Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges
Abstract
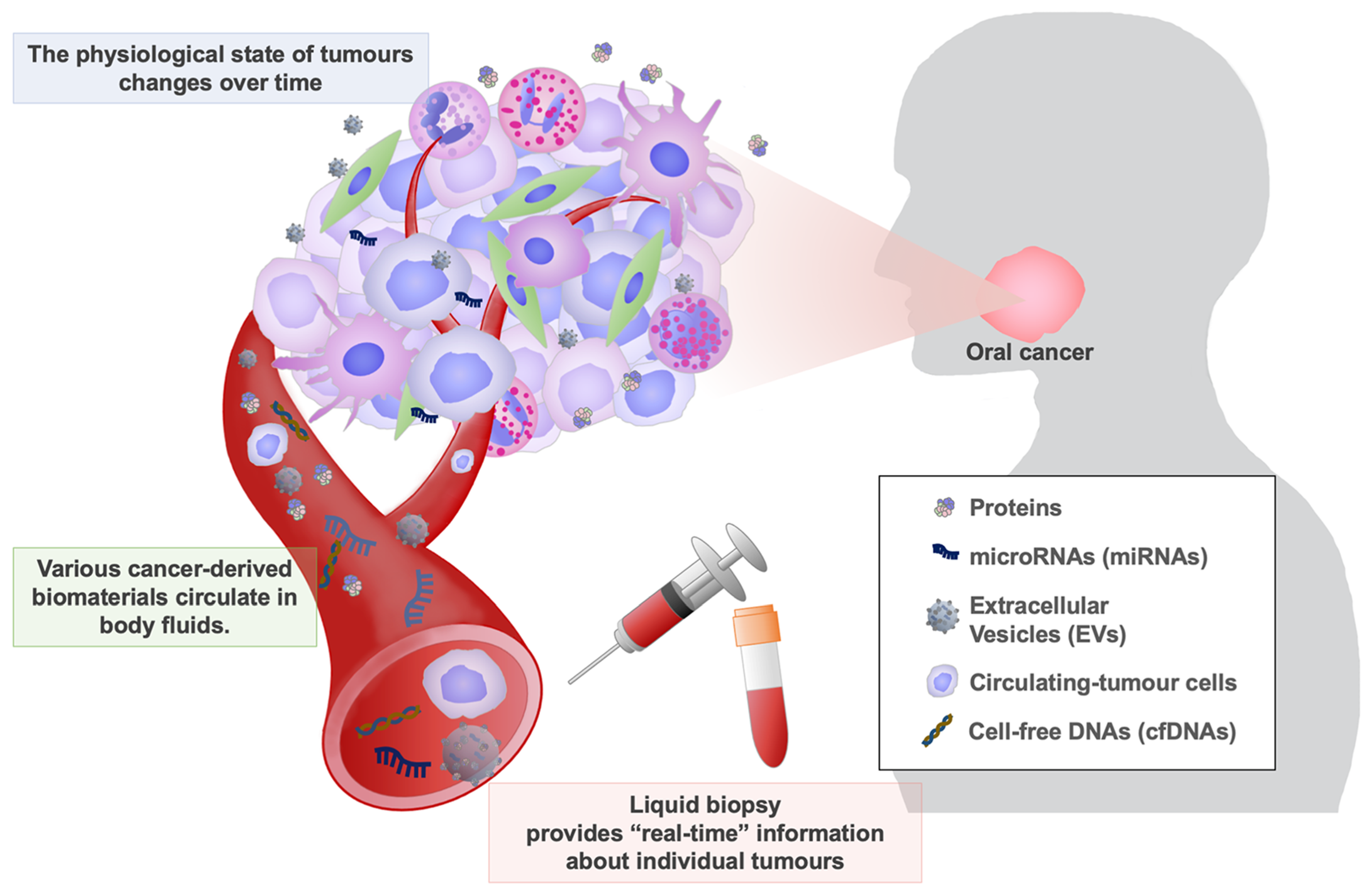
1. Introduction
2. Liquid Biopsy for Cancer Detection
2.1. Conventional Cancer Markers
2.2. miRNAs for Cancer Detection
2.3. Extracellular Vesicles for Cancer Detection
2.4. Circulating Cell-Free DNAs for Cancer Detection
3. Biomarkers for Predicting Therapeutic Efficacy
3.1. Mutation Status of Genes in Cancer Cells for Predicting Therapeutic Efficacy
3.2. EVs for Predicting Therapeutic Efficacy
3.3. Circulating Tumour Cells and ctDNAs for Predicting Therapeutic Efficacy
4. Liquid Biopsy for Prognostic Biomarkers of Oral Cancer
4.1. miRNAs and EVs for Predicting Patient Outcomes
5. Limitations and Challenges of Novel Biomarkers for Oral Cancer
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hudson, T.J.; Anderson, W.; Artez, A.; Barker, A.D.; Bell, C.; Bernabé, R.R.; Bhan, M.K.; Calvo, F.; Eerola, I.; Gerhard, D.S.; et al. International Network of Cancer Genome Projects. Nature 2010, 464, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Crosetto, N.; Bienko, M.; van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 2014, 16, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Giesen, C.; Wang, H.A.O.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schüffler, P.J.; Grolimund, D.; Buhmann, J.M.; Brandt, S.; et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S.; Nittner, D.; Burton, O.; Bassez, A.; Decaluwé, H.; Pircher, A.; Van den Eynde, K.; et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018, 24, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Jia, D. Application of Single-Cell Multi-Omics in Dissecting Cancer Cell Plasticity and Tumor Heterogeneity. Front. Mol. Biosci. 2021, 8, 757024. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Garattini, S.; Nerini, I.F.; D’Incalci, M. Not only tumor but also therapy heterogeneity. Ann. Oncol. 2017, 29, 13–18. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Aoki, M.; Shoji, H.; Kashiro, A.; Takeuchi, K.; Shimizu, Y.; Honda, K. Prospects for Comprehensive Analyses of Circulating Tumor Cells in Tumor Biology. Cancers. 2020, 12, 1135. [Google Scholar] [CrossRef]
- Biomarkers Definitions Working Group; Atkinson, A.J., Jr.; Colburn, W.A.; DeGruttola, V.G.; DeMets, D.L.; Downing, G.J.; Hoth, D.F.; Oates, J.A.; Peck, C.C.; Spilker, B.A.; et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- Wongsurakiat, P.; Wongbunnate, S.; Dejsomritrutai, W.; Charoenratanakul, S.; Tscheikuna, J.; Youngchaiyud, P.; Pushpakom, R.; Maranetra, N.; Nana, A.; Chierakul, N.; et al. Diagnostic value of bronchoalveolar lavage and postbronchoscopic sputum cytology in peripheral lung cancer. Respirology 1998, 3, 131–137. [Google Scholar] [CrossRef]
