1. Introduction
Medical image analysis is the initial step in medical image processing, which makes images more understandable and increases diagnostic effectiveness [
1]. Medical image segmentation is an essential and critical step in the field of biomedical image processing, and it has significantly improved the sustainability of medical care [
2]. It currently has a substantial study direction in the area of computer vision [
1]. In some medical applications, it is desired to classify image pixels into distinct regions, such as bones and blood vessels. In other applications, searching for pathological regions, such as cancer or tissue deformities, is more applicable [
3]. Additionally, to provide essential details regarding the size and volumes of body organs, a crucial task in medical image segmentation is identifying and removing redundant pixels or undesired background regions.
Several traditional machine learning and image processing approaches were used on the histogram characteristics [
4], including region cut-based segmentation [
5] and the segmentation method based on edge and region [
6]. At present, image segmentation techniques are progressing more quickly and precisely. However, segmentation approaches that utilize deep learning (DL) have become very popular in recent years in the field of medical image segmentation, localization, and detection [
2]. Deep learning techniques provide several benefits over traditional machine learning and computer vision techniques regarding segmentation accuracy and speed.
Deep learning approaches are known as universal learning approaches that allow a single model to be efficiently used in a variety of medical imaging modalities, such as magnetic resonance imaging (MRI), computed tomography (CT), and X-ray. According to [
2,
7], most publications have been published on segmentation tasks in various medical imaging modalities. With the rapid development of deep learning models, deep convolutional neural networks (DCNNs) [
