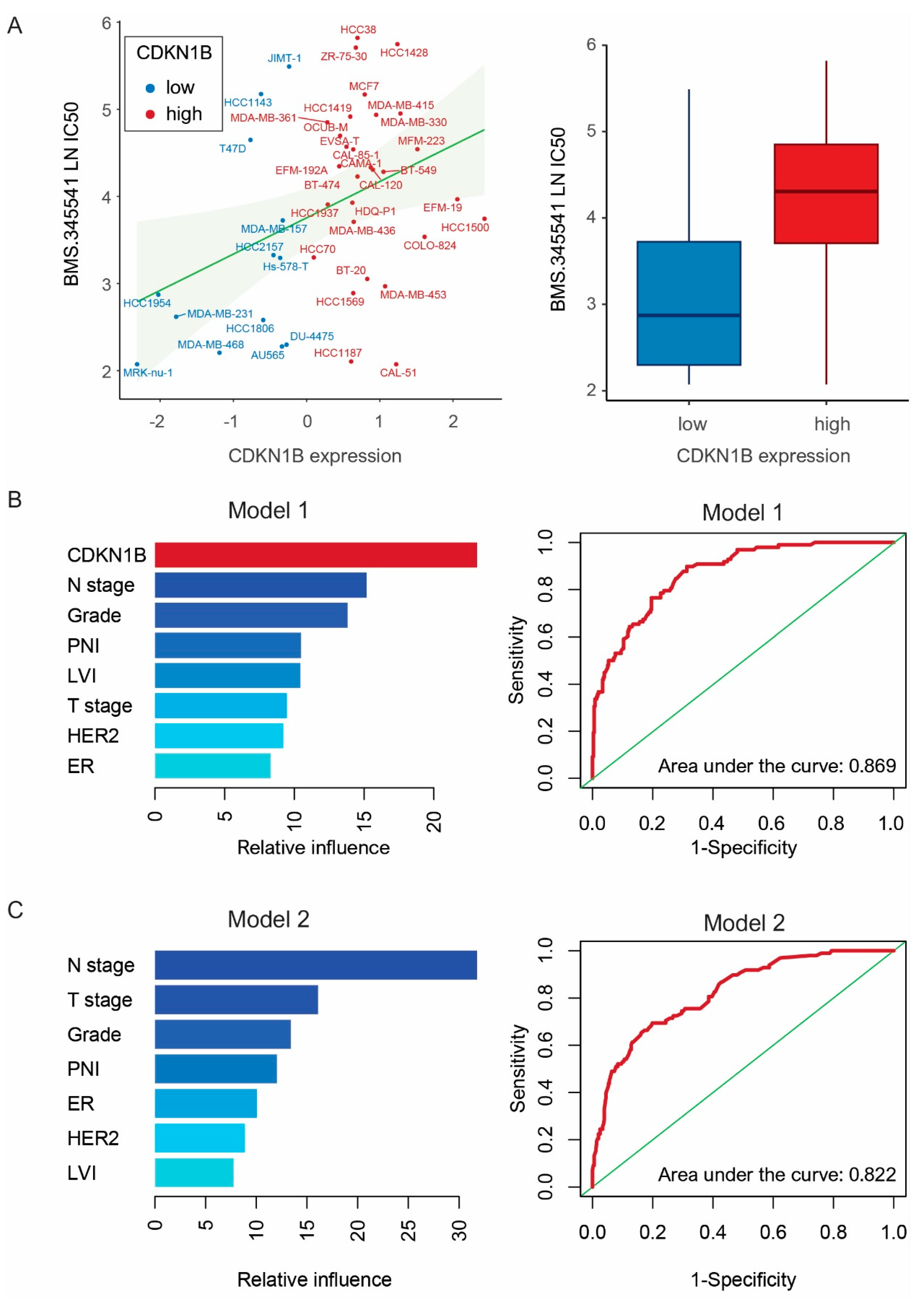
2.5. Genomics of Drug Sensitivity in Cancer Database and GBMs
2.6. Statistical Analysis
3. Results
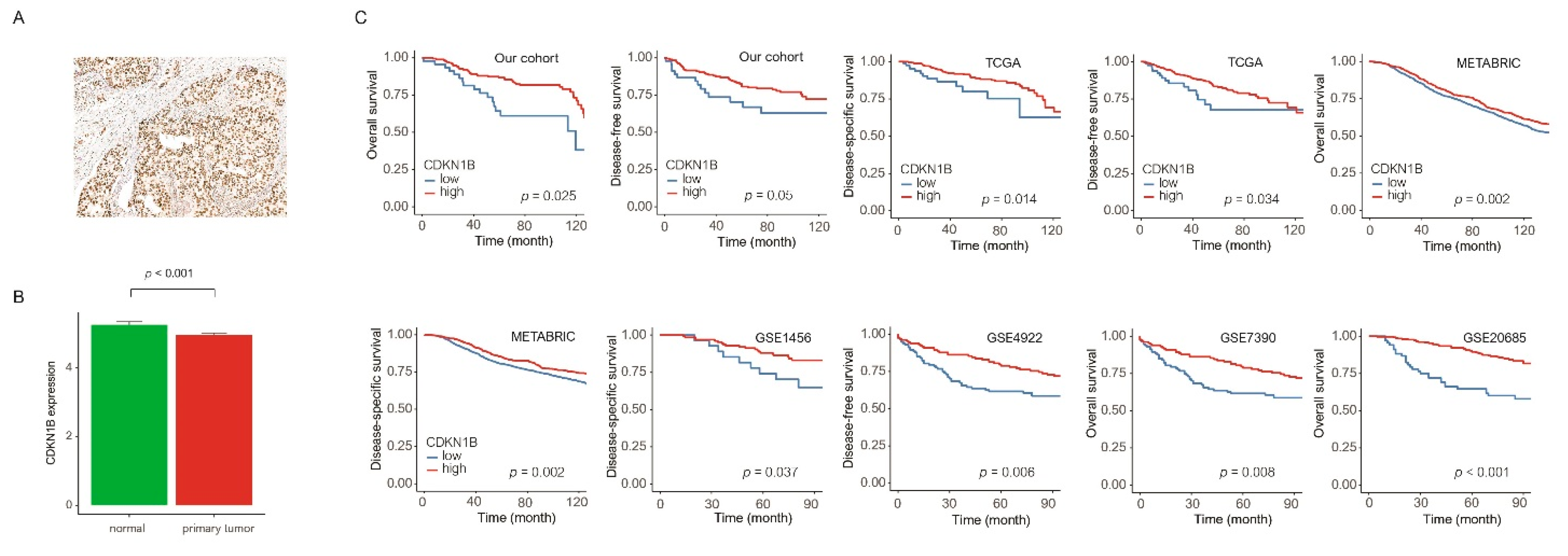
3.1. Clinicopathological Parameters and Survival Analysis of CDKN1B
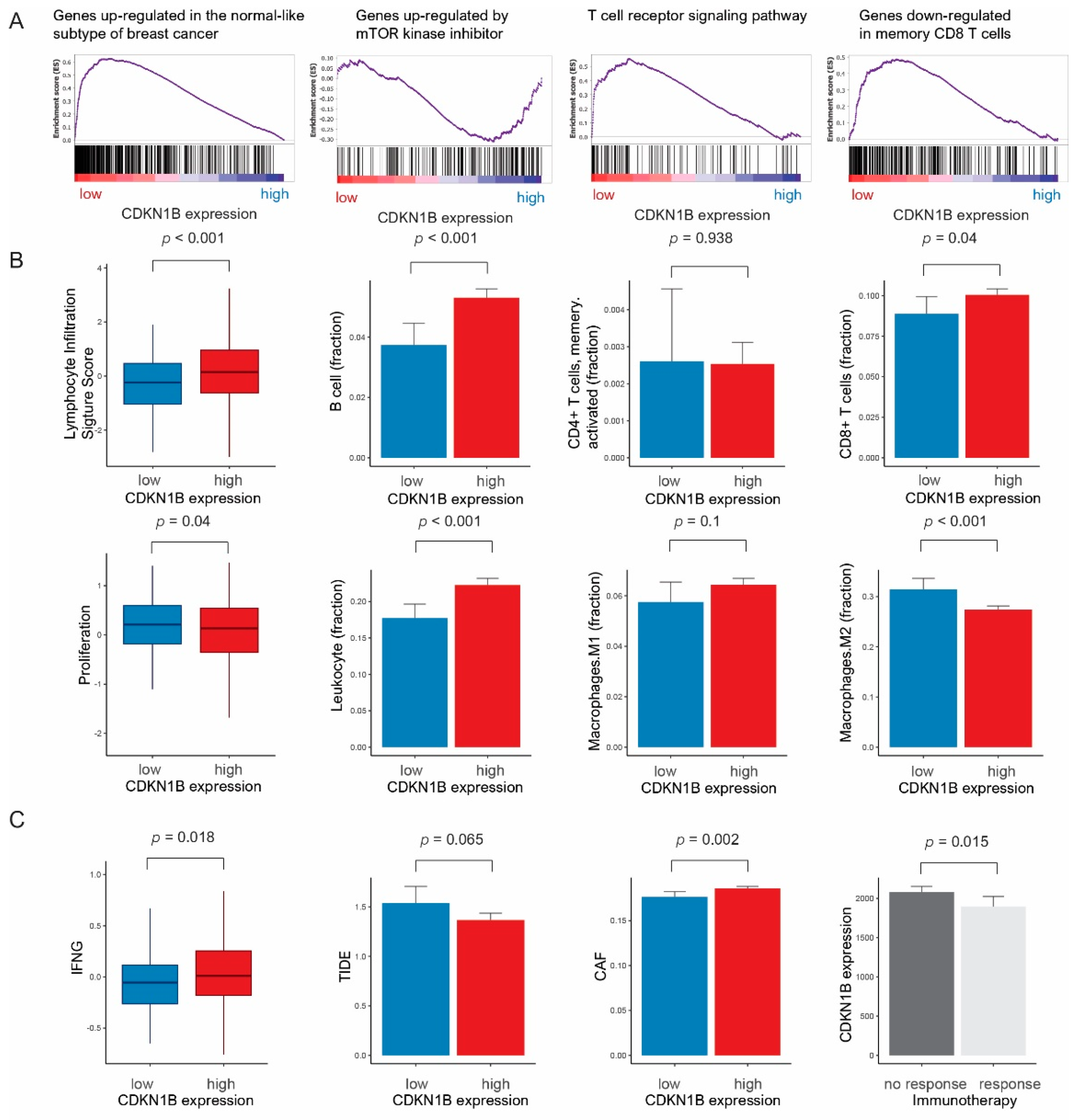
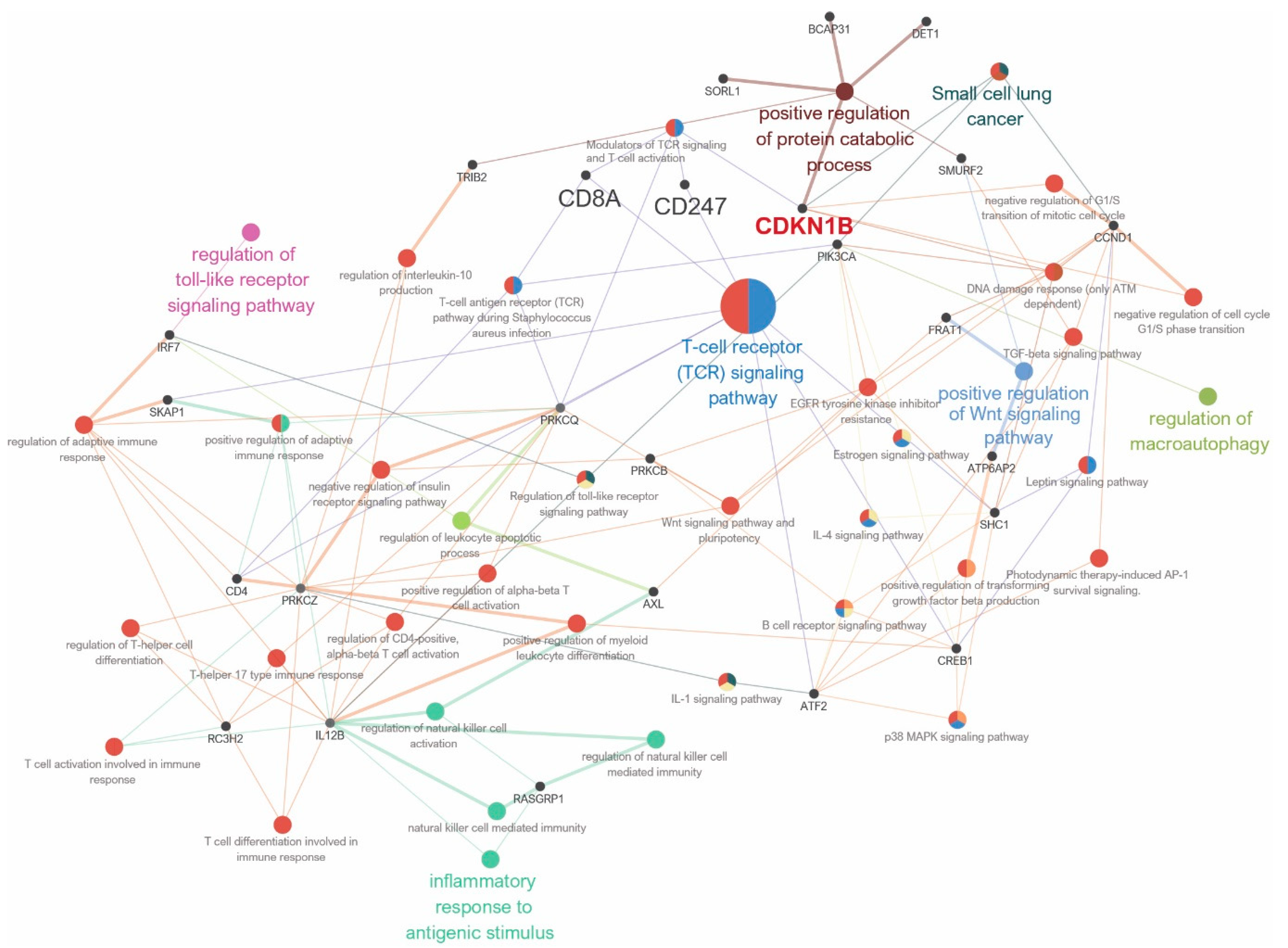
3.2. Gene Sets, Immune Response, and Pathway Network Analysis According to CDKN1B Expression
3.3. Drug Screening and Machine Learning Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Stingl, J.; Caldas, C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat. Rev. Cancer 2007, 7, 791–799. [Google Scholar] [CrossRef]
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar]
- Chu, I.M.; Hengst, L.; Slingerland, J.M. The Cdk inhibitor p27 in human cancer: Prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer 2008, 8, 253–267. [Google Scholar] [CrossRef]
- Razavipour, S.F.; Harikumar, K.B.; Slingerland, J.M. p27 as a Transcriptional Regulator: New Roles in Development and Cancer. Cancer Res. 2020, 80, 3451–3458. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Bangma, C.H.; Bjartell, A.; Catto, J.W.; Culig, Z.; Gronberg, H.; Luo, J.; Visakorpi, T.; Rubin, M.A. The mutational landscape of prostate cancer. Eur. Urol. 2013, 64, 567–576. [Google Scholar] [CrossRef]
- Yates, L.R.; Knappskog, S.; Wedge, D.; Farmery, J.H.R.; Gonzalez, S.; Martincorena, I.; Alexandrov, L.B.; Van Loo, P.; Haugland, H.K.; Lilleng, P.K.; et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017, 32, 169–184.e167. [Google Scholar] [CrossRef]
