Return of Participants’ Incidental Genetic Research Findings: Experience from a Case-Control Study of Asthma in an American Indian Community
Abstract
:1. Introduction
2. Materials and Methods
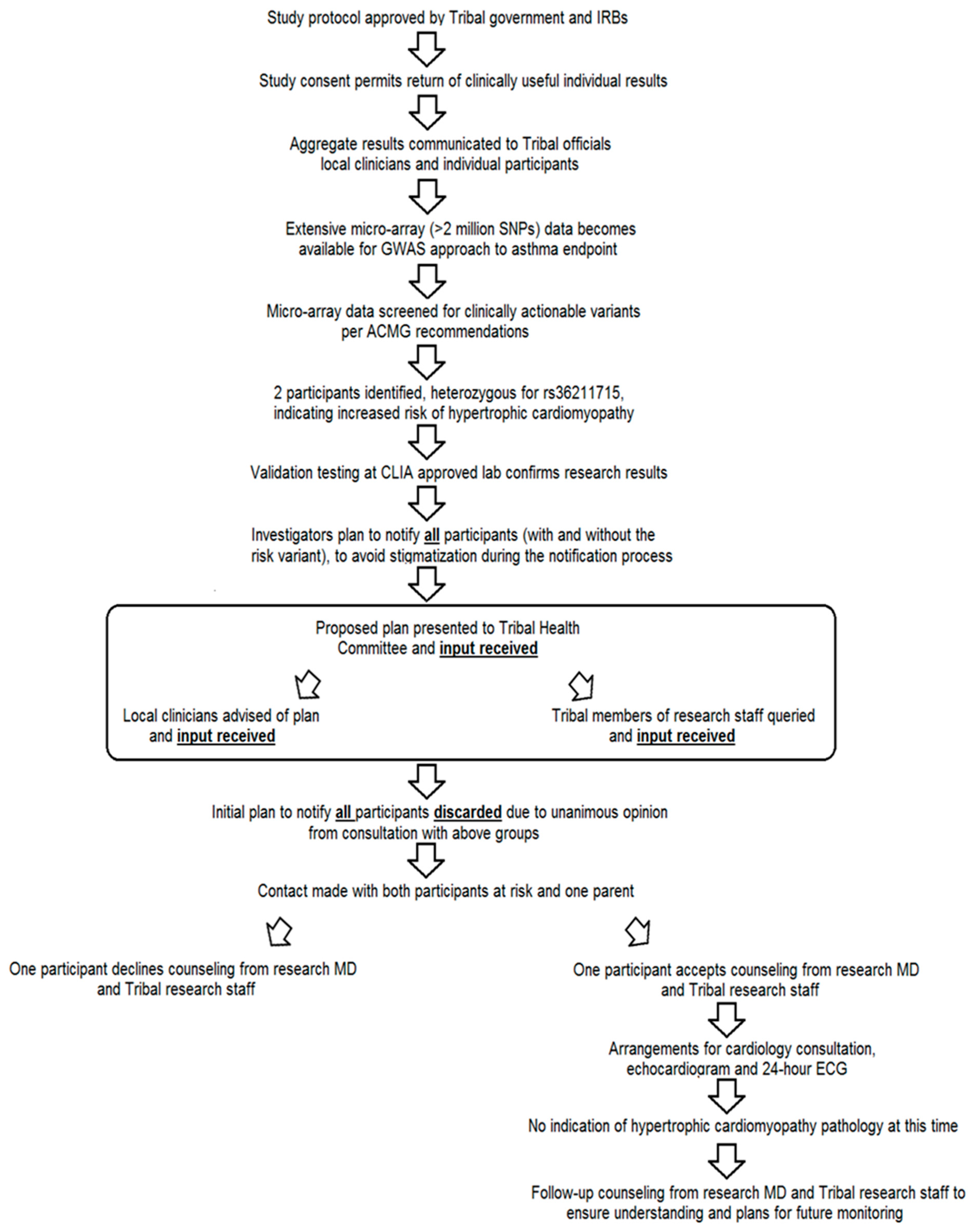
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Schupmann, W.; Miner, S.A.; Sullivan, H.K.; Glover, J.R.; Hall, J.E.; Schurman, S.H.; Berkman, B.E. Exploring the motivations of research participants who chose not to learn medically actionable secondary genetic findings about themselves. Genet. Med. 2021, 23, 2281–2288. [Google Scholar] [CrossRef]
- Renegar, G.; Webster, C.J.; Stuerzebecher, S.; Harty, L.; Ide, S.E.; Balkite, B.; Rogalski-Salter, T.A.; Cohen, D.; Spear, B.B.; Barnes, D.M.; et al. Returning genetic research results to individuals: Points-to-consider. Bioethics 2006, 20, 24–36. [Google Scholar] [CrossRef]
- Available online: https://allofus.nih.gov/news-events-and-media/announcements/nihs-all-us-research-program-returns-first-genetic-results-participants (accessed on 15 February 2022).