8] have attained great success in a broad range of computer vision, such as image classification and image segmentation, and they provide state-of-the-art performances in medical image segmentation [
9]. CNNs have robust methodologies that allow image segmentation to be treated as semantic segmentation, which refers to the ability to understand an image at the pixel level by assigning a class label to each pixel of the image [
10]. In [
11], the researchers introduced a fully convolutional network (FCN), which is the foundation of most contemporary techniques for semantic segmentation. For biomedical image segmentation, the researchers suggested an encoder–decoder network of fully convolutional networks called U-Net [
12]. Due to U-Net architecture’s excellent performance, since it was proposed in 2015, many scientists have adopted it as the backbone of their models. At present, U-Net is a popular network for biomedical image segmentation, and many variants of it exist, including Unet++ [
13], AttentionU-Net [
14] and MultiResUNet [
15]. The U-Net model structure consists of two paths: contracting and expansive. The former is the contracting path, also known as the encoder or down-sampling; it is similar to a regular convolution network and gives the classification information. The latter is the expansive path, also known as the decoder or up-sampling, which contains up-convolutions and concatenations, which enable the network to learn localized classification information [
16]. The concatenation in U-Net architecture, called skip connection [
17], combines the information of the two paths in the U-Net architecture.
These models are very sophisticated and demand more GPU power, which makes them computationally expensive and requires substantial expertise. Furthermore, these neural architectures are manually designed and have a propensity for having many parameters, making them prone to overfitting when utilizing insufficient training data, and they generally have a computational complexity [