- Viotto, D.; Russo, F.; Anania, I.; Segatto, I.; Rampioni Vinciguerra, G.L.; Dall’Acqua, A.; Bomben, R.; Perin, T.; Cusan, M.; Schiappacassi, M.; et al. CDKN1B mutation and copy number variation are associated with tumor aggressiveness in luminal breast cancer. J. Pathol. 2021, 253, 234–245. [Google Scholar] [CrossRef]
- Stendahl, M.; Nilsson, S.; Wigerup, C.; Jirstrom, K.; Jonsson, P.E.; Stal, O.; Landberg, G. p27Kip1 is a predictive factor for tamoxifen treatment response but not a prognostic marker in premenopausal breast cancer patients. Int. J. Cancer 2010, 127, 2851–2858. [Google Scholar] [CrossRef]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef]
- Pereira, B.; Chin, S.F.; Rueda, O.M.; Vollan, H.K.; Provenzano, E.; Bardwell, H.A.; Pugh, M.; Jones, L.; Russell, R.; Sammut, S.J.; et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016, 7, 11479. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Peng, X.; Chen, Z.; Farshidfar, F.; Xu, X.; Lorenzi, P.L.; Wang, Y.; Cheng, F.; Tan, L.; Mojumdar, K.; Du, D.; et al. Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers. Cell Rep. 2018, 23, 255–269.e254. [Google Scholar] [CrossRef]
- Pawitan, Y.; Bjohle, J.; Amler, L.; Borg, A.L.; Egyhazi, S.; Hall, P.; Han, X.; Holmberg, L.; Huang, F.; Klaar, S.; et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: Derived and validated in two population-based cohorts. Breast Cancer Res. 2005, 7, R953–R964. [Google Scholar] [CrossRef]