- Hu, T.; Wolfram, J.; Srivastava, S. Extracellular Vesicles in Cancer Detection: Hopes and Hypes. Trends Cancer 2020, 7, 122–133. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef]
- Mody, M.D.; Rocco, J.W.; Yom, S.S.; Haddad, R.I.; Saba, N.F. Head and neck cancer. Lancet 2021, 398, 2289–2299. [Google Scholar] [CrossRef]
- Bagan, J.; Sarrion, G.; Jimenez, Y. Oral cancer: Clinical features. Oral Oncol. 2010, 46, 414–417. [Google Scholar] [CrossRef]
- Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef]
- Tiwana, M.; Wu, J.; Hay, J.; Wong, F.; Cheung, W.; Olson, R. 25 Year survival outcomes for squamous cell carcinomas of the head and neck: Population-based outcomes from a Canadian province. Oral Oncol. 2014, 50, 651–656. [Google Scholar] [CrossRef]
- Nakamura, K.; Hiyake, N.; Hamada, T.; Yokoyama, S.; Mori, K.; Yamashiro, K.; Beppu, M.; Sagara, Y.; Sagara, Y.; Sugiura, T. Circulating microRNA Panel as a Potential Novel Biomarker for Oral Squamous Cell Carcinoma Diagnosis. Cancers 2021, 13, 449. [Google Scholar] [CrossRef]
- Romani, C.; Salviato, E.; Paderno, A.; Zanotti, L.; Ravaggi, A.; Deganello, A.; Berretti, G.; Gualtieri, T.; Marchini, S.; D’Incalci, M.; et al. Genome-wide study of salivary miRNAs identifies miR-423-5p as promising diagnostic and prognostic biomarker in oral squamous cell carcinoma. Theranostics 2021, 11, 2987–2999. [Google Scholar] [CrossRef] [PubMed]
- Bigagli, E.; Locatello, L.G.; Di Stadio, A.; Maggiore, G.; Valdarnini, F.; Bambi, F.; Gallo, O.; Luceri, C. Extracellular vesicles miR-210 as a potential biomarker for diagnosis and survival prediction of oral squamous cell carcinoma patients. J. Oral Pathol. Med. 2021, 51, 350–357. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2019, 121, 109553. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018, 18, 439. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, E.; Sakakura, H.; Mii, S.; Yamamoto, N.; Hibi, H.; Asai, M.; Takahashi, M. Detection of serum/salivary exosomal Alix in patients with oral squamous cell carcinoma. Oral Dis. 2020, 27, 439–447. [Google Scholar] [CrossRef]
- Yamana, K.; Inoue, J.; Yoshida, R.; Sakata, J.; Nakashima, H.; Arita, H.; Kawaguchi, S.; Gohara, S.; Nagao, Y.; Takeshita, H.; et al. Extracellular vesicles derived from radioresistant oral squamous cell carcinoma cells contribute to the acquisition of radioresistance via the miR-503-3p-BAK axis. J. Extracell. Vesicles. 2021, 10, e12169. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; López-Cedrún, J.L.; Lago-Lestón, R.M.; Abalo, A.; Rubin-Roger, G.; Salgado-Barreira, Á.; López-López, R.; Muinelo-Romay, L.; Suárez-Cunqueiro, M.M. Integrity and quantity of salivary cell-free DNA as a potential molecular biomarker in oral cancer: A preliminary study. J. Oral Pathol. Med. 2022, 51, 429–435. [Google Scholar] [CrossRef]
