Pharmacomicrobiomics of Antidepressants in Depression: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
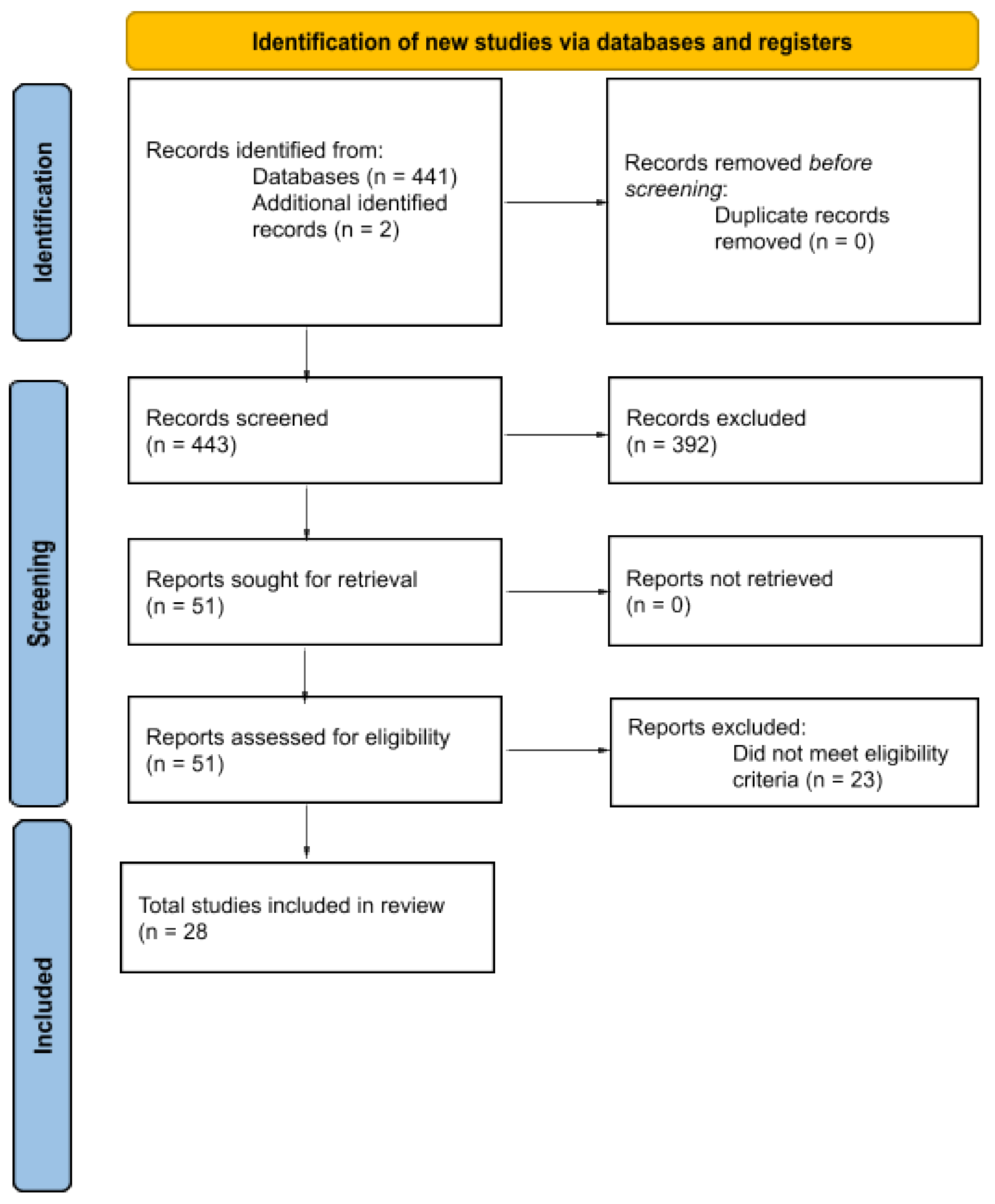
2.1. Information Sources and Search Strategy
2.2. Eligibility Criteria
2.3. Risk of Bias Assessment
2.4. Effect Measures and Synthesis Methods
3. Results
3.1. Animal Studies
3.2. Human Studies
3.3. Risk of Bias Assessment
4. Discussion
4.1. Limitations
4.2. Future Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carter, G.C.; Cantrell, R.A.; Victoria Zarotsky Haynes, V.S.; Phillips, G.; Alatorre, C.I.; Goetz, I.; Paczkowski, R.; Marangell, L.B. Comprehensive Review of Factors Implicated in the Heterogeneity of Response in Depression. Depress. Anxiety 2012, 29, 340–354. [Google Scholar] [CrossRef] [PubMed]
- Gaynes, B.N.; Warden, D.; Trivedi, M.H.; Wisniewski, S.R.; Fava, M.; Rush, A.J. What Did STAR*D Teach Us? Results from a Large-Scale, Practical, Clinical Trial for Patients with Depression. Psychiatry Serv. 2009, 60, 1439–1445. [Google Scholar] [CrossRef]
- Trevino, K.; McClintock, S.M.; Fischer, N.M.; Vora, A.; Husain, M.M. Defining treatment-resistant depression: A comprehensive review of the literature. Ann. Clin. Psychiatry Off. J. Am. Acad. Clin. Psychiatry 2014, 26, 222–232. [Google Scholar]
- Kessler, R.C. The potential of predictive analytics to provide clinical decision support in depression treatment planning. Curr. Opin. Psychiatry 2018, 31, 32–39. [Google Scholar] [CrossRef]
- Musliner, K.L.; Munk-Olsen, T.; Eaton, W.W.; Zandi, P.P. Heterogeneity in long-term trajectories of depressive symptoms: Patterns, predictors and outcomes. J. Affect. Disord. 2016, 192, 199–211. [Google Scholar] [CrossRef]
- Athreya, A.P.; Neavin, D.; Carrillo-Roa, T.; Skime, M.; Biernacka, J.; Frye, M.A.; Rush, A.J.; Wang, L.; Binder, E.B.; Iyer, R.K.; et al. Pharmacogenomics-Driven Prediction of Antidepressant Treatment Outcomes: A Machine-Learning Approach with Multi-trial Replication. Clin. Pharmacol. Ther. 2019, 106, 855–865. [Google Scholar] [CrossRef]
- Bobo, W.V.; Ommeren, B.V.; Athreya, A.P. Machine learning, pharmacogenomics, and clinical psychiatry: Predicting antidepressant response in patients with major depressive disorder. Expert. Rev. Clin. Phar. 2022, 15, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.C.; Stanton, J.D.; Bharthi, K.; Maruf, A.A.; Müller, D.J.; Bousman, C.A. Pharmacogenomic Testing and Depressive Symptom Remission: A Systematic Review and Meta-Analysis of Prospective, Controlled Clinical Trials. Clin. Pharmacol. Ther. 2022, 112, 1303–1317. [Google Scholar] [CrossRef]
- Weersma, R.K.; Zhernakova, A.; Fu, J. Interaction between drugs and the gut microbiome. Gut 2020, 69, 1510–1519. [Google Scholar] [CrossRef]
- Weiss, G.A.; Hennet, T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol. Life Sci. 2017, 74, 2959–2977. [Google Scholar] [CrossRef]
- Liang, S.; Wu, X.; Hu, X.; Wang, T.; Jin, F. Recognizing Depression from the Microbiota–Gut–Brain Axis. Int. J. Mol. Sci. 2018, 19, 1592. [Google Scholar] [CrossRef] [PubMed]
- Sanada, K.; Nakajima, S.; Kurokawa, S.; Barceló-Soler, A.; Ikuse, D.; Hirata, A.; Yoshizawa, A.; Tomizawa, Y.; Salas-Valero, M.; Noda, Y.; et al. Gut microbiota and major depressive disorder: A systematic review and meta-analysis. J. Affect. Disord. 2020, 266, 1–13. [Google Scholar] [CrossRef]
- Peirce, J.M.; Alviña, K. The role of inflammation and the gut microbiome in depression and anxiety. J. Neurosci. Res. 2019, 97, 1223–1241. [Google Scholar] [CrossRef]