- Ivshina, A.V.; George, J.; Senko, O.; Mow, B.; Putti, T.C.; Smeds, J.; Lindahl, T.; Pawitan, Y.; Hall, P.; Nordgren, H.; et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006, 66, 10292–10301. [Google Scholar] [CrossRef]
- Patil, P.; Bachant-Winner, P.O.; Haibe-Kains, B.; Leek, J.T. Test set bias affects reproducibility of gene signatures. Bioinformatics 2015, 31, 2318–2323. [Google Scholar] [CrossRef]
- Kao, K.J.; Chang, K.M.; Hsu, H.C.; Huang, A.T. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer 2011, 11, 143. [Google Scholar] [CrossRef]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Remmele, W.; Stegner, H.E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 1987, 8, 138–140. [Google Scholar]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J.; Panel, m. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef]
- Dahlin, A.M.; Henriksson, M.L.; Van Guelpen, B.; Stenling, R.; Oberg, A.; Rutegard, J.; Palmqvist, R. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod. Pathol. 2011, 24, 671–682. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Cancer cell cycles. Science 1996, 274, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Bencivenga, D.; Stampone, E.; Aulitto, A.; Tramontano, A.; Barone, C.; Negri, A.; Roberti, D.; Perrotta, S.; Della Ragione, F.; Borriello, A. A cancer-associated CDKN1B mutation induces p27 phosphorylation on a novel residue: A new mechanism for tumor suppressor loss-of-function. Mol. Oncol. 2021, 15, 915–941. