A Gel-Based Proteomic Analysis Reveals Synovial α-Enolase and Fibrinogen β-Chain Dysregulation in Knee Osteoarthritis: A Controlled Trial
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients Selection
2.2. Synovial Fluid Sample Collection and Processing
2.3. Two-Dimensional Gel Electrophoresis (2DE)
2.4. Mass Spectrometry Analysis and Bioinformatic Analysis
2.5. Statistical Analysis
3. Results and Discussion
3.1. Results
| OA-Group | Control-Group | p | |
|---|---|---|---|
| Patients (n) | 24 | 19 | |
| Age | |||
| Mean ± SD | 68.19 ± 6.18 | 24.65 ± 4.73 | 0.001 * |
| Range | 54–80 | 19–34 | - |
| Gender | |||
| Male, n (%) | 11 (45.83%) | 9 (47.37%) | 0.221 |
| Female, n (%) | 13 (54.17%) | 10 (52.63%) | 0.124 |
| BMI (kg/m2) | |||
| Mean ± SD | 27.75 ± 4.88 | 22.16 ± 1.7 | 0.002 * |
| Smoking status | |||
| Number of smokers, n (%) | 6 (20.69%) | 2 (12.5%) | 0.03 * |
| Kellgren Lawrence classification | |||
| Grade 3 | 11 (37–93%) | // | // |
| Grade 4 | 18 (62.07%) | // | // |
| CRP (mg/dL) | |||
| Mean ± SD | 1.73 ± 1.25 | 0.21 ± 0.43 | 0.001 * |
| Serum Fibrinogen (mg/dL) | |||
| Mean ± SD | 302.4 ± 78.5 | 245.6 ± 57.3 | 0.003 * |
| KSS | |||
| Mean ± SD | 42.09 ± 7.24 | 94.45 ± 4.35 | <0.001 * |
| IKDC | |||
| Mean ± SD | 0.346 ± 0.0825 | 0.964 ± 0.021 | <0.001 * |
| KOOS main outcome | |||
| Mean ± SD | 38.45 ± 7.6 | 95.7 ± 3.5 | <0.001 * |
| VAS | |||
| Mean ± SD | 5.78 ± 1.45 | 4.53 ± 1.65 | 0.07 |
3.2. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Mobasheri, A.; Bay-Jensen, A.-C.; van Spil, W.E.; Larkin, J.; Levesque, M.C. Osteoarthritis Year in Review 2016: Biomarkers (Biochemical Markers). Osteoarthr. Cartil. 2017, 25, 199–208. [Google Scholar] [CrossRef]
- Hunter, D.J.; Nevitt, M.; Losina, E.; Kraus, V. Biomarkers for Osteoarthritis: Current Position and Steps towards Further Validation. Best Pract. Res. Clin. Rheumatol. 2014, 28, 61–71. [Google Scholar] [CrossRef]
- Goldring, M.B.; Goldring, S.R. Osteoarthritis. J. Cell. Physiol. 2007, 213, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.J.; Ruiz-Romero, C. Metabolomic Characterization of Metabolic Phenotypes in OA. Nat. Rev. Rheumatol. 2012, 8, 130–132. [Google Scholar] [CrossRef] [PubMed]
- Cross, M.; Smith, E.; Hoy, D.; Nolte, S.; Ackerman, I.; Fransen, M.; Bridgett, L.; Williams, S.; Guillemin, F.; Hill, C.L.; et al. The global burden of hip and knee osteoarthritis: Estimates from the global burden of disease 2010 study. Ann. Rheum. Dis. 2014, 73, 1323–1330. [Google Scholar] [CrossRef]