- Simpson, C.A.; Diaz-Arteche, C.; Eliby, D.; Schwartz, O.S.; Simmons, J.G.; Cowan, C.S.M. The gut microbiota in anxiety and depression—A systematic review. Clin. Psychol. Rev. 2021, 83, 101943. [Google Scholar] [CrossRef]
- Madan, A.; Thompson, D.; Fowler, J.C.; Ajami, N.; Salas, R.; Frueh, B.; Bradshaw, M.; Weinstein, B.; Oldham, J.; Petrosino, J. The gut microbiota is associated with psychiatric symptom severity and treatment outcome among individuals with serious mental illness. J. Affect. Disord. 2020, 264, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.R.; Borre, Y.; O’Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatry Res. 2016, 82, 109–118. [Google Scholar] [CrossRef]
- Hao, W.Z.; Li, X.J.; Zhang, P.W.; Chen, J.X. A review of antibiotics, depression, and the gut microbiome. Psychiatry Res. 2020, 284, 112691. [Google Scholar] [CrossRef]
- Fan, X.; Deng, H.; Qiu, J.; Ji, H.; Shen, X. Antibiotics-induced depression in mice via the microbiota-gut-brain axis. J. Affect. Disord. 2022, 318, 152–158. [Google Scholar] [CrossRef]
- Misera, A.; Łoniewski, I.; Palma, J.; Kulaszyńska, M.; Czarnecka, W.; Kaczmarczyk, M.; Liśkiewicz, P.; Samochowiec, J.; Skonieczna-Żydecka, K. Clinical significance of microbiota changes under the influence of psychotropic drugs. An updated narrative review. Front. Microbiol. 2023, 14, 1125022. [Google Scholar] [CrossRef] [PubMed]
- Chait, Y.A.; Mottawea, W.; Tompkins, T.A.; Hammami, R. Unravelling the antimicrobial action of antidepressants on gut commensal microbes. Sci. Rep. 2020, 10, 17878. [Google Scholar] [CrossRef]
- Hua, H.; Huang, C.; Liu, H.; Xu, X.; Xu, X.; Wu, Z.; Liu, C.; Wang, Y.; Yang, C. Depression and antidepressant effects of ketamine and its metabolites: The pivotal role of gut microbiota. Neuropharmacology 2022, 220, 109272. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef]
- Zhu, H.-Z.; Liang, Y.-D.; Ma, Q.-Y.; Hao, W.-Z.; Li, X.-J.; Wu, M.-S.; Deng, L.-J.; Li, Y.-M.; Chen, J.-X. Xiaoyaosan improves depressive-like behavior in rats with chronic immobilization stress through modulation of the gut microbiota. Biomed. Pharmacother. 2019, 112, 108621. [Google Scholar] [CrossRef]
- Klünemann, M.; Andrejev, S.; Blasche, S.; Mateus, A.; Phapale, P.; Devendran, S.; Vappiani, J.; Simon, B.; Scott, T.A.; Kafkia, E.; et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 2021, 597, 533–538. [Google Scholar] [CrossRef]
- Lukić, I.; Getselter, D.; Ziv, O.; Oron, O.; Reuveni, E.; Koren, O.; Elliott, E. Antidepressants affect gut microbiota and Ruminococcus flavefaciens is able to abolish their effects on depressive-like behavior. Transl. Psychiatry 2019, 9, 133. [Google Scholar] [CrossRef]
- Dethloff, F.; Vargas, F.; Elijah, E.; Quinn, R.; Park, D.I.; Herzog, D.P.; Müller, M.B.; Gentry, E.C.; Knight, R.; Gonzalez, A.; et al. Paroxetine Administration Affects Microbiota and Bile Acid Levels in Mice. Front. Psychiatry 2020, 11, 518. [Google Scholar] [CrossRef]
- Kim, J.K.; Han, S.K.; Joo, M.K.; Kim, D.H. Buspirone alleviates anxiety, depression, and colitis; and modulates gut microbiota in mice. Sci. Rep. 2021, 11, 6094. [Google Scholar] [CrossRef]
- Lyte, M.; Daniels, K.M.; Schmitz-Esser, S. Fluoxetine-induced alteration of murine gut microbial community structure: Evidence for a microbial endocrinology-based mechanism of action responsible for fluoxetine-induced side effects. PeerJ 2019, 7, e6199. [Google Scholar] [CrossRef]