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, J.; Sokol, E.S.; Hartmaier, R.J.; Trabucco, S.E.; Frampton, G.M.; Goldberg, M.E.; Albacker, L.A.; Daemen, A.; Manning, G. The genomic landscape of metastatic breast cancer: Insights from 11,000 tumors. PLoS ONE 2020, 15, e0231999. [Google Scholar] [CrossRef]
- Berton, S.; Cusan, M.; Segatto, I.; Citron, F.; D’Andrea, S.; Benevol, S.; Avanzo, M.; Dall’Acqua, A.; Schiappacassi, M.; Bristow, R.G.; et al. Loss of p27(kip1) increases genomic instability and induces radio-resistance in luminal breast cancer cells. Sci. Rep. 2017, 7, 595. [Google Scholar] [CrossRef]
- Economopoulou, P.; Kotoula, V.; Koliou, G.A.; Papadopoulou, K.; Christodoulou, C.; Pentheroudakis, G.; Lazaridis, G.; Arapantoni-Dadioti, P.; Koutras, A.; Bafaloukos, D.; et al. Prognostic Impact of Src, CDKN1B, and JAK2 Expression in Metastatic Breast Cancer Patients Treated with Trastuzumab. Transl. Oncol. 2019, 12, 739–748. [Google Scholar] [CrossRef]
- Giraldo, N.A.; Sanchez-Salas, R.; Peske, J.D.; Vano, Y.; Becht, E.; Petitprez, F.; Validire, P.; Ingels, A.; Cathelineau, X.; Fridman, W.H.; et al. The clinical role of the TME in solid cancer. Br. J. Cancer 2019, 120, 45–53. [Google Scholar] [CrossRef]
- Manuel, M.; Tredan, O.; Bachelot, T.; Clapisson, G.; Courtier, A.; Parmentier, G.; Rabeony, T.; Grives, A.; Perez, S.; Mouret, J.F.; et al. Lymphopenia combined with low TCR diversity (divpenia) predicts poor overall survival in metastatic breast cancer patients. Oncoimmunology 2012, 1, 432–440. [Google Scholar] [CrossRef]