- Bizzoca, D.; Moretti, L.; Gnoni, A.; Moretti, F.L.; Scacco, S.; Banfi, G.; Piazzolla, A.; Solarino, G.; Moretti, B. The Usefulness of Synovial Fluid Proteome Analysis in Orthopaedics: Focus on Osteoarthritis and Periprosthetic Joint Infections. J. Funct. Morphol. Kinesiol. 2022, 7, 97. [Google Scholar] [CrossRef]
- Moretti, L.; Bizzoca, D.; Geronimo, A.; Moretti, F.L.; Monaco, E.; Solarino, G.; Moretti, B. Towards Precision Medicine for Osteoarthritis: Focus on the Synovial Fluid Proteome. Int. J. Mol. Sci. 2022, 23, 9731. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Nirujogi, R.; Ahmad, S.; Bhattacharjee, M.; Manda, S.S.; Renuse, S.; Kelkar, D.S.; Subbannayya, Y.; Raju, R.; Goel, R.; et al. Proteomic Analysis of Human Osteoarthritis Synovial Fluid. Clin. Proteom. 2014, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Hepinstall, M.S.; Rutledge, J.R.; Bornstein, L.J.; Mazumdar, M.; Westrich, G.H. Factors that impact expectations before total knee arthroplasty. J. Arthroplasty. 2011, 26, 870–876. [Google Scholar] [CrossRef]
- Corigliano, A.; Preianò, M.; Terracciano, R.; Savino, R.; De Gori, M.; Galasso, O.; Gasparini, G. C3f Is a Potential Tool for the Staging of Osteoarthritis. J. Biol. Regul. Homeost. Agents 2017, 31, 29–35. [Google Scholar]
- Mahendran, S.M.; Oikonomopoulou, K.; Diamandis, E.P.; Chandran, V. Synovial Fluid Proteomics in the Pursuit of Arthritis Mediators: An Evolving Field of Novel Biomarker Discovery. Crit. Rev. Clin. Lab. Sci. 2017, 54, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Wanner, J.P.; Subbaiah, R.; Skomorovska-Prokvolit, Y.; Shishani, Y.; Boilard, E.; Mohan, S.; Gillespie, R.; Miyagi, M.; Gobezie, R. Proteomic Profiling and Functional Characterization of Early and Late Shoulder Osteoarthritis. Arthritis Res. Ther. 2013, 15, R180. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, A.I.; Beekhuizen, M.; Rutgers, M.; van Osch, G.J.; Bekkers, J.E.; Bot, A.G.; Geurts, B.; Dhert, W.J.; Saris, D.B.; Creemers, L.B. Interleukin-6 Is Elevated in Synovial Fluid of Patients with Focal Cartilage Defects and Stimulates Cartilage Matrix Production in an In Vitro Regeneration Model. Arthritis Res. Ther. 2012, 14, R262. [Google Scholar] [CrossRef] [PubMed]
- Kokebie, R.; Aggarwal, R.; Lidder, S.; Hakimiyan, A.A.; Rueger, D.C.; Block, J.A.; Chubinskaya, S. The Role of Synovial Fluid Markers of Catabolism and Anabolism in Osteoarthritis, Rheumatoid Arthritis and Asymptomatic Organ Donors. Arthritis Res. Ther. 2011, 13, R50. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Satoh, T.; Chiba, J.; Ju, C.; Inoue, K.; Kagawa, J. Interleukin-6 and Interleukin-8 Levels in Serum and Synovial Fluid of Patients with Osteoarthritis. Cytokines. Cell. Mol. Ther. 2000, 6, 71–79. [Google Scholar] [CrossRef]