- Ramsteijn, A.S.; Jašarević, E.; Houwing, D.J.; Bale, T.L.; Olivier, J.D. Antidepressant treatment with fluoxetine during pregnancy and lactation modulates the gut microbiome and metabolome in a rat model relevant to depression. Gut Microbes 2020, 11, 735–753. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, H.; Cao, Y.; Wang, C.; Zhao, C.; Wang, H.; Cui, G.; Wang, M.; Pan, Y.; Shi, Y.; et al. Fluoxetine ameliorates dysbiosis in a depression model induced by chronic unpredicted mild stress in mice. Int. J. Med. Sci. 2019, 16, 1260–1270. [Google Scholar] [CrossRef]
- Zhang, W.; Qu, W.; Wang, H.; Yan, H. Antidepressants fluoxetine and amitriptyline induce alterations in intestinal microbiota and gut microbiome function in rats exposed to chronic unpredictable mild stress. Transl. Psychiatry 2021, 11, 131. [Google Scholar] [CrossRef] [PubMed]
- Siopi, E.; Chevalier, G.; Katsimpardi, L.; Saha, S.; Bigot, M.; Moigneu, C.; Eberl, G.; Lledo, P.-M. Changes in Gut Microbiota by Chronic Stress Impair the Efficacy of Fluoxetine. Cell Rep. 2020, 30, 3682–3690.e6. [Google Scholar] [CrossRef]
- Vuong, H.E.; Coley, E.J.; Kazantsev, M.; Cooke, M.E.; Rendon, T.K.; Paramo, J.; Hsiao, E.Y. Interactions between maternal fluoxetine exposure, the maternal gut microbiome and fetal neurodevelopment in mice. Behav. Brain Res. 2021, 410, 113353. [Google Scholar] [CrossRef]
- Getachew, B.; Aubee, J.I.; Schottenfeld, R.S.; Csoka, A.B.; Thompson, K.M.; Tizabi, Y. Ketamine interactions with gut-microbiota in rats: Relevance to its antidepressant and anti-inflammatory properties. BMC Microbiol. 2018, 18, 222. [Google Scholar] [CrossRef]
- Qu, Y.; Yang, C.; Ren, Q.; Ma, M.; Dong, C.; Hashimoto, K. Comparison of (R)-ketamine and lanicemine on depression-like phenotype and abnormal composition of gut microbiota in a social defeat stress model. Sci. Rep. 2017, 7, 15725. [Google Scholar] [CrossRef]
- Huang, N.; Hua, D.; Zhan, G.; Li, S.; Zhu, B.; Jiang, R.; Yang, L.; Bi, J.; Xu, H.; Hashimoto, K.; et al. Role of Actinobacteria and Coriobacteriia in the antidepressant effects of ketamine in an inflammation model of depression. Pharmacol. Biochem. Behav. 2019, 176, 93–100. [Google Scholar] [CrossRef]
- Wan, X.; Eguchi, A.; Fujita, Y.; Ma, L.; Wang, X.; Yang, Y.; Qu, Y.; Chang, L.; Zhang, J.; Mori, C.; et al. Effects of (R)-ketamine on reduced bone mineral density in ovariectomized mice: A role of gut microbiota. Neuropharmacology 2022, 213, 109139. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, R.; Wu, Z.; Zhou, L.; Xu, J.; Huang, C.; Yang, L.; Zhu, B.; Yan, E.; Liu, C.; et al. Gut microbiota is involved in the antidepressant-like effect of (S)-norketamine in an inflammation model of depression. Pharmacol. Biochem. Behav. 2021, 207, 173226. [Google Scholar] [CrossRef]
- Yang, C.; Qu, Y.; Fujita, Y.; Ren, Q.; Ma, M.; Dong, C.; Hashimoto, K. Possible role of the gut microbiota–brain axis in the antidepressant effects of (R)-ketamine in a social defeat stress model. Transl. Psychiatry 2017, 7, 1294. [Google Scholar] [CrossRef]
- Deng, Y.; Zhou, M.; Wang, J.; Yao, J.; Yu, J.; Liu, W.; Wu, L.; Wang, J.; Gao, R. Involvement of the microbiota-gut-brain axis in chronic restraint stress: Disturbances of the kynurenine metabolic pathway in both the gut and brain. Gut Microbes 2021, 13, 1869501. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Huang, Y.; Tan, X.; Chai, T.; Wu, J.; Zhang, H.; Li, Y.; Hu, X.; Zheng, P.; Ji, P.; et al. Characterization of gut microbiome in mice model of depression with divergent response to escitalopram treatment. Transl. Psychiatry 2021, 11, 303. [Google Scholar] [CrossRef] [PubMed]
- Schmidtner, A.K.; Slattery, D.A.; Gläsner, J.; Hiergeist, A.; Gryksa, K.; Malik, V.A.; Hellmann-Regen, J.; Heuser, I.; Baghai, T.C.; Gessner, A.; et al. Minocycline alters behavior, microglia and the gut microbiome in a trait-anxiety-dependent manner. Transl. Psychiatry 2019, 9, 223. [Google Scholar] [CrossRef]
