Effectiveness of Radiomic ZOT Features in the Automated Discrimination of Oncocytoma from Clear Cell Renal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
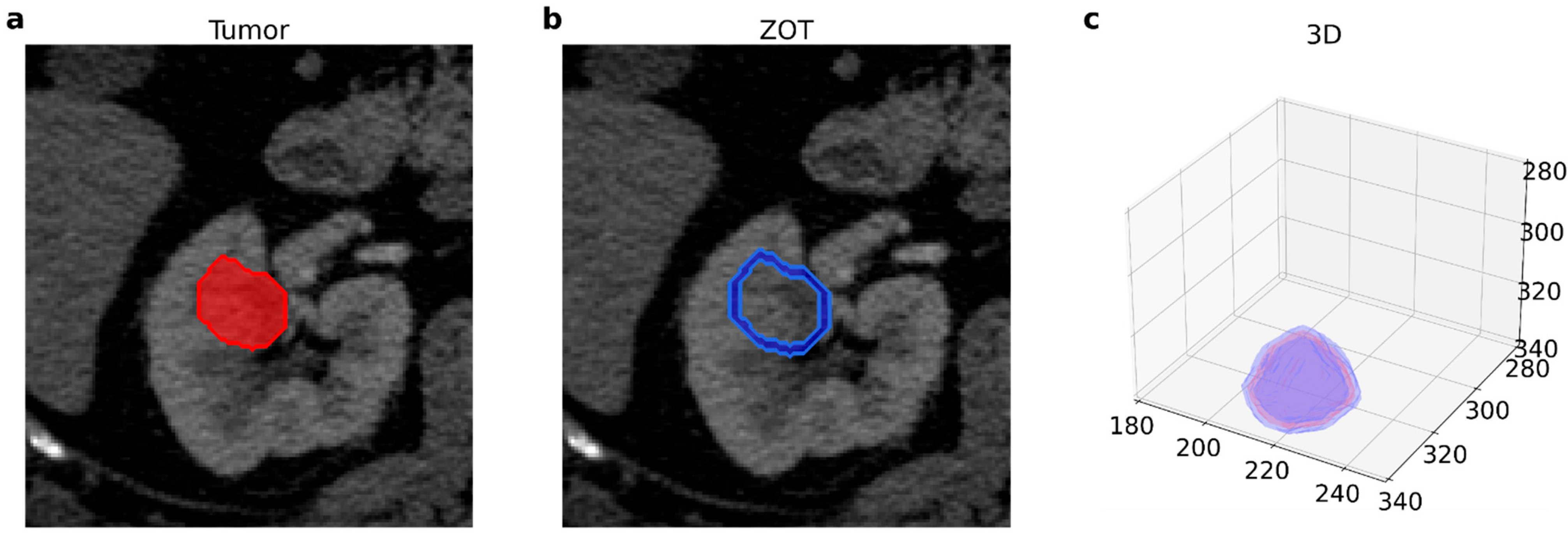
2.2. Image Acquisition and Radiomic Features Extraction
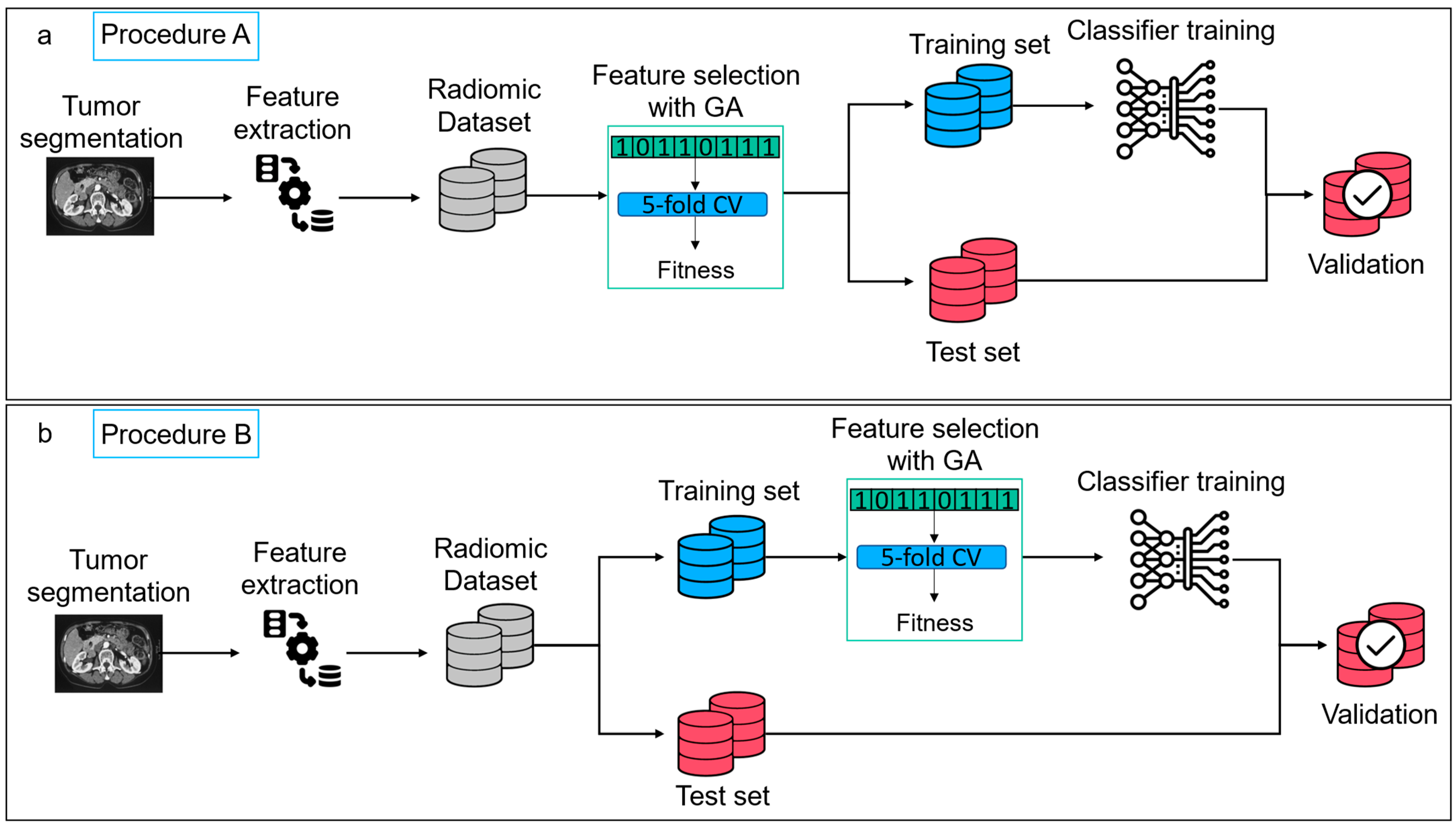
2.3. Feature Selection
2.4. Classification Model
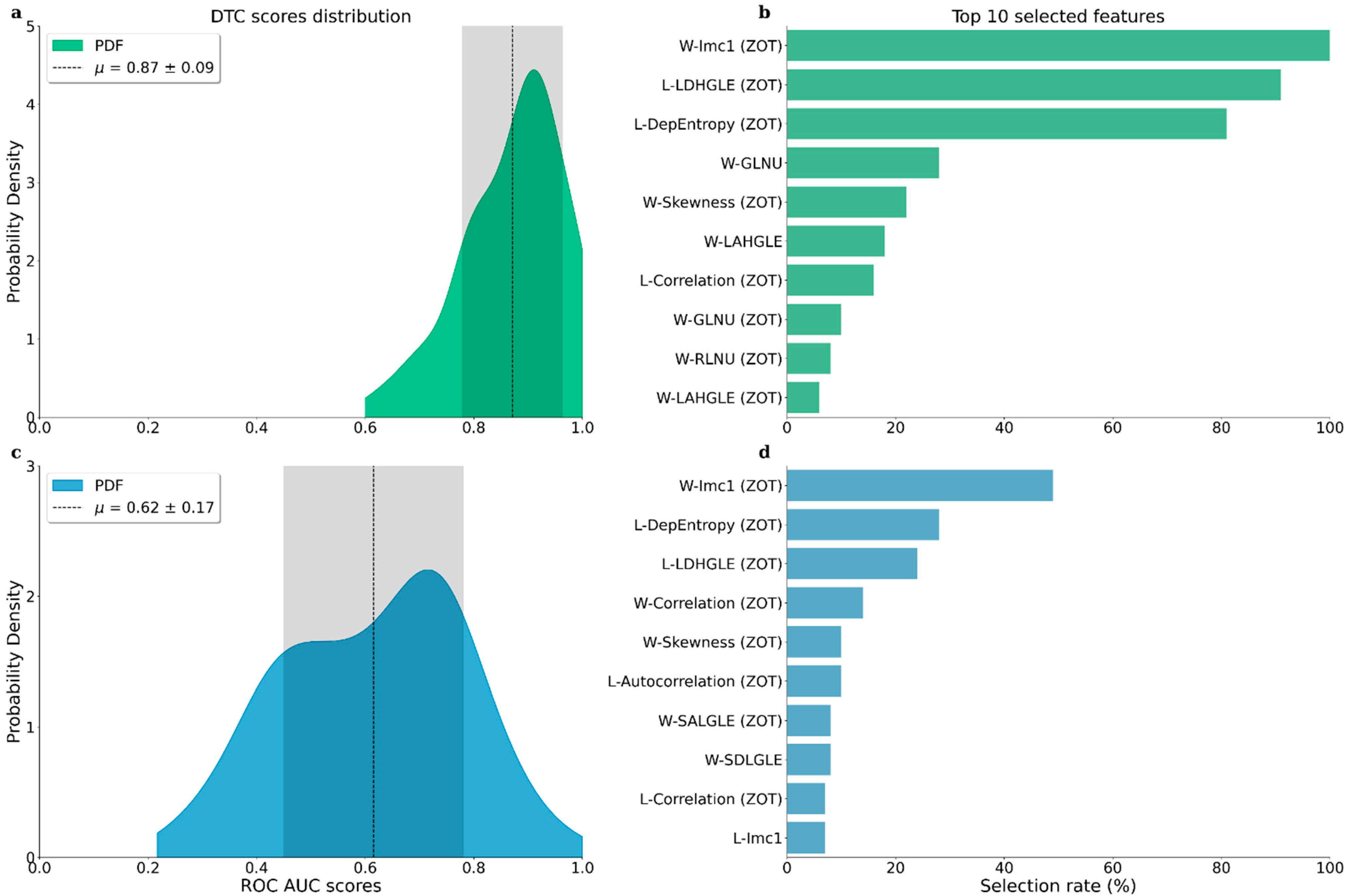
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Jonasch, E.; Walker, C.L.; Rathmell, W.K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 2021, 17, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Volpe, A.; Panzarella, T.; Rendon, R.A.; Haider, M.A.; Kondylis, F.I.; Jewett, M.A.S. The natural history of incidentally detected small renal masses. Cancer 2004, 100, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Gentili, F.; Bronico, I.; Maestroni, U.; Ziglioli, F.; Silini, E.M.; Buti, S.; de Filippo, M. Small renal masses (≤ 4 cm): Differentiation of oncocytoma from renal clear cell carcinoma using ratio of lesion to cortex attenuation and aorta–lesion attenuation difference (ALAD) on contrast-enhanced CT. Radiol. Med. 2020, 125, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Cai, F.; Shang, Y.N.; Ma, E.S.; Huang, Z.G.; Wang, W.; Lu, Y.; Zhang, X.Z. Differentiation of renal oncocytoma and renal clear cell carcinoma using relative CT enhancement ratio. Chin. Med. J. 2015, 128, 175–179. [Google Scholar] [CrossRef]
- Grajo, J.R.; Terry, R.S.; Ruoss, J.; Noennig, B.J.; Pavlinec, J.G.; Bozorgmehri, S.; Crispen, P.L.; Su, L.M. Using Aorta-Lesion-Attenuation Difference on Preoperative Contrast-enhanced Computed Tomography Scan to Differentiate between Malignant and Benign Renal Tumors. Urology 2019, 125, 123–130. [Google Scholar] [CrossRef]
- Lee-Felker, S.A.; Felker, E.R.; Tan, N.; Margolis, D.J.; Young, J.R.; Sayre, J.; Raman, S.S. Qualitative and quantitative MDCT features for differentiating clear cell renal cell carcinoma from other solid renal cortical masses. AJR Am. J. Roentgenol. 2014, 203, W516–W524. [Google Scholar] [CrossRef]