- Liao, W.; Li, Z.; Li, T.; Zhang, Q.; Zhang, H.; Wang, X. Proteomic Analysis of Synovial Fluid in Osteoarthritis Using SWATH-mass Spectrometry. Mol. Med. Rep. 2018, 17, 2827–2836. [Google Scholar] [CrossRef]
- Insall, J.N.; Dorr, L.D.; Scott, R.D.; Scott, W.N. Rationale of the Knee Society clinical rating system. Clin. Orthop. Relat. Res. 1989, 248, 13–14. [Google Scholar] [CrossRef]
- Higgins, L.D.; Taylor, M.K.; Park, D.; Ghodadra, N.; Marchant, M.; Pietrobon, R.; Cook, C. Reliability and validity of the International Knee Documentation Committee (IKDC) Subjective Knee Form. Jt. Bone Spine 2007, 74, 594–599. [Google Scholar] [CrossRef]
- Lyman, S.; Lee, Y.Y.; McLawhorn, A.S.; Islam, W.; MacLean, C.H. What Are the Minimal and Substantial Improvements in the HOOS and KOOS and JR Versions after Total Joint Replacement? Clin. Orthop. Relat. Res. 2018, 476, 2432–2441. [Google Scholar] [CrossRef]
- Bizzoca, D.; Rocchetti, M.T.; Scacco, S.; Taurino, F.; Vicenti, G.; Spadaccino, F.; Moretti, L.; Ranieri, E.; Gesualdo, L.; Moretti, F.; et al. Beyond pre-analytical and analytical concerns in the study of synovial fluid proteome: Description of an optimized Gel-based protocol. J. Biol. Regul. Homeost. Agents 2021, 35, 827–832. [Google Scholar] [CrossRef]
- Rocchetti, M.T.; Papale, M.; d’Apollo, A.M.; Suriano, I.V.; Di Palma, A.M.; Vocino, G.; Montemurno, E.; Varraso, L.; Grandaliano, G.; Di Paolo, S.; et al. Association of urinary laminin G-like 3 and free K light chains with disease activity and histological injury in IgA nephropathy. Clin. J. Am. Soc. Nephrol. 2013, 8, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Zhang, Z.; Liao, W.; Xu, D.; Fu, M.; Kang, Y. Screening of gene signatures for rheumatoid arthritis and osteoarthritis based on bioinformatics analysis. Mol. Med. Rep. 2016, 14, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Dong, Z.; Cheng, L.; Guo, Z.; Lv, Z. Identifying the gender-specific genes and pathways in osteoarthritis by bioinformatics. Osteoarthr. Cartil. 2020, 28, S207–S208. [Google Scholar] [CrossRef]
- Shorter, E.; Avelar, R.; Zachariou, M.; Spyrou, G.M.; Raina, P.; Smagul, A.; Ashraf Kharaz, Y.; Peffers, M.; Goljanek-Whysall, K.; de Magalhães, J.P.; et al. Identifying Novel Osteoarthritis-Associated Genes in Human Cartilage Using a Systematic Meta-Analysis and a Multi-Source Integrated Network. Int. J. Mol. Sci. 2022, 23, 4395. [Google Scholar] [CrossRef] [PubMed]