18]. Therefore, it is not trivial to manually design the architecture of these models and select their appropriate hyperparameters based on past design experience.
In this study, we propose an automated design method for a state-of-the-art convolutional neural network, called GA-UNet, which is based on the genetic algorithm (GA). This is a metaheuristic method that generates a CNN architecture with a U-shape and automatically selects its hyperparameters that can achieve competitive performance with much shorter architecture compared with the original U-Net [
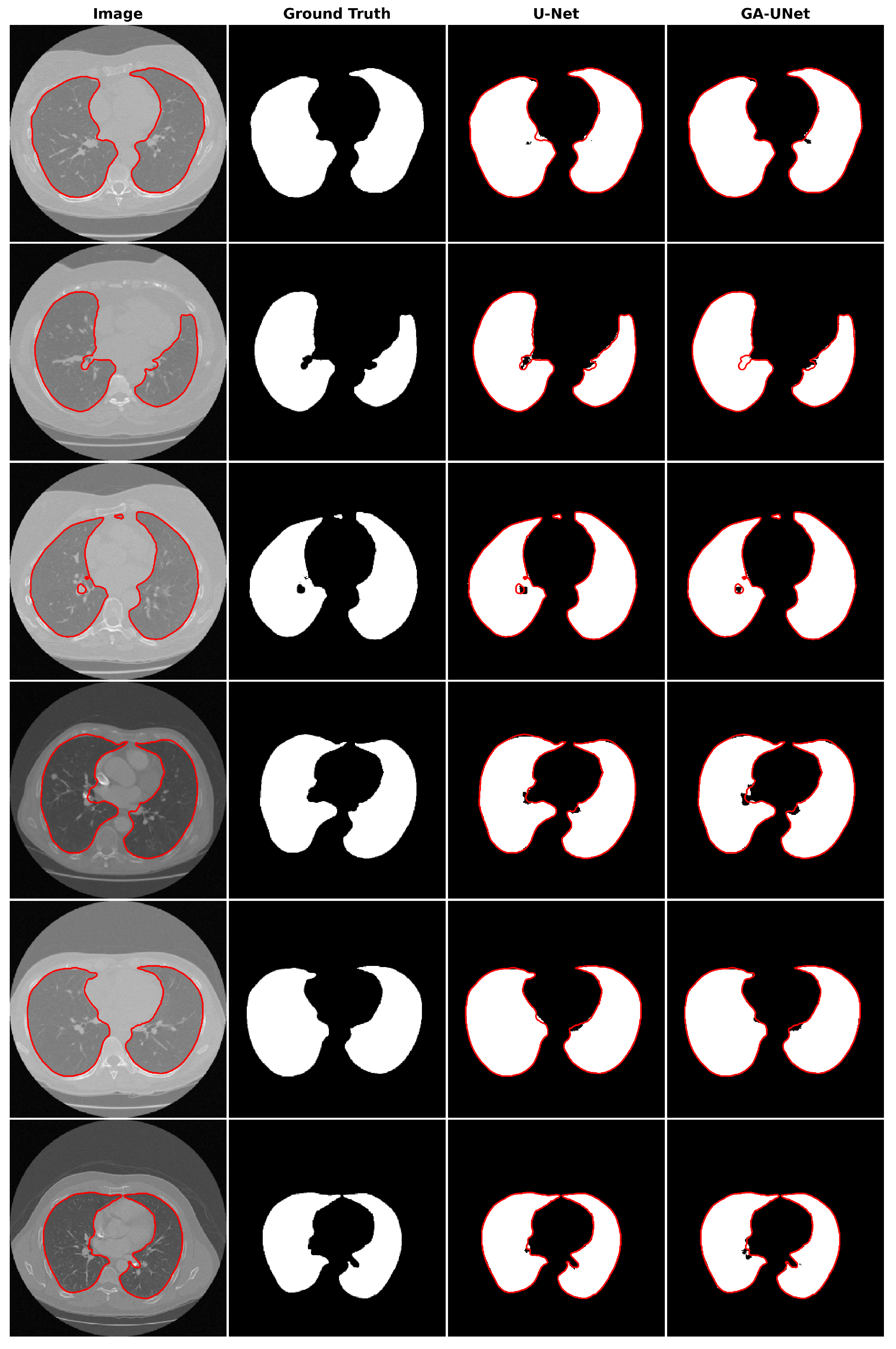
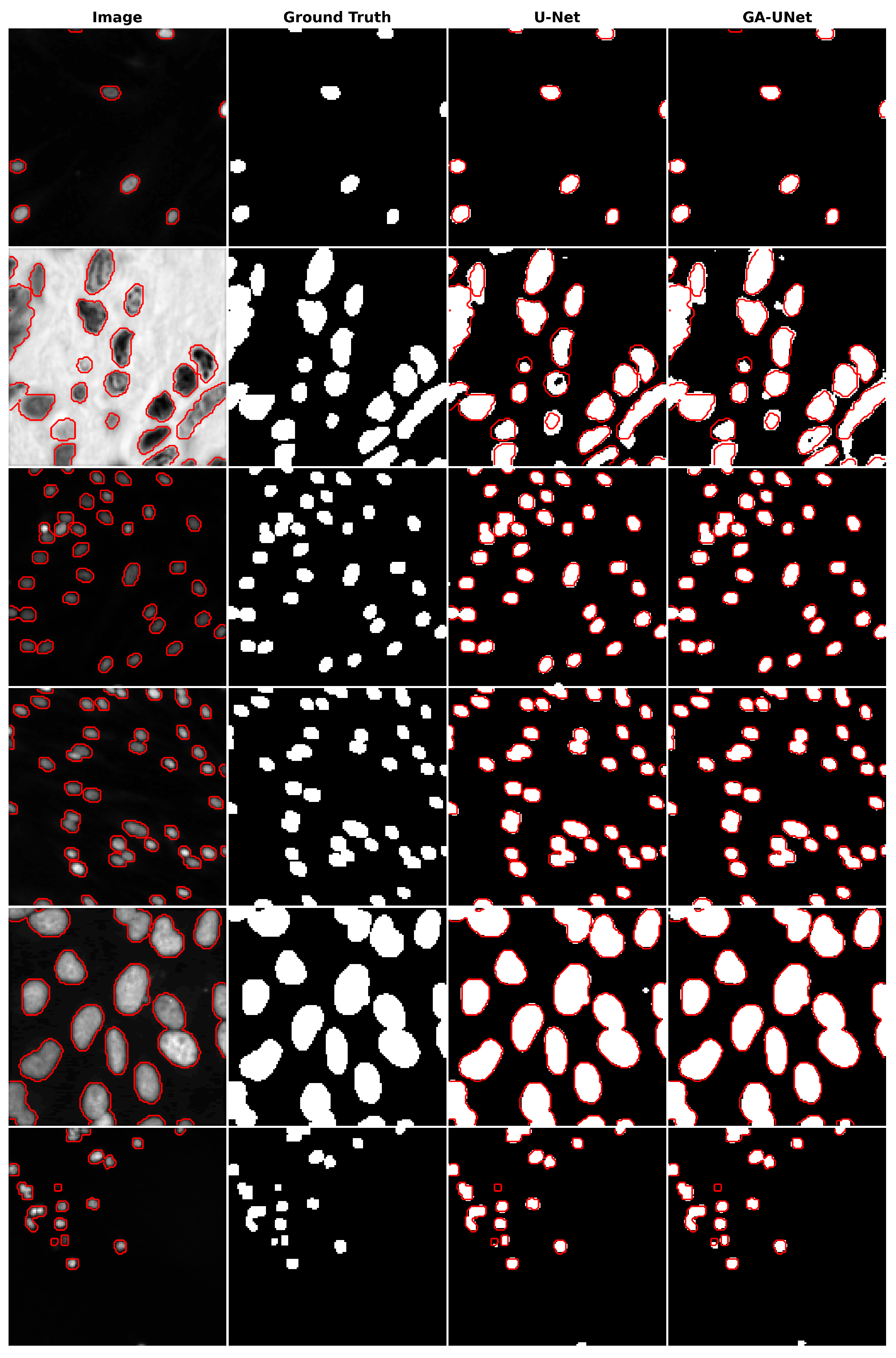
12] architecture, which was manually designed. The proposed GA-UNet starts by designing a search space and then automates the design for network architecture using genetic algorithm operators, including (crossover, mutation, and selection), searching for a competitive architecture with few parameters. To validate the effectiveness of GA-UNet, we have conducted evaluations on three distinct image segmentation datasets: the lung segmentation dataset (lung segmentation) [
19], the nuclei segmentation in a microscope images dataset (DSB 2018) [
20], and the liver segmentation dataset (liver segmentation) [
21]. Our experimental results show that the architectures proposed by the genetic algorithm, GA-UNet, succeed in reaching a U-Net architecture not only with a competitive performance against the manually designed U-Net [
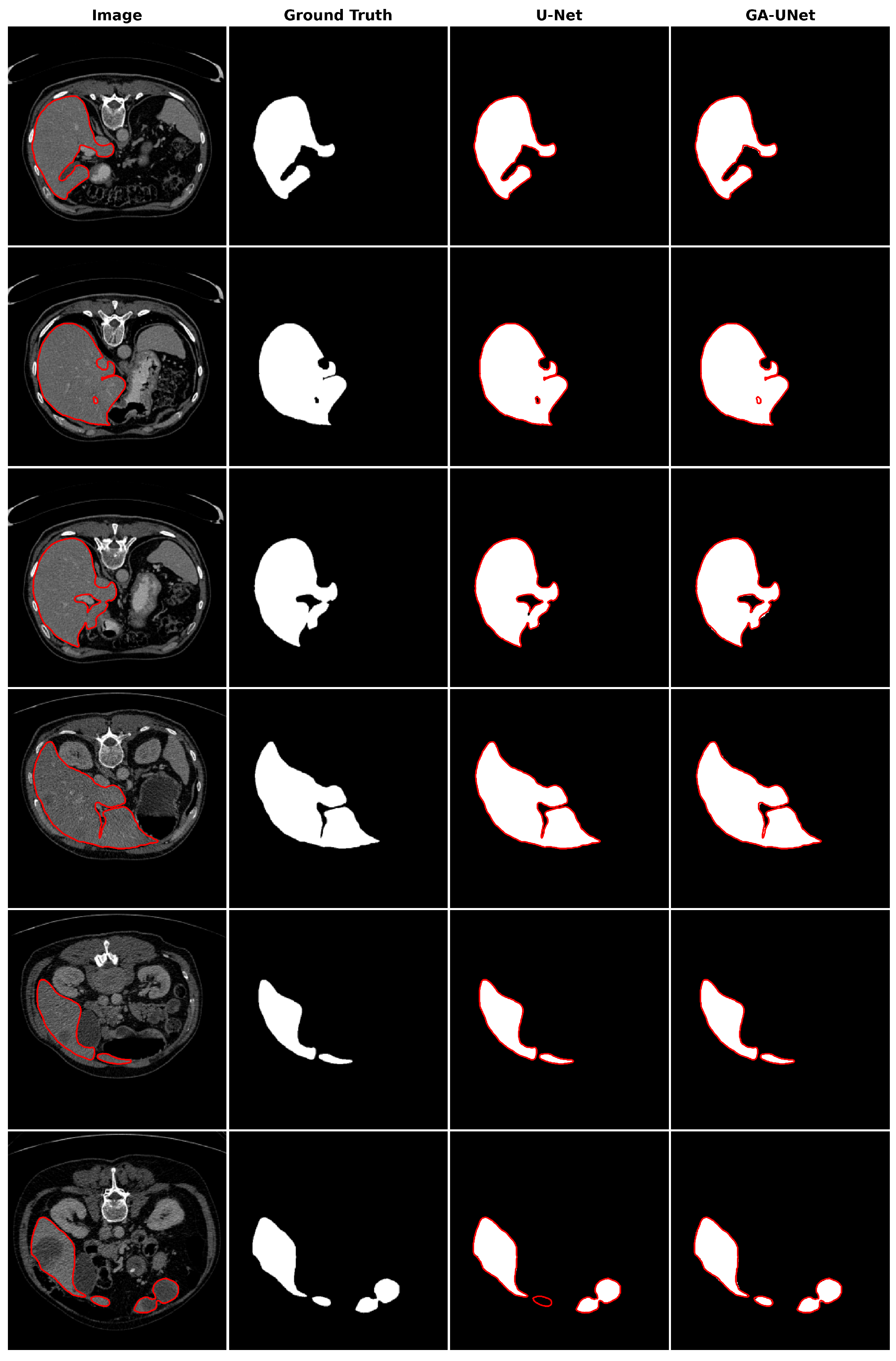
12], but they are even able to find a much shorter CNN architecture (0.24%, 0.48%, and 0.67% of the number of parameters in the original U-Net architecture for the lung image segmentation dataset, the DSB 2018 dataset, and the liver image segmentation dataset, respectively), which means reducing the computational effort without affecting their performance.