- Paño, B.; Soler, A.; Goldman, D.A.; Salvador, R.; Buñesch, L.; Sebastià, C.; Nicolau, C. Usefulness of multidetector computed tomography to differentiate between renal cell carcinoma and oncocytoma. A model validation. Br. J. Radiol. 2020, 93, 20200064. [Google Scholar] [CrossRef]
- Gaudiano, C.; Schiavina, R.; Vagnoni, V.; Busato, F.; Borghesi, M.; Bandini, M.; Di Carlo, M.; Brunocilla, E.; Martorana, G.; Golfieri, R. Can the multiphasic computed tomography be useful in the clinical management of small renal masses? Acta Radiol. Stockh. Swed. 1987 2017, 58, 625–633. [Google Scholar] [CrossRef]
- Kumar, V.; Gu, Y.; Basu, S.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.; Dekker, A.; Fenstermacher, D.; Goldgof, D.B.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging. 2012, 30, 1234–1248. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Yip, S.S.; Aerts, H.J.W.L. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef]
- Filitto, G.; Coppola, F.; Curti, N.; Giampieri, E.; Dall’Olio, D.; Merlotti, A.; Cattabriga, A.; Cocozza, M.A.; Tomassoni, M.T.; Remondini, D.; et al. Automated Prediction of the Response to Neoadjuvant Chemoradiotherapy in Patients Affected by Rectal Cancer. Cancers 2022, 14, 2231. [Google Scholar] [CrossRef]
- Carlini, G.; Curti, N.; Strolin, S.; Giampieri, E.; Sala, C.; Dall’Olio, D.; Merlotti, A.; Fanti, S.; Remondini, D.; Nanni, C.; et al. Prediction of Overall Survival in Cervical Cancer Patients Using PET/CT Radiomic Features. Appl. Sci. 2022, 12, 5946. [Google Scholar] [CrossRef]
- He, M.; Zhang, P.; Ma, X.; He, B.; Fang, C.; Jia, F. Radiomic Feature-Based Predictive Model for Microvascular Invasion in Patients With Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 574228. Available online: https://www.frontiersin.org/articles/10.3389/fonc.2020.574228 (accessed on 17 November 2022). [CrossRef]