- Sayal, L.; Hamadah, O.; Almasri, A.; Idrees, M.; Thomson, P.; Kujan, O. Saliva-based cell-free DNA and cell-free mitochondrial DNA in head and neck cancers have promising screening and early detection role. J. Oral Pathol. Med. 2022. [Google Scholar] [CrossRef]
- Yang, K.; Yuan, C.; Tang, H.; Chen, D. Diagnostic values of serum tumor markers Cyfra21-1, SCCAg, ferritin, CEA, CA19-9, and AFP in oral/oropharyngeal squamous cell carcinoma. OncoTargets Ther. 2016, 9, 3381–3386. [Google Scholar] [CrossRef]
- Kurokawa, H.; Tsuru, S.; Okada, M.; Nakamura, T.; Kajiyama, M. Evaluation of tumor markers in patients with squamous cell carcinoma in the oral cavity. Int. J. Oral Maxillofac. Surg. 1993, 22, 35–38. [Google Scholar] [CrossRef]
- Shitrit, D.; Zingerman, B.; Shitrit, A.B.-G.; Shlomi, D.; Kramer, M.R. Diagnostic Value of CYFRA 21-1, CEA, CA 19-9, CA 15-3, and CA 125 Assays in Pleural Effusions: Analysis of 116 Cases and Review of the Literature. Oncologist 2005, 10, 501–507. [Google Scholar] [CrossRef]
- Hammarström, S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999, 9, 67–81. [Google Scholar] [CrossRef]
- Mizejewski, G.J. Physiology of Alpha-Fetoprotein as a Biomarker for Perinatal Distress: Relevance to Adverse Pregnancy Outcome. Exp. Biol. Med. 2007, 232, 993–1004. [Google Scholar] [CrossRef]
- Hernández, J.; Thompson, I.M. Prostate-specific antigen: A review of the validation of the most commonly used cancer biomarker. Cancer 2004, 101, 894–904. [Google Scholar] [CrossRef]
- Maruo, T.; Yoshida, S.; Samoto, T.; Tateiwa, Y.; Peng, X.; Takeuchi, S.; Motoyama, S. Factors Regulating SCC Antigen Expression in Squamous Cell Carcinoma of the Uterine Cervix. Tumor Biol. 1998, 19, 494–504. [Google Scholar] [CrossRef]
- Hamada, E.; Taniguchi, T.; Baba, S.; Maekawa, M. Investigation of unexpected serum CA19-9 elevation in Lewis-negative cancer patients. Ann. Clin. Biochem. Int. J. Biochem. Lab. Med. 2012, 49, 266–272. [Google Scholar] [CrossRef]
- Munkley, J. The glycosylation landscape of pancreatic cancer (Review). Oncol. Lett. 2019, 17, 2569–2575. [Google Scholar] [CrossRef]
- Silverman, N.A.; Alexander, J.C., Jr.; Chretien, P.B. CEA levels in head and neck cancer. Cancer 1976, 37, 2204–2211. [Google Scholar] [CrossRef]