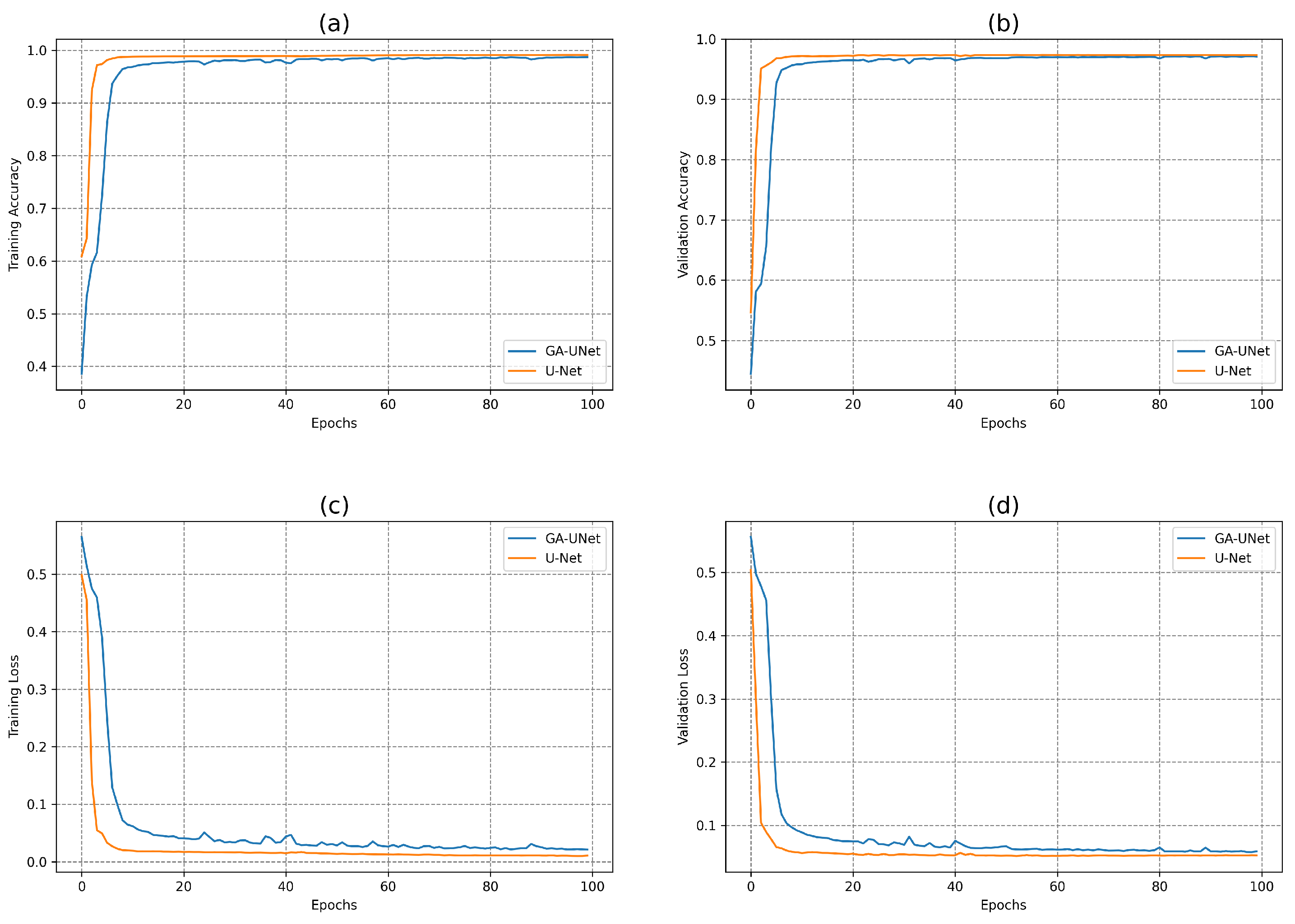
We also evaluated the performance of our proposed GA-UNet method for lung segmentation in comparison to several existing CNN-based techniques, such as [
22,
23,
24,
25]. The experimental results illustrates the outperformance of our method against these methods using multiple metrics such as the Dice similarity coefficient, precision, recall, and accuracy. The results underline the effectiveness of our proposed architecture over current state-of-the-art methods.
The rest of this work can be summarized as follows:
Section 2 discusses the literature review. The proposed methodology is proposed in
Section 3.
Section 4 describes the materials of the experiments. The results are described in
Section 5. Then, conclusions and limitations are drawn in
Section 6. Finally, in
Section 7, we discuss our future works.
3. Methodology
In this section, the details of the proposed GA-UNet method for lung segmentation [
19], the DSB 2018 dataset [
20], and liver segmentation [