- Collignon, J.; Lousberg, L.; Schroeder, H.; Jerusalem, G. Triple-negative breast cancer: Treatment challenges and solutions. Breast Cancer 2016, 8, 93–107. [Google Scholar] [CrossRef]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Clinical relevance of tumor microenvironment: Immune cells, vessels, and mouse models. Hum. Cell 2020, 33, 930–937. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Huleatt, J.W.; Cresswell, J.; Bottomly, K.; Crispe, I.N. P27kip1 regulates the cell cycle arrest and survival of activated T lymphocytes in response to interleukin-2 withdrawal. Immunology 2003, 108, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Patsoukis, N.; Sari, D.; Boussiotis, V.A. PD-1 inhibits T cell proliferation by upregulating p27 and p15 and suppressing Cdc25A. Cell Cycle 2012, 11, 4305–4309. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Amiri, K.I.; Burke, J.R.; Schmid, J.A.; Richmond, A. BMS-345541 targets inhibitor of kappaB kinase and induces apoptosis in melanoma: Involvement of nuclear factor kappaB and mitochondria pathways. Clin. Cancer Res. 2006, 12, 950–960. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-kappaB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- Buontempo, F.; Chiarini, F.; Bressanin, D.; Tabellini, G.; Melchionda, F.; Pession, A.; Fini, M.; Neri, L.M.; McCubrey, J.A.; Martelli, A.M. Activity of the selective IkappaB kinase inhibitor BMS-345541 against T-cell acute lymphoblastic leukemia: Involvement of FOXO3a. Cell Cycle 2012, 11, 2467–2475. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef]