- Zhang, X.; Ruan, S.; Xiao, W.; Shao, J.; Tian, W.; Liu, W.; Zhang, Z.; Wan, D.; Huang, J.; Huang, Q.; et al. Contrast-enhanced CT radiomics for preoperative evaluation of microvascular invasion in hepatocellular carcinoma: A two-center study. Clin. Transl. Med. 2020, 10, e111. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of Multiparametric MRI for Pretreatment Prediction of Pathologic Complete Response to Neoadjuvant Chemotherapy in Breast Cancer: A Multicenter Study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef]
- Ji, G.W.; Zhang, Y.D.; Zhang, H.; Zhu, F.P.; Wang, K.; Xia, Y.X.; Zhang, Y.D.; Jiang, W.J.; Li, X.C.; Wang, X.H. Biliary Tract Cancer at CT: A Radiomics-based Model to Predict Lymph Node Metastasis and Survival Outcomes. Radiology 2019, 290, 90–98. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Erdim, C.; Ates, E.; Kaya, O.K.; Kilickesmez, O. Radiomics of Renal Masses: Systematic Review of Reproducibility and Validation Strategies. Am. J. Roentgenol. 2020, 214, 129–136. [Google Scholar] [CrossRef]
- Li, Y.; Huang, X.; Xia, Y.; Long, L. Value of radiomics in differential diagnosis of chromophobe renal cell carcinoma and renal oncocytoma. Abdom. Radiol. 2020, 45, 3193–3201. [Google Scholar] [CrossRef]
- Wang, P.; Pei, X.; Yin, X.P.; Retn, J.L.; Wang, Y.; Ma, L.Y.; Du, X.G.; Gao, B.L. Radiomics models based on enhanced computed tomography to distinguish clear cell from non-clear cell renal cell carcinomas. Sci. Rep. 2021, 11, 13729. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, Q.; Tao, C.; Liu, J.; Nie, P.; Dong, C. A CT-based radiomics nomogram for differentiation of small masses (<4 cm) of renal oncocytoma from clear cell renal cell carcinoma. Abdom. Radiol. N. Y. 2021, 46, 5240–5249. [Google Scholar] [CrossRef]
- Baghdadi, A.; Aldhaam, N.A.; Elsayed, A.S.; Hussetin, A.A.; Cavuoto, L.A.; Kauffman, E.; Guru, K.A. Automated differentiation of benign renal oncocytoma and chromophobe renal cell carcinoma on computed tomography using deep learning. BJU Int. 2020, 125, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Soule, E.; Cui, E.; Shah, S.; Lall, C.; Sundaram, C.; Sandrasegaran, K. Usefulness of CT texture analysis in differentiating benign and malignant renal tumours. Clin. Radiol. 2020, 75, 108–115. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef]