21] will be provided. We first describe the decoding strategy, define a specific search space and then realize the automated design for network architectures using genetic algorithm operations including crossover, mutation, and selection, searching for competitive architectures.
3.1. Search Space Encoding
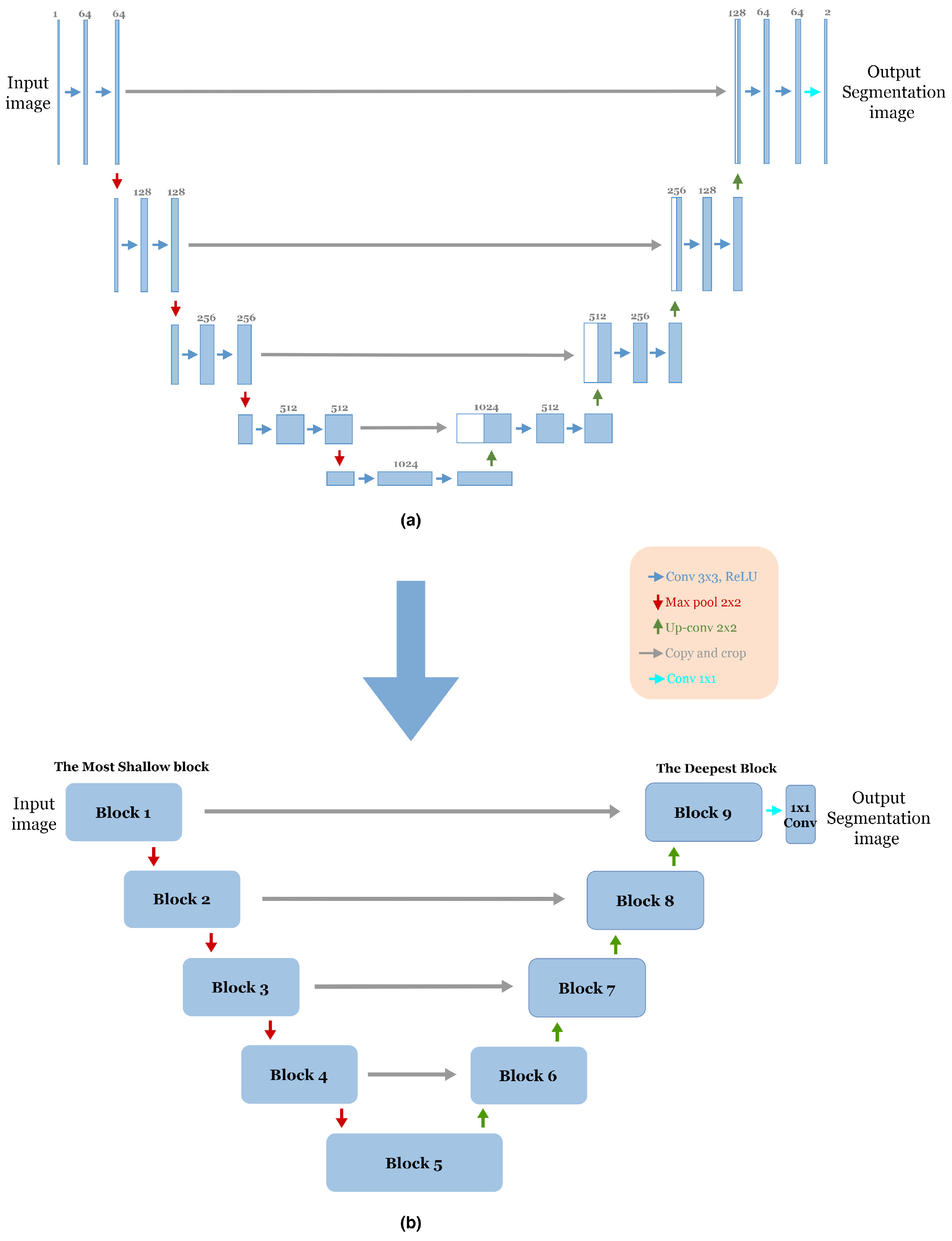
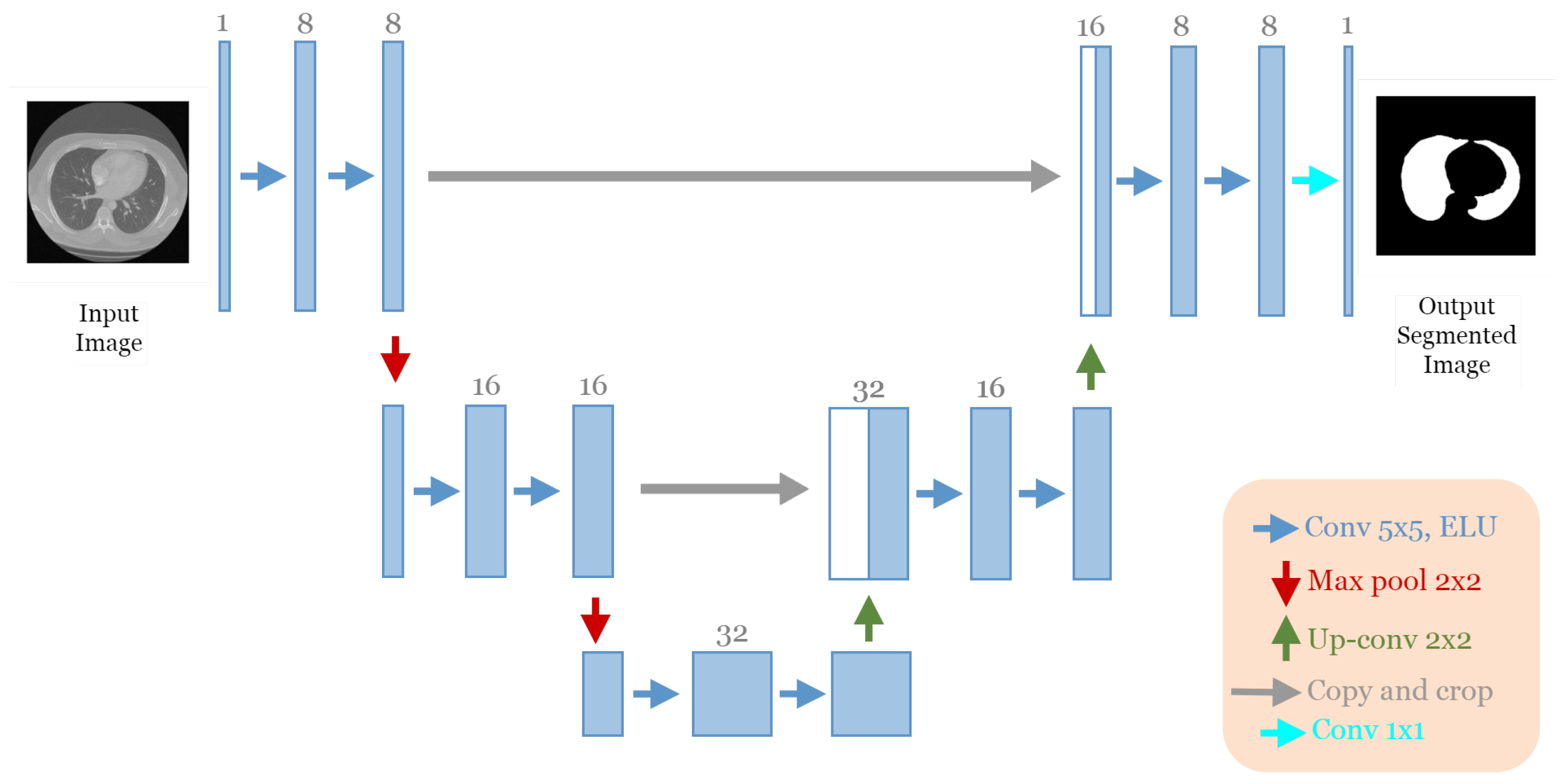
To reduce the complexity of U-Net architecture, we adopted a block structure as shown in
Figure 1b. We regrouped into one block all layers between two successive maxpooling or up-sampling layers. Therefore, each block
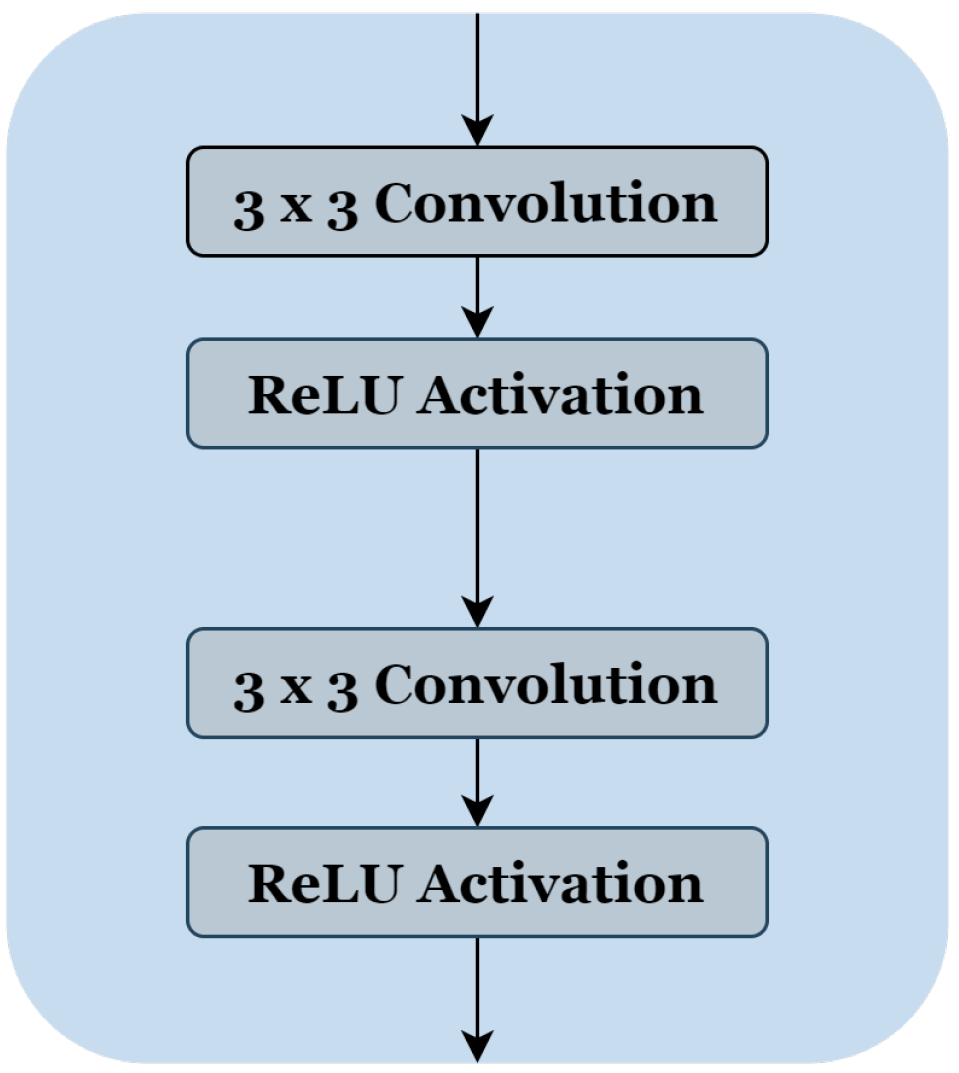
Figure 2 consists of two 3 × 3 convolution layers with a Rectified Linear Unit (ReLU) activation function. The number of blocks is similar in the down-sampling path and the up-sampling path, with one single block in the bottleneck.
In this work, we will realize an automated design for network architecture by adjusting the number of blocks, their internal structure, and other parameters, using the genetic algorithm to find a satisfactory