| Parameter | CDKN1B Expression | p-Value 1 | |
|---|---|---|---|
| Low (n = 85), n (%) | High (n = 371), n (%) | ||
| Age (years) | 50.4 ± 10.0 | 49.3 ± 9.9 | 0.333 |
| T stage | |||
| 1 | 30 (35.3%) | 178 (48.0%) | 0.034 3 |
| 2 | 48 (56.5%) | 178 (48.0%) | |
| 3 | 7 (8.2%) | 15 (4.0%) | |
| N stage | |||
| 0 | 39 (45.9%) | 194 (52.3%) | 0.021 3 |
| 1 | 22 (25.9%) | 113 (30.5%) | |
| 2 | 15 (17.6%) | 45 (12.1%) | |
| 3 | 9 (10.6%) | 19 (5.1%) | |
| Histological grade | |||
| 1 | 13 (15.3%) | 74 (19.9%) | 0.325 3 |
| 2 | 45 (52.9%) | 171 (46.1%) | |
| 3 | 27 (31.8%) | 126 (34.0%) | |
| Lymphatic invasion | |||
| Negative | 44 (51.8%) | 186 (50.1%) | 0.88 |
| Positive | 41 (48.2%) | 185 (49.9%) | |
| Vascular invasion | 0.001 | ||
| Negative | 72 (84.7%) | 354 (95.4%) | |
| Positive | 13 (15.3%) | 17 (4.6%) | |
| Perineural invasion | 0.008 | ||
| Negative | 58 (68.2%) | 304 (81.9%) | |
| Positive | 27 (31.8%) | 67 (18.1%) | |
| ER | 0.356 | ||
| Negative | 28 (32.9%) | 101 (27.2%) | |
| Positive | 57 (67.1%) | 270 (72.8%) | |
| PR | 0.44 | ||
| Negative | 37 (43.5%) | 142 (38.3%) | |
| Positive | 48 (56.5%) | 229 (61.7%) | |
| HER2 | 0.016 | ||
| Negative | 69 (81.2%) | 249 (67.1%) | |
| Positive | 16 (18.8%) | 122 (32.9%) | |
| Covariate | Disease-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | p-Value | HR | 95%CI | p-Value | |||
| Our cohort | ||||||||
| Multivariate 1 | 0.530 | 0.330 | 0.852 | 0.009 | 0.498 | 0.315 | 0.788 | 0.003 |
| Covariate | Disease-free survival | Disease-specific survival | ||||||