- Fischbach, W.; Meyer, T.; Barthel, K. Squamous cell carcinoma antigen in the diagnosis and treatment follow-up of oral and facial squamous cell carcinoma. Cancer 1990, 65, 1321–1324. [Google Scholar] [CrossRef]
- Doweck, I.; Barak, M.; Greenberg, E.; Uri, N.; Kellner, J.; Lurie, M.; Gruener, N. Cyfra 21-1: A New Potential Tumor Marker for Squamous Cell Carcinoma of Head and Neck. Arch. Otolaryngol. Neck Surg. 1995, 121, 177–181. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of MicroRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Geekiyanage, H.; Rayatpisheh, S.; Wohlschlegel, J.A.; Brown, R.; Ambros, V. Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proc. Natl. Acad. Sci. USA 2020, 117, 24213–24223. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Kato, K.; Oono, K.; Tsuchiya, N.; Sudo, K.; Shimomura, A.; Tamura, K.; Shiino, S.; Kinoshita, T.; Daiko, H.; et al. Prediction of tissue-of-origin of early stage cancers using serum miRNomes. JNCI Cancer Spectr. 2022, 7, pkac080. [Google Scholar] [CrossRef]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef]
- Shiino, S.; Matsuzaki, J.; Shimomura, A.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Aoki, Y.; Yoshida, M.; Tamura, K.; Kato, K.; et al. Serum miRNA–based Prediction of Axillary Lymph Node Metastasis in Breast Cancer. Clin. Cancer Res. 2019, 25, 1817–1827. [Google Scholar] [CrossRef]
- Usuba, W.; Urabe, F.; Yamamoto, Y.; Matsuzaki, J.; Sasaki, H.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Kato, K.; et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2018, 110, 408–419. [Google Scholar] [CrossRef]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.-I.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef]
- Asano, N.; Matsuzaki, J.; Ichikawa, M.; Kawauchi, J.; Takizawa, S.; Aoki, Y.; Sakamoto, H.; Yoshida, A.; Kobayashi, E.; Tanzawa, Y.; et al. A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat. Commun. 2019, 10, 1299. [Google Scholar] [CrossRef]
- Urabe, F.; Matsuzaki, J.; Yamamoto, Y.; Kimura, T.; Hara, T.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Sakamoto, H.; et al. Large-scale Circulating microRNA Profiling for the Liquid Biopsy of Prostate Cancer. Clin. Cancer Res. 2019, 25, 3016–3025. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kondo, S.; Matsuzaki, J.; Esaki, M.; Okusaka, T.; Shimada, K.; Murakami, Y.; Enomoto, M.; Tamori, A.; Kato, K.; et al. Highly Sensitive Circulating MicroRNA Panel for Accurate Detection of Hepatocellular Carcinoma in Patients with Liver Disease. Hepatol. Commun. 2019, 4, 284–297. [Google Scholar] [CrossRef]