architecture. The first objective is to select the most relevant parameters that influence the performance of the discovered architecture. The following parameters are chosen to be optimized in this work:
- (1)
Number of blocks ‘B’;
- (2)
Number of filters ‘C’;
- (3)
Filter size ‘F’;
- (4)
Activation ‘A’;
- (5)
Pooling ‘P’;
- (6)
Batch-normalization ‘BN’;
- (7)
Dropout ‘D’;
- (8)
Optimizer ‘O’;
- (9)
Learning rate ‘LR’;
- (10)
Batch size ‘BA’.
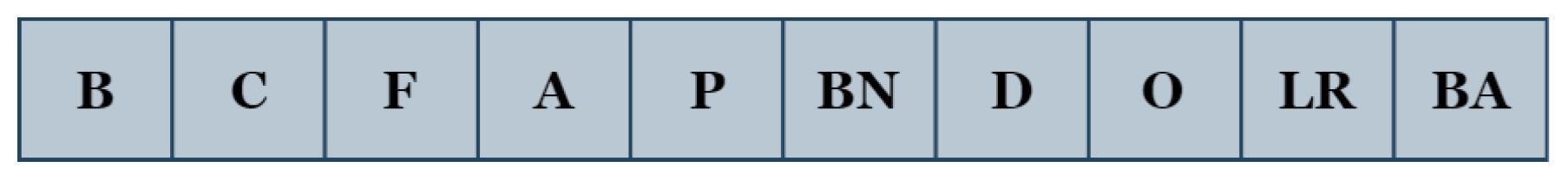
We develop a simple yet effective encoding approach in which practically all the aforementioned parameters (B, C, F, A, P, BN, D, O, LR, and BA) are encoded as an integer vector. The block ‘B’ contains two operations: convolution and activation. The convolution operation has two parameters: the number of filters ‘C’ and the filter size ‘F’. The operations of activation, pooling, batch-normalization, and dropout are denoted by ‘A’, ‘P’, ‘BN’, and ‘D’, respectively. The optimizer, learning rate, and batch size are represented as ‘O’, ‘LR’, and ‘BA’.