- Renzulli, M.; Mottola, M.; Coppola, F.; Cocozza, M.A.; Malavasi, S.; Cattabriga, A.; Vara, G.; Ravaioli, M.; Cescon, M.; Vasuri, F.; et al. Automatically Extracted Machine Learning Features from Preoperative CT to Early Predict Microvascular Invasion in HCC: The Role of the Zone of Transition (ZOT). Cancers 2022, 14, 1816. [Google Scholar] [CrossRef]
- Zhang, L.; Yankelevitz, D.F.; Henschke, C.I.; Jirapatnakul, A.C.; Reeves, A.P.; Carter, D. Zone of Transition: A Potential Source of Error in Tumor Volume Estimation. Radiology 2010, 256, 633–639. [Google Scholar] [CrossRef]
- Bianchi, L.; Schiavina, R.; Bortolani, B.; Cercenelli, L.; Gaudiano, C.; Carpani, G.; Rustici, A.; Droghetti, M.; Mottaran, A.; Boschi, S.; et al. Interpreting nephrometry scores with three-dimensional virtual modelling for better planning of robotic partial nephrectomy and predicting complications. Urol. Oncol. 2021, 39, e1–e836. [Google Scholar] [CrossRef]
- Bianchi, L.; Schiavina, R.; Bortolani, B.; Cercenelli, L.; Gaudiano, C.; Mottaran, A.; Droghetti, M.; Chessa, F.; Boschi, S.; Molinaroli, E.; et al. Novel Volumetric and Morphological Parameters Derived from Three-dimensional Virtual Modeling to Improve Comprehension of Tumor’s Anatomy in Patients with Renal Cancer. Eur. Urol. Focus 2021, 8, 1300–1308. [Google Scholar] [CrossRef]
- Bianchi, L.; Barbaresi, U.; Cercenelli, L.; Bortolani, B.; Gaudiano, C.; Chessa, F.; Angiolini, A.; Lodi, S.; Porreca, A.; Bianchi, F.M.; et al. The Impact of 3D Digital Reconstruction on the Surgical Planning of Partial Nephrectomy: A Case-control Study. Still Time for a Novel Surgical Trend? Clin. Genitourin. Cancer 2020, 18, e669–e678. [Google Scholar] [CrossRef]