- Yang, Q.; Luo, L.; Sun, T.; Yang, L.; Cheng, L.-F.; Wang, Y.; Liu, Q.-Q.; Liu, A.; Liu, H.-Y.; Zhao, M.-G.; et al. Chronic minocycline treatment exerts antidepressant effect, inhibits neuroinflammation, and modulates gut microbiota in mice. Psychopharmacology 2020, 237, 3201–3213. [Google Scholar] [CrossRef]
- Serrano-Contreras, J.I.; García-Pérez, I.; Meléndez-Camargo, M.E.; Zepeda-Vallejo, L.G. NMR-based metabonomic analysis of normal rat urine and faeces in response to (±)-venlafaxine treatment. J. Pharmaceut Biomed. 2016, 123, 82–92. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y.-X.; Wang, Z.; Wang, X.-Q.; Zhang, J.-J.; Jiang, R.-H.; Wang, X.-Q.; Zhu, S.-W.; Wang, K.; Liu, Z.-J.; et al. Clinical characteristic and fecal microbiota responses to probiotic or antidepressant in patients with diarrhea-predominant irritable bowel syndrome with depression comorbidity: A pilot study. Chin. Med. J. 2019, 132, 346–351. [Google Scholar] [CrossRef]
- Dong, Z.; Shen, X.; Hao, Y.; Li, J.; Xu, H.; Yin, L.; Kuang, W. Gut microbiome: A potential indicator for predicting treatment outcomes in major depressive disorder. Front. Neurosci. 2022, 16, 813075. [Google Scholar] [CrossRef]
- Shen, Y.; Yang, X.; Li, G.; Gao, J.; Liang, Y. The change of gut microbiota in MDD patients under SSRIs treatment. Sci. Rep. 2021, 11, 14918. [Google Scholar] [CrossRef]
- Ye, X.; Wang, D.; Zhu, H.; Wang, D.; Li, J.; Tang, Y.; Wu, J. Gut Microbiota Changes in Patients with Major Depressive Disorder Treated with Vortioxetine. Front. Psychiatry 2021, 12, 641491. [Google Scholar] [CrossRef]
- Tomizawa, Y.; Kurokawa, S.; Ishii, D.; Miyaho, K.; Ishii, C.; Sanada, K.; Fukuda, S.; Mimura, M.; Kishimoto, T. Effects of Psychotropics on the Microbiome in Patients with Depression and Anxiety: Considerations in a Naturalistic Clinical Setting. Int. J. Neuropsychopharmacol. 2020, 24, 97–107. [Google Scholar] [CrossRef]
- Bharwani, A.; Bala, A.; Surette, M.; Bienenstock, J.; Vigod, S.N.; Taylor, V.H. Gut Microbiome Patterns Associated with Treatment Response in Patients with Major Depressive Disorder: Changements du microbiote intestinal associés à la réponse au traitement chez des patients souffrant de trouble dépressif majeur. Can. J. Psychiatry 2020, 65, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Dong, T.S.; Krause-Sorio, B.; Siddarth, P.; Milillo, M.M.; Lagishetty, V.; Datta, T.; Aguilar-Faustino, Y.; Jacobs, J.P.; Lavretsky, H. The intestinal microbiota as a predictor for antidepressant treatment outcome in geriatric depression: A prospective pilot study. Int. Psychogeriatr. 2022, 34, 33–45. [Google Scholar] [CrossRef]
- Zhu, F.; Tu, H.; Chen, T. The Microbiota–Gut–Brain Axis in Depression: The Potential Pathophysiological Mechanisms and Microbiota Combined Antidepression Effect. Nutrients 2022, 14, 2081. [Google Scholar] [CrossRef]
- Macedo, D.; Chaves-Filho, A.J.M.; Soares de Sousa, C.N.; Quevedo, J.; Barichello, T.; Júnior, H.V.N.; de Lucena, D.F. Antidepressants, antimicrobials or both? Gut microbiota dysbiosis in depression and possible implications of the antimicrobial effects of antidepressant drugs for antidepressant effectiveness. J. Affect. Disord. 2017, 208, 22–32. [Google Scholar] [CrossRef]
- Javdan, B.; Lopez, J.G.; Chankhamjon, P.; Lee, Y.J.; Hull, R.; Wu, Q.; Wang, X.; Chatterjee, S.; Donia, M.S. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell 2020, 181, 1661–1679.e22. [Google Scholar] [CrossRef] [PubMed]