When designing the search space, there are two main things to consider: making the search space concise but flexible, and limiting the number of architecture parameters.
Table 1 shows all the selected parameters, their codes, and range of values: (1) The number of blocks ‘B’ has a search space from (3 to 7), indicating the reproduced architecture can be with a minimum of three blocks to seven blocks as maximum. (2) The number of filters ‘C’ of the first block takes the range (2, 4, or 8 filters). (3) The value ranges of the filter size ‘F’ are from (3 to 7), where 3 represents the filter size (3 × 3), 4 represents (4 × 4), and so on, respectively, for each value. (4) The activation function ‘A’ takes the values ranges from (1, 2, or 3), representing ReLU [
26], ELU [
38], and LeakyReLU [
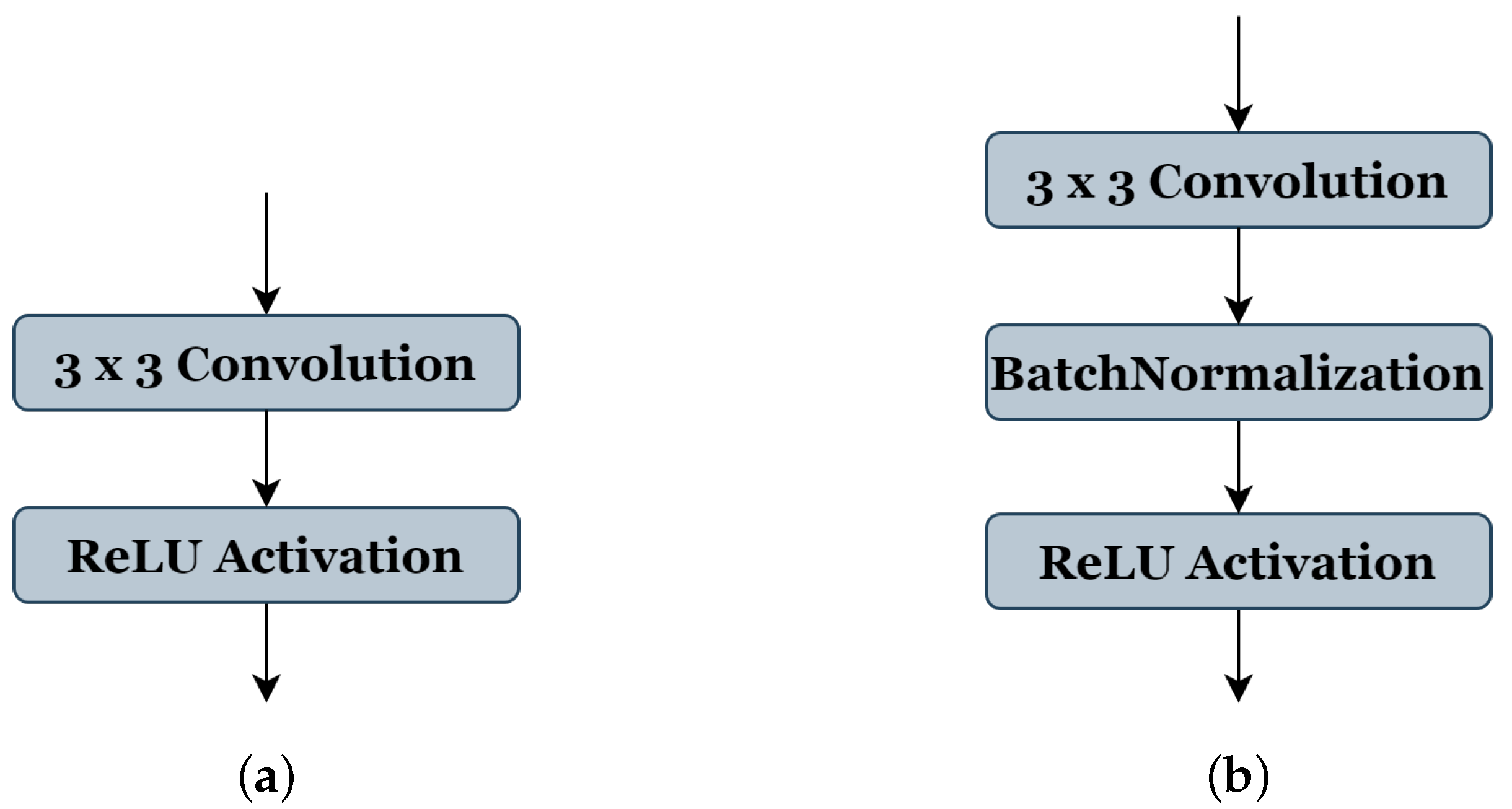
39], respectively. (5) The type of pooling operation ‘P’ could take a value of (1) representing ’Maxpooling’ or (2) representing ‘Averagepooling’, all using the same stride of 2 × 2. (6) The batch-normalization operation ‘BN’ accepts a value (0 or 1) to indicate whether this operation should be used (see
Figure 3b) or not (see
Figure 3a). (7) Similar to the ‘BN’ operation, the dropout operation ‘D’ takes the value (0) or (1) to express if this operation could be used or not; in this work, the probability of the random drop is (0.3). (8) The search space of optimizer type ‘O’ is from (0 to 4), representing the SGD [
40], RMSprop, Adam, and Adamax [
41], respectively. (9) The learning rate ‘LR’ takes a value between
and
. (10) The value range of batch size ‘BA’ is (4, 8, 16, or 32). During the genetic algorithm optimization, we use 20 epochs for the training phase of each candidate architecture.