- Gedeon, A.K.; Colley, A.; Jamieson, R.; Thompson, E.M.; Rogers, J.; Sillence, D.; Tiller, G.E.; Mulley, J.C.; Gécz, J. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat. Genet. 1999, 22, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Ishii, H.; Takahashi, K.; Takai, S.; Ozawa, H. Suitable reference gene selection for gene expression studies in knee osteoarthritis synovium using quantitative PCR analysis. Connect. Tissue Res. 2018, 59, 356–368. [Google Scholar] [CrossRef]
- Pereira, D.; Ramos, E.; Branco, J. Osteoarthritis. Acta Med. Port. 2015, 28, 99–106. [Google Scholar] [CrossRef]
- Marshall, D.A.; Vanderby, S.; Barnabe, C.; MacDonald, K.V.; Maxwell, C.; Mosher, D.; Wasylak, T.; Lix, L.; Enns, E.; Frank, C.; et al. Estimating the Burden of Osteoarthritis to Plan for the Future. Arthritis Care Res. 2015, 67, 1379–1386. [Google Scholar] [CrossRef]
- Bae, S.; Kim, H.; Lee, N.; Won, C.; Kim, H.R.; Hwang, Y.I.; Song, Y.W.; Kang, J.S.; Lee, W.J. α-Enolase expressed on the surfaces of monocytes and macrophages induces robust synovial inflammation in rheumatoid arthritis. J. Immunol. 2012, 189, 365–372. [Google Scholar] [CrossRef]
- Capello, M.; Ferri-Borgogno, S.; Cappello, P.; Novelli, F. α-Enolase: A promising therapeutic and diagnostic tumour target. FEBS J. 2011, 278, 1064–1074. [Google Scholar] [CrossRef]
- Pancholi, V. Multifunctional alpha-enolase: Its role in diseases. Cell. Mol. Life Sci. 2001, 58, 902–920. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Luo, L.; Gui, T.; Yu, F.; Yan, L.; Yao, L.; Zhong, L.; Yu, W.; Han, B.; Patel, J.M.; et al. Targeting cartilage EGFR pathway for osteoarthritis treatment. Sci. Transl. Med. 2021, 13, eabb3946. [Google Scholar] [CrossRef] [PubMed]
- Gui, T.; Wei, Y.; Luo, L.; Li, J.; Zhong, L.; Yao, L.; Beier, F.; Nelson, C.L.; Tsourkas, A.; Liu, X.S.; et al. Activating EGFR Signaling Attenuates Osteoarthritis Development Following Loading Injury in Mice. J. Bone Min. Res. 2022, 37, 2498–2511. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Ma, X.; Sun, H.; Gui, T.; Li, J.; Yao, L.; Zhong, L.; Yu, W.; Han, B.; Nelson, C.L.; et al. EGFR Signaling Is Required for Maintaining Adult Cartilage Homeostasis and Attenuating Osteoarthritis Progression. J. Bone Min. Res. 2022, 37, 1012–1023. [Google Scholar] [CrossRef]
- Lu, Q.; Liu, P.; Miao, Z.; Luo, D.; Li, S.; Lu, M. SIRT1 restoration enhances chondrocyte autophagy in osteoarthritis through PTEN-mediated EGFR ubiquitination. Cell. Death Discov. 2022, 8, 203. [Google Scholar] [CrossRef]