- Peters, E.J.; Slager, S.L.; Kraft, J.B.; Jenkins, G.D.; Reinalda, M.S.; McGrath, P.J.; Hamilton, S.P. Pharmacokinetic Genes Do Not Influence Response or Tolerance to Citalopram in the STAR*D Sample. PLoS ONE 2008, 3, e1872. [Google Scholar] [CrossRef]
- Hodgson, K.; Tansey, K.; Dernovšek, M.Z.; Hauser, J.; Henigsberg, N.; Maier, W.; Mors, O.; Placentino, A.; Rietschel, M.; Souery, D.; et al. Genetic differences in cytochrome P450 enzymes and antidepressant treatment response. J. Psychopharmacol. 2014, 28, 133–141. [Google Scholar] [CrossRef]
- Florio, V.; Porcelli, S.; Saria, A.; Serretti, A.; Conca, A. Escitalopram plasma levels and antidepressant response. Eur. Neuropsychopharmacol. 2017, 27, 940–944. [Google Scholar] [CrossRef]
- Liu, L.; Wang, H.; Chen, X.; Zhang, Y.; Zhang, H.; Xie, P. Gut microbiota and its metabolites in depression: From pathogenesis to treatment. Ebiomedicine 2023, 90, 104527. [Google Scholar] [CrossRef]
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef]
- Forouzan, S.; McGrew, K.; Kosten, T.A. Drugs and bugs: Negative affect, psychostimulant use and withdrawal, and the microbiome. Am. J. Addict. 2021, 30, 525–538. [Google Scholar] [CrossRef]
- Lewis, G.; Marston, L.; Duffy, L.; Freemantle, N.; Gilbody, S.; Hunter, R.; Kendrick, T.; Kessler, D.; Mangin, D.; King, M.; et al. Maintenance or Discontinuation of Antidepressants in Primary Care. N. Engl. J. Med. 2021, 385, 1257–1267. [Google Scholar] [CrossRef]
- Ostaff, M.J.; Stange, E.F.; Wehkamp, J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 2013, 5, 1465–1483. [Google Scholar] [CrossRef]
- Liu, D.; Ray, B.; Neavin, D.R.; Zhang, J.; Athreya, A.P.; Biernacka, J.M.; Bobo, W.V.; Hall-Flavin, D.K.; Skime, M.K.; Zhu, H.; et al. Beta-defensin 1, aryl hydrocarbon receptor and plasma kynurenine in major depressive disorder: Metabolomics-informed genomics. Transl. Psychiatry 2018, 8, 10. [Google Scholar] [CrossRef]
- Sittipo, P.; Shim, J.W.; Lee, Y.K. Microbial Metabolites Determine Host Health and the Status of Some Diseases. Int. J. Mol. Sci. 2019, 20, 5296. [Google Scholar] [CrossRef]
- Pu, J.; Liu, Y.; Gui, S.; Tian, L.; Yu, Y.; Wang, D.; Zhong, X.; Chen, W.; Chen, X.; Chen, Y.; et al. Effects of pharmacological treatment on metabolomic alterations in animal models of depression. Transl. Psychiatry 2022, 12, 175. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Wallace, C.J.K.; Milev, R. The effects of probiotics on depressive symptoms in humans: A systematic review. Ann. Gen. Psychiatry 2017, 16, 14. [Google Scholar] [CrossRef]
- Alli, S.R.; Gorbovskaya, I.; Liu, J.C.W.; Kolla, N.J.; Brown, L.; Müller, D.J. The Gut Microbiome in Depression and Potential Benefit of Prebiotics, Probiotics and Synbiotics: A Systematic Review of Clinical Trials and Observational Studies. Int. J. Mol. Sci. 2022, 23, 4494. [Google Scholar] [CrossRef]
- Nikolova, V.L.; Cleare, A.J.; Young, A.H.; Stone, J.M. Updated Review and Meta-Analysis of Probiotics for the Treatment of Clinical Depression: Adjunctive vs. Stand-Alone Treatment. J. Clin. Med. 2021, 10, 647. [Google Scholar] [CrossRef]
- Ma, J.; Lyu, Y.; Liu, X.; Jia, X.; Cui, F.; Wu, X.; Deng, S.; Yue, C. Engineered probiotics. Microb. Cell Fact. 2022, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Cordaillat-Simmons, M.; Rouanet, A.; Pot, B. Live biotherapeutic products: The importance of a defined regulatory framework. Exp. Mol. Med. 2020, 52, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
| Author [Ref] | Pubmed ID | Model and Sample Size | Study Design | Study Duration | Stool Collection Time | Outcome Measure | Characteristics |