- Knoppers, B.M.; Joly, Y.; Simard, J.; Durocher, F. The emergence of an ethical duty to disclose genetic research results: International perspectives. Eur. J. Hum. Genet. 2006, 14, 1170–1178, Erratum in Eur. J. Hum. Genet. 2006, 14, 1322. [Google Scholar] [CrossRef]
- Lynch, J.A.; Sharp, R.R.; Aufox, S.A.; Bland, S.T.; Blout, C.; Bowen, D.J.; Buchanan, A.H.; Halverson, C.; Harr, M.; Hebbring, S.J.; et al. Understanding the Return of Genomic Sequencing Results Process: Content Review of Participant Summary Letters in the eMERGE Research Network. J. Pers. Med. 2020, 10, 38. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine. Returning Individual Research Results to Participants: Guidance for a New Research Paradigm; The National Academies Press: Cambridge, MA, USA, 2018. [Google Scholar] [CrossRef]
- Gottesman, O.; Kuivaniemi, H.; Tromp, G.; Faucett, W.A.; Li, R.; Manolio, T.A.; Sanderson, S.C.; Kannry, J.; Zinberg, R.; Basford, M.A.; et al. The Electronic Medical Records and Genomics (eMERGE) Network: Past, present, and future. Genet. Med. 2013, 15, 761–771. [Google Scholar] [CrossRef]
- Best, L.G.; Azure, C.; Segarra, A.; Enright, K.J.; Hamley, S.; Jerome, D.; O’Leary, M.A.; O’Leary, R.A.; Parisien, A.; Trottier, K.; et al. Genetic variants and risk of asthma in an American Indian population. Ann. Allergy Asthma Immunol. 2017, 119, 31–36.e1. [Google Scholar] [CrossRef]
- Kalia, S.S.; Adelman, K.; Bale, S.J.; Chung, W.K.; Eng, C.; Evans, J.P.; Herman, G.E.; Hufnagel, S.B.; Klein, T.E.; Korf, B.R.; et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017, 19, 249–255, Erratum in Genet Med. 2017, 19, 484. [Google Scholar] [CrossRef]
- Available online: https://apps.bea.gov/iTable/index_nipa.cfm (accessed on 30 January 2022).
- Available online: https://www.census.gov/quickfacts/fact/table/ziebachcountysouthdakota,corsoncountysouthdakota (accessed on 30 January 2022).
- Available online: https://www.ncbi.nlm.nih.gov/clinvar/docs/acmg/ (accessed on 30 January 2022).
- Available online: https://www.fda.gov/media/99200/download (accessed on 30 January 2022).
- Available online: https://www.ncbi.nlm.nih.gov/snp/rs36211715?horizontal_tab=true#publications (accessed on 30 January 2022).