The number of architecture-based parameters within this search space is significantly correlated by the number of blocks ‘B’, the number of filters ‘C’, and filter size ‘F’. Therefore, we have employed a range of values in our search space to confine the number of architecture-based parameters. Unlike the state-of-the-art medical image segmentation architectures, which may comprise millions of parameters, most of the network architectures within our search space have fewer than 0.6 million parameters.
With this coding scheme, the individuals of the genetic algorithm consist of 10 parameters (genes), whose structure is presented in
Figure 4.
3.2. Evolutionary Algorithm
Our proposed approach GA-UNet is an iterative evolutionary technique designed to produce a progressively evolving population. The population comprises individuals, each of whom represents a distinct architecture. Each individual’s fitness is determined by evaluating the performance of the corresponding architecture in specific applications.
Figure 5 and Algorithm 1 illustrate the proposed approach through a flowchart and pseudocode, respectively.
Algorithm 1 starts with a random initialization of a population
of L individuals. After, we perform the number of generations G; at each generation, three evolutionary operations are applied (crossover, mutation, and selection) in order to improve the fitness of individuals. Next, the newly generated individuals are evaluated by training their corresponding architectures on the provided dataset. The process is carried out iteratively until the desired number of generations (G) is attained.
| Algorithm 1: General framework of the proposed method |
![Jpm 13 01298 i001]() |
We begin by setting the population size L, the maximal number of generation G for the genetic algorithm, and K, the number of the best parent individuals. A population is initialized randomly with the predefined population size L, using the proposed encoding strategy (line 1). Then, we set a counter t for the current generation to zero (line 2); at each generation, we evaluate all the individuals of population by training their corresponding architectures on the given dataset (line 4). After that, we select the number of the best parents, K, based on their fitness and classify them according to their fitness values from high to low. Next, we applied a genetic operation such as crossover and mutation operators to generate a new offspring population (line 6). Then, using the environmental selection method, we select from the current population a population of individuals surviving into the next generation (line 7). This population is composed explicitly of the K selected best parent individuals and the generated offspring population . Finally, the counter t is incremented by one (line 8), and the same process will be repeated until the counter exceeds the maximal number of generation G.
3.2.1. Fitness Evaluation
To evaluate the fitness in this work, Algorithm 2 details the procedure for evaluating individuals in the population
. By giving a population
containing L individuals, training data
, and validation data
in the beginning, F is set as an empty vector to memorize the fitness of all individuals (line 1). Algorithm 2 evaluates all the L individuals in the same manner (line 2→6) and transforms each individual of the population
to its corresponding architecture. Then, we trained and validated the architecture on the training
and the validation data
, respectively. The performance of every architecture is evaluated, and its validation accuracy score
will be calculated; the fitness value F is equal to
. At last, Algorithm 2 returns the population
of L individuals with their fitness.
| Algorithm 2: Fitness Evaluation |
![Jpm 13 01298 i002]() |
3.2.2. Environmental Selection
Typically, genetic algorithms choose the next population by tournament or roulette selection. Both selection methods may miss the best individuals, which can lead to performance degradation [
32]. This can have a significant impact on the optimization results. In contrast, if we select
K parent individuals for the next generation, we have a higher probability of trapping the algorithm to a local optimum [
18]. Algorithm 3 shows the details of the environmental selection. Firstly, given the current population
, the
K best parent individuals of
were selected and the generated offspring population,
, become the next population
. Secondly, the
individuals are selected from the generated offspring population
and then placed into
. The next population
size is retained at the same as the current population
.
| Algorithm 3: Environmental Selection |
![Jpm 13 01298 i003]() |
3.2.3. Offspring Generating
In this work, we apply two genetic operators, crossover and mutation, to generate the offspring population
. Algorithm 4 shows the process of generating offspring population
, which contains two parts. The first is a crossover (lines 1 to 9), and the second is a mutation (lines 10 to 14). During the crossover part, the Algorithm 4 starts by selecting
K best fitness parent individuals in population
into
. After that, two successive parent individuals from
are selected based on the better fitness score (line 4), and they exchange information with each other. This exchange can increase the performance of the algorithm. Otherwise, each parent individual is divided into two parts; these parts are swapped to create two new offspring (lines 5 to 8). In the second part, each offspring generated
p experiences the mutation, and a random perturbation
r is added to the last point
i of the offspring
p. This operation can increase the diversity of the population and prevent the algorithm from being trapped at the local optimum.
| Algorithm 4: Offspring Generating |
![Jpm 13 01298 i004]() |
6. Conclusions
This paper proposes an automated architecture design approach based on the U-shaped encoder–decoder structure called GA-UNet. This approach utilizes the genetic algorithm (GA) to generate competitive architectures with a restricted number of parameters through a compact but dynamic search space. We evaluated the performance of GA-UNet on three datasets: lung segmentation, cell nuclei segmentation (DSB 2018), and liver segmentation datasets.
The experimental findings indicate that the discovered architectures generated by our proposed approach, GA-UNet, achieve competitive performance compared with the baseline model, such as U-Net [
12], with a considerable reduction in parameter numbers and computation complexity. It requires only 0.24%, 0.48% and 0.67% of the number of parameters present in the original U-Net architecture for the lung image segmentation dataset, the DSB 2018 dataset and the liver image segmentation dataset, respectively; this translates into a reduction in computational effort without compromising on performance, which makes our proposed methodology, GA-UNet, more suitable for deployment in resource-limited environments or real-world implementations. We further examined the performance of our proposed GA-UNet method for lung segmentation by contrasting it with several existing CNN-based techniques, such as those detailed in [
22,
23,
24,
25]. The empirical findings demonstrate, via multiple metrics, the outperformance of our approach in comparison to these techniques.
Despite these promising results, there are a few limitations to note. Firstly, the GA-UNet performance may depend on the initial parameters of the genetic algorithm, such as population size and number of generations, which we intend to adjust in future research. Secondly, the proposed search space design might not capture all potential architectural solutions, needing further investigation.