- Jiang, L.; Zhou, X.; Xu, K.; Hu, P.; Bao, J.; Li, J.; Zhu, J.; Wu, L. miR-7/EGFR/MEGF9 axis regulates cartilage degradation in osteoarthritis via PI3K/AKT/mTOR signalling pathway. Bioengineered 2021, 12, 8622–8634. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhang, A.; Wang, J.; Han, K.; Gao, H. Estrogen alleviates post-traumatic osteoarthritis progression and decreases p-EGFR levels in female mouse cartilage. BMC Musculoskelet. Disord. 2022, 23, 685. [Google Scholar] [CrossRef]
- Qin, L.; Beier, F. EGFR Signaling: Friend or Foe for Cartilage? JBMR Plus. 2019, 3, e10177. [Google Scholar] [CrossRef]
- Damgaard, D.; Bawadekar, M.; Senolt, L.; Stensballe, A.; Shelef, M.A.; Nielsen, C.H. Relative efficiencies of peptidyl arginine deiminase 2 and 4 in generating target sites for anti-citrullinated protein antibodies in fibrinogen, alpha-enolase and histone H3. PLoS ONE 2018, 13, e0203214. [Google Scholar] [CrossRef]
- Beydoun, H.A.; Archer, D.F.; Zonderman, A.B.; Beydoun, M.A. Interrelationships of Sex, Physician-Diagnosed Arthritis, Chronic Inflammation, and Physical Functioning in the Third National Health and Nutrition Examination Surveys. Gerontol. Geriatr. Med. 2015, 1. [Google Scholar] [CrossRef]
- Antony, B.; Venn, A.; Cicuttini, F.; March, L.; Blizzard, L.; Dwyer, T.; Cross, M.; Jones, G.; Ding, C. Association of Body Composition and Hormonal and Inflammatory Factors with Tibial Cartilage Volume and Sex Difference in Cartilage Volume in Young Adults. Arthritis Care Res. 2016, 68, 517–525. [Google Scholar] [CrossRef]
- Skriner, K.; Adolph, K.; Jungblut, P.R.; Burmester, G.R. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006, 54, 3809–3814. [Google Scholar] [CrossRef]
- Ebrahimi-Rad, M.; Khatami, S.; Akhbari, H.; Mahmoudzadeh-Niknam, H.; Valadbeigi, S.; Mahmoudi, M.; Jamshidi, A.; Riazi-Rad, F.; Saghiri, R. Evaluation of autoantibodies against vimentin and α-enolase in rheumatoid arthritis patients. Rheumatology 2020, 58, 350–356. [Google Scholar] [CrossRef]
- Schadler, P.; Lohberger, B.; Thauerer, B.; Faschingbauer, M.; Kullich, W.; Stradner, M.H.; Husic, R.; Leithner, A.; Steinecker-Frohnwieser, B. Fatty Acid-Binding Protein 4 (FABP4) Is Associated with Cartilage Thickness in End-Stage Knee Osteoarthritis. Cartilage 2021, 13 (Suppl. S2), 1165S–1173S. [Google Scholar] [CrossRef]
- Belcaro, G.; Cesarone, M.R.; Errichi, S.; Zulli, C.; Errichi, B.M.; Vinciguerra, G.; Ledda, A.; Di Renzo, A.; Stuard, S.; Dugall, M.; et al. Variations in C-reactive protein, plasma free radicals and fibrinogen values in patients with osteoarthritis treated with Pycnogenol. Redox Rep. 2008, 13, 271–276. [Google Scholar] [CrossRef]