| HR | 95%CI | p value | HR | 95%CI | p value | |||
| TCGA | ||||||||
| Multivariate 2 | 0.560 | 0.318 | 0.989 | 0.046 | 0.509 | 0.267 | 0.971 | 0.04 |
| Covariate | Disease-specific survival | Overall survival | ||||||
| HR | 95%CI | p value | HR | 95%CI | p value | |||
| METABRIC | ||||||||
| Multivariate 3 | 0.829 | 0.689 | 0.999 | 0.048 | 0.857 | 0.746 | 0.984 | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-S.; Noh, Y.-K.; Min, K.-W.; Kim, D.-H. Low CDKN1B Expression Associated with Reduced CD8+ T Lymphocytes Predicts Poor Outcome in Breast Cancer in a Machine Learning Analysis. J. Pers. Med. 2024, 14, 30. https://doi.org/10.3390/jpm14010030
Kim H-S, Noh Y-K, Min K-W, Kim D-H. Low CDKN1B Expression Associated with Reduced CD8+ T Lymphocytes Predicts Poor Outcome in Breast Cancer in a Machine Learning Analysis. Journal of Personalized Medicine. 2024; 14(1):30. https://doi.org/10.3390/jpm14010030
Chicago/Turabian StyleKim, Hyung-Suk, Yung-Kyun Noh, Kyueng-Whan Min, and Dong-Hoon Kim. 2024. "Low CDKN1B Expression Associated with Reduced CD8+ T Lymphocytes Predicts Poor Outcome in Breast Cancer in a Machine Learning Analysis" Journal of Personalized Medicine 14, no. 1: 30. https://doi.org/10.3390/jpm14010030
APA StyleKim, H.-S., Noh, Y.-K., Min, K.-W., & Kim, D.-H. (2024). Low CDKN1B Expression Associated with Reduced CD8+ T Lymphocytes Predicts Poor Outcome in Breast Cancer in a Machine Learning Analysis. Journal of Personalized Medicine, 14(1), 30. https://doi.org/10.3390/jpm14010030