- Chong, H.H.; Yang, L.; Sheng, R.F.; Yu, Y.-L.; Wu, D.-J.; Rao, S.-X.; Yang, C.; Zetng, M.S. Multi-scale and multi-parametric radiomics of gadoxetate disodium-enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma ≤5 cm. Eur. Radiol. 2021, 31, 4824–4838. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.L.; Liu, Q.P.; Sun, S.W.; Zhang, J.; Zhu, F.P.; Yang, G.; Yan, X.; Zhang, Y.D.; Liu, X.S. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J. Hepatol. 2019, 70, 1133–1144. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets Tan, R.G.H.; Fillion Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Chen, M.; Yin, F.; Yu, Y.; Zhang, H.; Wen, G. CT-based multi-phase Radiomic models for differentiating clear cell renal cell carcinoma. Cancer Imaging 2021, 21, 42. [Google Scholar] [CrossRef]
- van Oostenbrugge, T.J.; Spenkelink, I.M.; Bokacheva, L.; Rusinetk, H.; van Amerongen, M.J.; Langenhuijsen, J.F.; Mulders, P.F.; Fütterer, J.J. Kidney tumor diffusion-weighted magnetic resonance imaging derived ADC histogram parameters combined with patient characteristics and tumor volume to discriminate oncocytoma from renal cell carcinoma. Eur. J. Radiol. 2021, 145, 110013. [Google Scholar] [CrossRef]
- Gao, R.; Qin, H.; Lin, P.; Ma, C.; Li, C.; Wen, R.; Huang, J.; Wan, D.; Wen, D.; Liang, Y.; et al. Development and Validation of a Radiomic Nomogram for Predicting the Prognosis of Kidney Renal Clear Cell Carcinoma. Front. Oncol. 2021, 11, 613668. Available online: https://www.frontiersin.org/articles/10.3389/fonc.2021.613668 (accessed on 11 July 2022). [CrossRef]
- Bianchi, L.; Schiavina, R.; Borghesi, M.; Chessa, F.; Casablanca, C.; Angiolini, A.; Ercolino, A.; Pultrone, C.V.; Mineo Bianchi, F.; Barbaresi, U.; et al. Which patients with clinical localized renal mass would achieve the trifecta after partial nephrectomy? The impact of surgical technique. Minerva Urol. E Nefrol. Ital. J. Urol. Nephrol. 2020, 72, 339–349. [Google Scholar] [CrossRef]
- Uroweb—European Association of Urology. EAU Guidelines on RCC—INTRODUCTION—Uroweb. Available online: https://uroweb.org/guidelines/renal-cell-carcinoma (accessed on 30 September 2022).