|---|---|---|---|---|---|---|---|
| Klunemann et al. [25] | 34497420 | Caenorhabditis elegans Each experiment = 50 | Interventional observational, Controlled | 1 h | N/A | Behavior | C. elegans N2 wild type Escherichia coli IAI1 |
| Lukić et al. [26] | 30967529 | Mouse CTL = 9 FLU = 11 ESC = 12 VEN = 12 DUL = 11 DES = 12 | Interventional observational, Controlled | 3–4 wks | 3 wks | Gut microbiota changes | Male BALB/c OlaHsd Ruminococcus flavefaciens 17 Adlercreutzia equolifaciens FJC-B9 |
| Dethloff et al. [27] | 32581888 | Mouse PARO = 17 VEH = 17 | Interventional observational, Controlled | 1 and 2 wks | 7 and 14 days (PARO = 17, VEH = 17) | Gut microbiota changes and gut metabolite changes | DBA/2J mice Depressed mouse model |
| Kim et al. [28] | 33731795 | Mouse CTL = 12 IS-treated = 12 EC-treated = 12 | Interventional observational, Controlled | 7 days | 48 h after treatment | Gut microbiota changes and behavior | Male C57BL/6N mice IS or EC-induced model |
| Lyte et al. [29] | 30643701 | Mouse FLU = 10 CTL = 10 | Interventional observational, Controlled | 29 days | Baseline, days 15 and 29 | Gut microbiota changes and behavior | Male CF-1 mice |
| Ramsteijn et al. [30] | 31971855 | Rat sMV-MC = 13 cMV-MC = 20 sMV-FLU = 25 cMV-FLU = 34 | Interventional observational, Controlled | 35 days | GD0 (before conception), GD7, GD14, PND2, PND7, PND14, and PND21 | Gut microbiota and gut metabalomic changes | Serotonin transporter knockout (SERT−/−, Slc6a41Hubr) Wistar rats heterozygous SERT knockout (SERT+/−) female rats |
| Sun et al. [31] | 31588192 | Mouse CTL = 10 CUMS + VEH = 10 CUMS + FLU = 10 | Interventional observational, Controlled | 9 wks | 7 wks | Gut microbiota changes and behavior | Male C57/6 mice |
| Zhang et al. [32] | 33602895 | Rat HC = 12 CUMS = 6 CUMS + AMI = 6 CUMS + FLU = 7 | Interventional observational, Controlled | 15 wks | 9 (CUMS = 12 and HC = 12) and 15 (HC = 3, CUMS = 3, CUMS + AMI = 3, and CUMS + FLU = 3) wks | Gut microbiota changes, ARG changes, and behavior | Male pathogen-free Sprague–Dawley rats |
| Siopi et al. [33] | 32187541 | Mouse CTL = 10+ UCMS = 10+ CTL-tr = 10+ UCMS-tr = 10+ (all experiments had at least 10 animals in each group but varied) | Interventional observational, Controlled | 8–9 wks | 8–9 wks | Gut microbiota changes and behavior | Male C57BL/6J mice |
| Vuong et al. [34] | 33979656 | Mouse VEH = 3–6 FLU = 3–6 ABX + FLU = 3–6 ABX + VEH = 3–6 | Interventional observational, Controlled | 2 wks | E3.5, E6.5, E8.5, E11.5 and E14.5 | Gut microbiota and brain gene transcription changes | Female SPF C57BL/6 J mice |
| Getachew et al. [35] | 30579332 | Rat KET = 5 SAL = 5 | Interventional observational, Controlled | 8 days | Day 8 | Gut microbiota changes | Male Wistar rats |
| Qu et al. [36] | 29147024 | Mouse R-KET = 6 LAN = 6 SAL = 6 CTL = 6 | Interventional observational, Controlled | 15 days | Day 15 (3 days post-treatment) | Gut microbiota changes | Male C57BL/6 mice Male CD1 (ICR) mice |
| Huang et al. [37] | 30528936 | Mouse LPS + KET = 8 LPS + SAL = 8 CTL = 8 | Interventional observational, Controlled | ~25 h | ~25 h after treatment | Gut microbiota changes and behavior | Male C57BL/6 mice LPS-induced inflammatory depression model |
| Wan et al. [38] | 35594949 | Mouse CTL = ~9 OVX + SAL = ~9 OVX + KET = ~9 | Interventional observational, Controlled | 6 wks | Day 43 | Gut microbiota changes, bone marrow changes, and serum metabolite changes | Female C57BL/6 mice |
| Wang et al. [39] | 34217782 | Mouse CTL = ~9 SAL = ~9 RnKET = ~9 SnKET = ~9 | Interventional observational, Controlled | 28 h | 28 h | Gut microbiota changes | Male C57BL/6 mice |