- Masson-Bessieère, C.; Sebbag, M.; Girbal-Neuhauser, E.; Nogueira, L.; Vincent, C.; Senshu, T.; Serre, G. The Major Synovial Targets of the Rheumatoid Arthritis-Specific Antifilaggrin Autoantibodies Are Deiminated Forms of the α- and β-Chains of Fibrin1. J. Immunol. 2001, 166, 4177–4184. [Google Scholar] [CrossRef]
- Kandel, L.; Agar, G.; Elkayam, O.; Sharipov, A.; Slevin, O.; Rivkin, G.; Dahan, M.; Aloush, V.; Pyeser, A.B.; Brin, Y.; et al. A novel approach for knee osteoarthritis using high molecular weight hyaluronic acid conjugated to plasma fibrinogen—Interim findings of a double-blind clinical study. Heliyon 2020, 6, e04475. [Google Scholar] [CrossRef]
- Vicenti, G.; Bizzoca, D.; Nappi, V.S.; Moretti, F.; Carrozzo, M.; Belviso, V.; Moretti, B. Biophysical stimulation of the knee with PEMFs: From bench to bedside. J. Biol. Regul. Homeost. Agents 2018, 32 (Suppl. S1), 23–28. [Google Scholar]
- Moretti, L.; Bizzoca, D.; Giancaspro, G.A.; Cassano, G.D.; Moretti, F.; Setti, S.; Moretti, B. Biophysical Stimulation in Athletes’ Joint Degeneration: A Narrative Review. Medicina 2021, 57, 1206. [Google Scholar] [CrossRef]
- Scanzello, C.R.; McKeon, B.; Swaim, B.H.; DiCarlo, E.; Asomugha, E.U.; Kanda, V.; Nair, A.; Lee, D.M.; Richmond, J.C.; Katz, J.N.; et al. Synovial inflammation in patients undergoing arthroscopic meniscectomy: Molecular characterization and relationship to symptoms. Arthritis Rheum. 2011, 63, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Martel-Pelletier, J.; Pelletier, J.P.; Abram, F.; Raynauld, J.P.; Cicuttini, F.; Jones, G. Tear as an osteoarthritis risk factor in a largely non-osteoarthritic cohort: A cross-sectional study. J. Rheumatol. 2007, 34, 776–784. [Google Scholar] [PubMed]
- Vicenti, G.; Bizzoca, D.; Solarino, G.; Moretti, F.; Ottaviani, G.; Simone, F.; Zavattini, G.; Maccagnano, G.; Noia, G.; Moretti, B. The role of biophysical stimulation with pemfs in fracture healing: From bench to bedside. J. Biol. Regul. Homeost. Agents 2020, 34 (Suppl. S1), 131–135. [Google Scholar] [PubMed]
- Vicenti, G.; Bizzoca, D.; Cotugno, D.; Carrozzo, M.; Riefoli, F.; Rifino, F.; Belviso, V.; Elia, R.; Solarino, G.; Moretti, B. The use of a gentamicin-coated titanium nail, combined with RIA system, in the management of non-unions of open tibial fractures: A single centre prospective study. Injury 2020, 51 (Suppl. S3), S86–S91. [Google Scholar] [CrossRef] [PubMed]
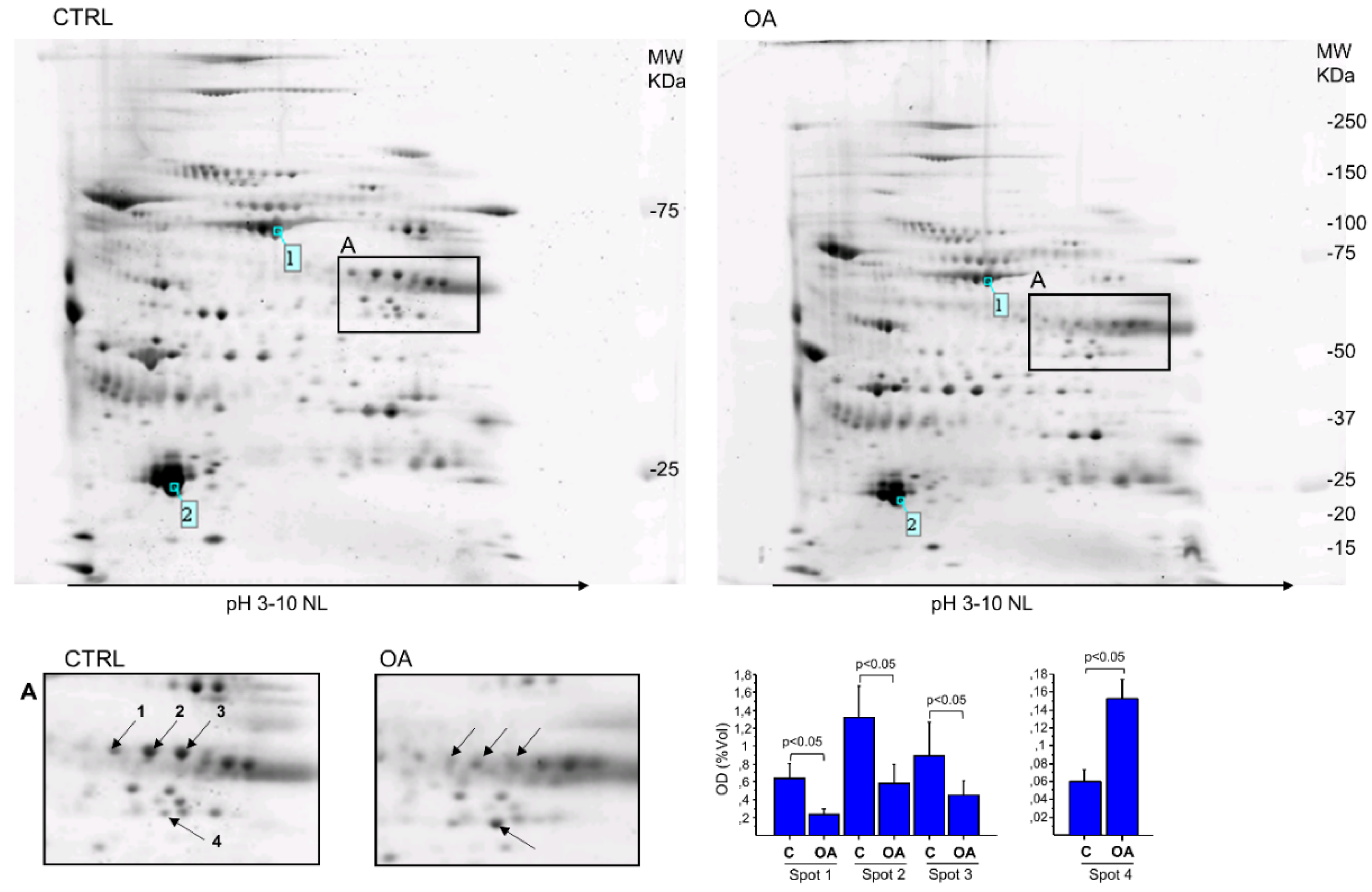