- Bianchi, L.; Chessa, F.; Piazza, P.; Ercolino, A.; Mottaran, A.; Recenti, D.; Serra, C.; Gaudiano, C.; Cappelli, A.; Modestino, F.; et al. Percutaneous ablation or minimally invasive partial nephrectomy for cT1a renal masses? A propensity score-matched analysis. Int. J. Urol. Off. J. Jpn. Urol. Assoc. 2022, 29, 222–228. [Google Scholar] [CrossRef]
- Bianchi, L.; Mineo Bianchi, F.; Chessa, F.; Barbaresi, U.; Casablanca, C.; Piazza, P.; Mottaran, A.; Droghetti, M.; Roveroni, C.; Balestrazzi, E.; et al. Percutaneous tumor ablation versus partial nephrectomy for small renal mass: The impact of histologic variant and tumor size. Minerva Urol. Nephrol. 2021, 73, 581–590. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carlini, G.; Gaudiano, C.; Golfieri, R.; Curti, N.; Biondi, R.; Bianchi, L.; Schiavina, R.; Giunchi, F.; Faggioni, L.; Giampieri, E.; et al. Effectiveness of Radiomic ZOT Features in the Automated Discrimination of Oncocytoma from Clear Cell Renal Cancer. J. Pers. Med. 2023, 13, 478. https://doi.org/10.3390/jpm13030478
Carlini G, Gaudiano C, Golfieri R, Curti N, Biondi R, Bianchi L, Schiavina R, Giunchi F, Faggioni L, Giampieri E, et al. Effectiveness of Radiomic ZOT Features in the Automated Discrimination of Oncocytoma from Clear Cell Renal Cancer. Journal of Personalized Medicine. 2023; 13(3):478. https://doi.org/10.3390/jpm13030478
Chicago/Turabian StyleCarlini, Gianluca, Caterina Gaudiano, Rita Golfieri, Nico Curti, Riccardo Biondi, Lorenzo Bianchi, Riccardo Schiavina, Francesca Giunchi, Lorenzo Faggioni, Enrico Giampieri, and et al. 2023. "Effectiveness of Radiomic ZOT Features in the Automated Discrimination of Oncocytoma from Clear Cell Renal Cancer" Journal of Personalized Medicine 13, no. 3: 478. https://doi.org/10.3390/jpm13030478
APA StyleCarlini, G., Gaudiano, C., Golfieri, R., Curti, N., Biondi, R., Bianchi, L., Schiavina, R., Giunchi, F., Faggioni, L., Giampieri, E., Merlotti, A., Dall’Olio, D., Sala, C., Pandolfi, S., Remondini, D., Rustici, A., Pastore, L. V., Scarpetti, L., Bortolani, B., ... Castellani, G. (2023). Effectiveness of Radiomic ZOT Features in the Automated Discrimination of Oncocytoma from Clear Cell Renal Cancer. Journal of Personalized Medicine, 13(3), 478. https://doi.org/10.3390/jpm13030478