| Yang et al. [40] | 29249803 | Mouse CTL = 6 CSDS + SAL = 6 CSDS + SKET = 6 CSDS + RKET = 6 | Interventional observational, Controlled | 16 days | Day 16 | Gut microbiota changes | Male C57BL/6 mice |
| Deng et al. [41] | 33535879 | Mouse CTL + PBS = 10 CTL + CIT = 10 CRS + PBS = 10 CRS + CIT = 10 | Interventional observational, Controlled | 5–6 wks | 5–6 wks | Gut microbiota changes | Male C57BL/6 J mice CRS model |
| Duan et al. [42] | 34016954 | Mouse CTL = 8 CUMS + VEH = 8 ESC responder = 7 ESC nonresponder = 9 | Interventional observational, Controlled | 4 wks | Baseline and 4 wks of ESC | Gut microbiota changes, plasma metabolite changes, and behavior | Male C57BL/6 mice CUMS model |
| Schmidtner et al. [43] | 31519869 | Rat Sample sizes varied by experiment and was a minimum of 6 up to 15 | Interventional observational, Controlled | 22 days | Day 22 | Gut microbiota changes and gut metabalomic changes | Male and female Wistar rats |
| Yang et al. [44] | 32671421 | Mouse CTL = 6–8 CUMS = 6–8 CUMS + MIN = 6–8 CUMS + IMI = 6–8 | Interventional observational, Controlled | 44 days | Day 44 | Gut microbiota changes, adverse events, and gut metabolite changes | Male C57BL/6 mice |
| Serrano-Contreras et al. [45] | 26895493 | Rat VEN (22 mg/kg) + VEH = 18 VEN (112 mg/kg) + VEH = 18 VEH = 18 | Interventional observational, Controlled | 24 h | 0 to 24 h after treatment | Gut metabolite changes | Female Wistar rats |
| Author [Ref.] | Pubmed ID | Model/Sample Size | Study Design | Study Duration | Stool Collection Times | Outcome Measure | Characteristics | Drug(s) |
|---|---|---|---|---|---|---|---|---|
| Zhang et al. [46] | 30681503 | Human DUL = 6 | Interventional observational, uncontrolled | 8 wks | Baseline and 8 wks | Gut microbiota changes, gut metabolite changes, plasma metabolite changes, and behavior | IBS and MDD 18–65 yo | DUL |
| Dong et al. [47] | 35937875 | Human MDD= 63 (20 males, 43 females) HC= 30 (10 males, 20 females) | Interventional observational case-controlled | 8 wks | Stool collected at baseline and after 8 wks treatment | Gut microbiota changes and behavior | 18–45 y/o First episode MDD Hospitalized BMI= 18.5–22.9 | CIT, ESC, PARO, or VEN |
| Shen et al. [48] | 34290352 | Human MDD = 30 HC = 30 | Interventional observational case-controlled | 4–6 wks | Baseline and 4–6 wks after treatment | Gut microbiota changes and behavior | 18–65 y/o Tx-naive, first episode MDD (no tx with antidepressant or antipsychotics) no recent effects on microbiome (ex: antibiotic usage) | ESC |
| Ye et al. [49] | 34025474 | Human HC = 28 MDD = 26 | Interventional observational case-controlled | 8 wks | Baseline, 4 wk, and 8 wks | Gut microbiota changes and behavior | 18–50 yo | VOR |
| Tomizawa et al. [50] | 32975292 | Human depressive = 32 anxious = 8 | Interventional observational, uncontrolled | 3 wks | Baseline, 2wks, and 3 wks | Gut microbiota changes | Adult inpatient and outpatients with MDD and/or ANX 17 males and 23 females 33 stool samples at endpoint | AMI, AMO, SERT, PARO, ESC, DUL, MIL, VEN, MIR, APs, and anxiolytics |
| Bharwani et al. [51] | 31958990 | Human MDD = 15 | Interventional observational, uncontrolled | 6 months | Baseline, 3, and 6 month | Gut microbiota changes and behavior | 18–60 y/o MDD dx Medication free at baseline | CIT and ESC |
| Lee et al. [52] | 33757609 | Human LVM = 4 (Remitters = 2) Placebo = 8 (Remitters = 3) Total Remitters = 5 Total Nonremitters = 7 | Interventional observational controlled | 12 wks | Baseline and 12 wks | Gut microbiota changes and behavior | Geriatric MDD dx Greater than 60 yo | LVM or Placebo |