| Spot N° | Protein Name | Gene Name | Accession Number * | Theoretical MW (kDa)/pI | Experimental MW (kDa)/pI | Mascot Score | SC % | Unique Peptide | Fold Change (%) |
|---|---|---|---|---|---|---|---|---|---|
| OA/CTRL | |||||||||
| 1 | Fibrinogen beta chain | FGB | P02675 | 56.7/8.54 | ~57/8.8 | 106 | 25 | 13 | 0.37 (−63%) |
| 2 | Fibrinogen beta chain | FGB | P02675 | 56.7/8.54 | ~57/9.0 | 105 | 34 | 15 | 0.30 (−70%) |
| 3 | Fibrinogen beta chain | FGB | P02675 | 56.7/8.54 | ~57/9.2 | 67 | 25 | 12 | 0.30 (−70%) |
| 4 | Alpha-enolase | ENO1 | P06733 | 47.4/7.01 | ~48/8.8 | 74 | 31 | 11 | 2.5 (+153%) |
| FGB | ENO1 | |||
|---|---|---|---|---|
| R | p | R | p | |
| Clinical scales | ||||
| KSS | −0.61 | 0.02 * | 0.33 | 0.108 |
| IKDC | −0.32 | 0.12 | 0.43 | 0.09 |
| KOOS main outcome | −0.44 | 0.08 | 0.68 | 0.01 * |
| VAS | −0.76 | 0.001 * | 0.72 | 0.005 * |
| Serum biomarkers | ||||
| CRP | −0.23 | 0.45 | 0.27 | 0.32 |
| Fibrinogen | −0.77 | 0.001 * | 0.756 | 0.001 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocchetti, M.T.; Bizzoca, D.; Moretti, L.; Ragni, E.; Moretti, F.L.; Vicenti, G.; Solarino, G.; Rizzello, A.; Petruzzella, V.; Palese, L.L.; et al. A Gel-Based Proteomic Analysis Reveals Synovial α-Enolase and Fibrinogen β-Chain Dysregulation in Knee Osteoarthritis: A Controlled Trial. J. Pers. Med. 2023, 13, 916. https://doi.org/10.3390/jpm13060916
Rocchetti MT, Bizzoca D, Moretti L, Ragni E, Moretti FL, Vicenti G, Solarino G, Rizzello A, Petruzzella V, Palese LL, et al. A Gel-Based Proteomic Analysis Reveals Synovial α-Enolase and Fibrinogen β-Chain Dysregulation in Knee Osteoarthritis: A Controlled Trial. Journal of Personalized Medicine. 2023; 13(6):916. https://doi.org/10.3390/jpm13060916
Chicago/Turabian StyleRocchetti, Maria Teresa, Davide Bizzoca, Lorenzo Moretti, Enrico Ragni, Francesco Luca Moretti, Giovanni Vicenti, Giuseppe Solarino, Alessandro Rizzello, Vittoria Petruzzella, Luigi Leonardo Palese, and et al. 2023. "A Gel-Based Proteomic Analysis Reveals Synovial α-Enolase and Fibrinogen β-Chain Dysregulation in Knee Osteoarthritis: A Controlled Trial" Journal of Personalized Medicine 13, no. 6: 916. https://doi.org/10.3390/jpm13060916
APA StyleRocchetti, M. T., Bizzoca, D., Moretti, L., Ragni, E., Moretti, F. L., Vicenti, G., Solarino, G., Rizzello, A., Petruzzella, V., Palese, L. L., Scacco, S., Banfi, G., Moretti, B., & Gnoni, A. (2023). A Gel-Based Proteomic Analysis Reveals Synovial α-Enolase and Fibrinogen β-Chain Dysregulation in Knee Osteoarthritis: A Controlled Trial. Journal of Personalized Medicine, 13(6), 916. https://doi.org/10.3390/jpm13060916