- Tanjore, R.R.; Sikindlapuram, A.D.; Calambur, N.; Thakkar, B.; Kerkar, P.; Nallari, P. Genotype-phenotype correlation of R870H mutation in hypertrophic cardiomyopathy. Clin. Genet. 2006, 69, 434–436. [Google Scholar] [CrossRef]
- Erdmann, J.; Daehmlow, S.; Wischke, S.; Senyuva, M.; Werner, U.; Raible, J.; Tanis, N.; Dyachenko, S.; Hummel, M.; Hetzer, R.; et al. Mutation spectrum in a large cohort of unrelated consecutive patients with hypertrophic cardiomyopathy. Clin. Genet. 2003, 64, 339–349. [Google Scholar] [CrossRef]
- Fananapazir, L.; Dalakas, M.C.; Cyran, F.; Cohn, G.; Epstein, N.D. Missense mutations in the beta-myosin heavy-chain gene cause central core disease in hypertrophic cardiomyopathy. Proc. Natl. Acad. Sci. USA 1993, 90, 3993–3997. [Google Scholar] [CrossRef]
- Nishi, H.; Kimura, A.; Harada, H.; Koga, Y.; Adachi, K.; Matsuyama, K.; Koyanagi, T.; Yasunaga, S.; Imaizumi, T.; Toshima, H.; et al. A myosin missense mutation, not a null allele, causes familial hypertrophic cardiomyopathy. Circulation 1995, 91, 2911–2915. [Google Scholar] [CrossRef] [PubMed]
- Bashyam, M.D.; Savithri, G.R.; Gopikrishna, M.; Narasimhan, C. A p.R870H mutation in the beta-cardiac myosin heavy chain 7 gene causes familial hypertrophic cardiomyopathy in several members of an Indian family. Can. J. Cardiol. 2007, 23, 788–790. [Google Scholar] [CrossRef] [PubMed]
- Fabsitz, R.R.; McGuire, A.; Sharp, R.R.; Puggal, M.A.; Beskow, L.M.; Biesecker, L.G.; Bookman, E.; Burke, W.; Burchard, E.G.; Church, G.; et al. Ethical and practical guidelines for reporting genetic research results to study participants: Updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ. Cardiovasc. Genet. 2010, 3, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Beskow, L.M.; Burke, W. Offering individual genetic research results: Context matters. Sci. Transl. Med. 2010, 2, 38cm20. [Google Scholar] [CrossRef] [PubMed]
- Arar, N.; Seo, J.; Lee, S.; Abboud, H.E.; Copeland, L.A.; Noel, P.; Parchman, M. Preferences regarding genetic research results: Comparing veterans and nonveterans responses. Public Health Genom. 2010, 13, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Carey, D.J.; Fetterolf, S.N.; Davis, F.D.; Faucett, W.A.; Kirchner, H.L.; Mirshahi, U.; Murray, M.F.; Smelser, D.T.; Gerhard, G.S.; Ledbetter, D.H. The Geisinger MyCode community health initiative: An electronic health record-linked biobank for precision medicine research. Genet. Med. 2016, 18, 906–913. [Google Scholar] [CrossRef]
- Kullo, I.J.; Olson, J.; Fan, X.; Jose, M.; Safarova, M.; Breitkopf, C.R.; Winkler, E.; Kochan, D.C.; Snipes, S.; Pacyna, J.E.; et al. The Return of Actionable Variants Empirical (RAVE) Study, a Mayo Clinic Genomic Medicine Implementation Study: Design and Initial Results. Mayo Clin. Proc. 2018, 93, 1600–1610. [Google Scholar] [CrossRef]
- Middleton, A.; Wright, C.F.; Morley, K.I.; Bragin, E.; Firth, H.V.; Hurles, M.E.; Parker, M. Potential research participants support the return of raw sequence data. J. Med. Genet. 2015, 52, 571–574. [Google Scholar] [CrossRef]
- Haga, S.B.; Zhao, J.Q. Stakeholder views on returning research results. Adv. Genet. 2013, 84, 41–81. [Google Scholar]
- Best, L.G.; University of North Dakota, Grand Forks, ND, USA. Personal communication, 2022.
- Long, C.R.; Purvis, R.S.; Flood-Grady, E.; Kimminau, K.S.; Rhyne, R.L.; Burge, M.R.; Stewart, M.K.; Jenkins, A.J.; James, L.P.; McElfish, P.A. Health researchers’ experiences, perceptions and barriers related to sharing study results with participants. Health Res. Policy Syst. 2019, 17, 25. [Google Scholar] [CrossRef]
- McElfish, P.A.; Purvis, R.S.; Long, C.R. Researchers’ experiences with and perceptions of returning results to participants: Study protocol. Contemp. Clin. Trials Commun. 2018, 11, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Shalowitz, D.I.; Miller, F.G. Communicating the results of clinical research to participants: Attitudes, practices, and future directions. PLoS Med. 2008, 5, e91. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.M.; Evans, B.J. Return of results and data to study participants. Science 2018, 362, 159–160. [Google Scholar] [CrossRef] [PubMed]
- Evans, B.J. Minimizing liability risks under the ACMG recommendations for reporting incidental findings in clinical exome and genome sequencing. Genet. Med. 2013, 15, 915–920. [Google Scholar] [CrossRef]
- Clayton, E.W.; McCullough, L.B.; Biesecker, L.G.; Joffe, S.; Ross, L.F.; Wolf, S.M.; Explora, F.T.C.S. Clinical Sequencing Exploratory Research (CSER) Consortium Pediatrics Working Group. Addressing the ethical challenges in genetic testing and sequencing of children. Am. J. Bioeth. 2014, 14, 3–9. [Google Scholar] [CrossRef]
- Green, R.C.; Berg, J.S.; Grody, W.W.; Kalia, S.S.; Korf, B.R.; Martin, C.L.; McGuire, A.L.; Nussbaum, R.L.; O’Daniel, J.M.; Ormond, K.E.; et al. American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013, 15, 565–574, Erratum in Genet Med. 2017, 19, 606. [Google Scholar] [CrossRef]
- Miller, D.T.; Lee, K.; Abul-Husn, N.S.; Amendola, L.M.; Brothers, K.; Chung, W.K.; Gollob, M.H.; Gordon, A.S.; Harrison, S.M.; Hershberger, R.E.; et al. Electronic address: Documents@acmg.net. ACMG SF v3.1 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2022, 24, 1407–1414. [Google Scholar] [CrossRef]
- Available online: https://healthynv.org/about/ (accessed on 15 February 2022).