| Study | Bias Due to Confounding | Bias in Selection of Participants into the Study | Bias in Classification of Interventions | Bias Due to Deviations from Intended Interventions | Bias Due to Missing Data | Bias in Measurement of Outcomes | Bias in Selection of the Reported Result | Overall Bias |
|---|---|---|---|---|---|---|---|---|
| Kluneman et al. [25] | Low | Low | Low | Low | Low | Moderate | Low | Moderate |
| Lukić et al. [26] | Moderate | Low | Low | Low | Low | Moderate | Low | Moderate |
| Dethloff et al. [27] | Moderate | Low | Low | Low | Moderate | Low | Low | Moderate |
| Kim et al. [28] | Moderate | Low | Low | Low | Low | Moderate | Low | Moderate |
| Lyte et al. [29] | Moderate | Low | Low | Low | Low | Moderate | Low | Moderate |
| Ramsteijn et al. [30] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Sun et al. [31] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Zhang et al. [32] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Siopi et al. [33] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Vuong et al. [34] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Getachew et al. [35] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Qu et al. [36] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Huang et al. [37] | Moderate | Low | Low | Low | Low | Moderate | Low | Moderate |
| Wan et al. [38] | Moderate | Low | Low | Low | Low | Moderate | Low | Moderate |
| Wang et al. [39] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Yang et al. [40] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Deng et al. [41] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Duan et al. [42] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Schmidtner et al. [43] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Yang et al. [44] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Serrano-Contreras et al. [45] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Zhang et al. [46] | Serious | Low | Low | Low | Low | Low | Low | Serious |
| Dong et al. [47] | Serious | Low | Low | Low | Low | Moderate | Low | Serious |
| Shen et al. [48] | Serious | Low | Low | Low | Low | Low | Low | Serious |
| Ye et al. [49] | Serious | Moderate | Low | Low | Low | Low | Low | Serious |
| Tomizawa et al. [50] | Serious | Low | Low | Low | Low | Serious | Low | Serious |
| Bharwani et al. [51] | Moderate | Low | Low | Low | Low | Low | Low | Moderate |
| Lee et al. [52] | Serious | Low | Low | Low | Moderate | Low | Low | Serious |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, L.C.; Bobo, W.V.; Gall, C.A.; Müller, D.J.; Bousman, C.A. Pharmacomicrobiomics of Antidepressants in Depression: A Systematic Review. J. Pers. Med. 2023, 13, 1086. https://doi.org/10.3390/jpm13071086
Brown LC, Bobo WV, Gall CA, Müller DJ, Bousman CA. Pharmacomicrobiomics of Antidepressants in Depression: A Systematic Review. Journal of Personalized Medicine. 2023; 13(7):1086. https://doi.org/10.3390/jpm13071086
Chicago/Turabian StyleBrown, Lisa C., William V. Bobo, Cory A. Gall, Daniel J. Müller, and Chad A. Bousman. 2023. "Pharmacomicrobiomics of Antidepressants in Depression: A Systematic Review" Journal of Personalized Medicine 13, no. 7: 1086. https://doi.org/10.3390/jpm13071086
APA StyleBrown, L. C., Bobo, W. V., Gall, C. A., Müller, D. J., & Bousman, C. A. (2023). Pharmacomicrobiomics of Antidepressants in Depression: A Systematic Review. Journal of Personalized Medicine, 13(7), 1086. https://doi.org/10.3390/jpm13071086