- Medical Research Council (MRC). Human Tissue and Biological Samples for Use in Research—Operational and Ethical Guidelines; Medical Research Council: London, UK, 2001. Available online: http://www.mrc.ac.uk/pdf-tissue_guide_fin.pdf (accessed on 21 March 2022).
- Laurie, G. Genetic databases: Assessing the benefits and the impact on human and patient rights--a WHO report. Eur. J. Health Law. 2004, 11, 87–92. [Google Scholar] [CrossRef]
- Council for International Organizations of Medical Sciences. International ethical guidelines for biomedical research involving human subjects. Bull. Med. Ethics 2002, 182, 17–23. [Google Scholar]
- Weiner, C. Anticipate and communicate: Ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts (December 2013 report of the Presidential Commission for the Study of Bioethical Issues). Am. J. Epidemiol. 2014, 180, 562–564. [Google Scholar] [CrossRef]
- Gliwa, C.; Yurkiewicz, I.R.; Lehmann, L.S.; Hull, S.C.; Jones, N.; Berkman, B.E. Institutional review board perspectives on obligations to disclose genetic incidental findings to research participants. Genet. Med. 2016, 18, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.C.F.; Knoppers, B.M.; Green, R.C. An international policy on returning genomic research results. Genome Med. 2021, 13, 115. [Google Scholar] [CrossRef] [PubMed]
- Miller, F.G.; Mello, M.M.; Joffe, S. Incidental findings in human subjects research: What do investigators owe research participants? J. Law. Med. Ethics 2008, 36, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.M.; Annas, G.J.; Elias, S. Point-counterpoint. Patient autonomy and incidental findings in clinical genomics. Science 2013, 340, 1049–1050. [Google Scholar] [CrossRef]
- Raymond, M.B.; Cooper, K.E.; Parker, L.S.; Bonham, V.L. Practices and Attitudes toward Returning Genomic Research Results to Low-Resource Research Participants. Public Health Genom. 2021, 24, 241–252. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Best, L.G.; O’Leary, M.; O’Leary, R.; Lawrence, W.; Torgerson, D.G. Return of Participants’ Incidental Genetic Research Findings: Experience from a Case-Control Study of Asthma in an American Indian Community. J. Pers. Med. 2023, 13, 1407. https://doi.org/10.3390/jpm13091407
Best LG, O’Leary M, O’Leary R, Lawrence W, Torgerson DG. Return of Participants’ Incidental Genetic Research Findings: Experience from a Case-Control Study of Asthma in an American Indian Community. Journal of Personalized Medicine. 2023; 13(9):1407. https://doi.org/10.3390/jpm13091407
Chicago/Turabian StyleBest, Lyle G., Marcia O’Leary, Rae O’Leary, Wendy Lawrence, and Dara G. Torgerson. 2023. "Return of Participants’ Incidental Genetic Research Findings: Experience from a Case-Control Study of Asthma in an American Indian Community" Journal of Personalized Medicine 13, no. 9: 1407. https://doi.org/10.3390/jpm13091407
APA StyleBest, L. G., O’Leary, M., O’Leary, R., Lawrence, W., & Torgerson, D. G. (2023). Return of Participants’ Incidental Genetic Research Findings: Experience from a Case-Control Study of Asthma in an American Indian Community. Journal of Personalized Medicine, 13(9), 1407. https://doi.org/10.3390/jpm13091407