- Mehterov, N.; Sacconi, A.; Pulito, C.; Vladimirov, B.; Haralanov, G.; Pazardjikliev, D.; Nonchev, B.; Berindan-Neagoe, I.; Blandino, G.; Sarafian, V. A novel panel of clinically relevant miRNAs signature accurately differentiates oral cancer from normal mucosa. Front. Oncol. 2022, 12, 1072579. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III–dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Reiter, J.G.; Baretti, M.; Gerold, J.M.; Makohon-Moore, A.P.; Daud, A.; Iacobuzio-Donahue, C.A.; Azad, N.S.; Kinzler, K.W.; Nowak, M.A.; Vogelstein, B. An analysis of genetic heterogeneity in untreated cancers. Nat. Rev. Cancer 2019, 19, 639–650. [Google Scholar] [CrossRef]
- Giaccone, G.; Herbst, R.S.; Manegold, C.; Scagliotti, G.; Rosell, R.; Miller, V.; Natale, R.B.; Schiller, J.H.; Von Pawel, J.; Pluzanska, A.; et al. Gefitinib in Combination with Gemcitabine and Cisplatin in Advanced Non–Small-Cell Lung Cancer: A Phase III Trial—INTACT 1. J. Clin. Oncol. 2004, 22, 777–784. [Google Scholar] [CrossRef]
- Herbst, R.S.; Giaccone, G.; Schiller, J.H.; Natale, R.B.; Miller, V.; Manegold, C.; Scagliotti, G.; Rosell, R.; Oliff, I.; Reeves, J.A.; et al. Gefitinib in Combination with Paclitaxel and Carboplatin in Advanced Non–Small-Cell Lung Cancer: A Phase III Trial—INTACT 2. J. Clin. Oncol. 2004, 22, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Miller, V.; Zakowski, M.; Doherty, J.; Politi, K.; Sarkaria, I.; Singh, B.; Heelan, R.; Rusch, V.; Fulton, L.; et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA 2004, 101, 13306–13311. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.-L.; Thongprasert, S.; Yang, C.-H.; Chu, D.-T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef]
- Sharma, S.V.; Bell, D.W.; Settleman, J.; Haber, D.A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar] [CrossRef]
- Pao, W.; Miller, V.A.; Politi, K.A.; Riely, G.J.; Somwar, R.; Zakowski, M.F.; Kris, M.G.; Varmus, H. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005, 2, e73. [Google Scholar] [CrossRef]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Egebjerg, K.; Harwood, C.D.; Woller, N.C.; Kristensen, C.A.; Mau-Sørensen, M. HER2 Positivity in Histological Subtypes of Salivary Gland Carcinoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2021, 11, 693394. [Google Scholar] [CrossRef]
- Takahashi, H.; Tada, Y.; Saotome, T.; Akazawa, K.; Ojiri, H.; Fushimi, C.; Masubuchi, T.; Matsuki, T.; Tani, K.; Osamura, R.Y.; et al. Phase II Trial of Trastuzumab and Docetaxel in Patients with Human Epidermal Growth Factor Receptor 2–Positive Salivary Duct Carcinoma. J. Clin. Oncol. 2019, 37, 125–134. [Google Scholar] [CrossRef]
- Li, Q.; Lv, M.; Lv, L.; Cao, N.; Zhao, A.; Chen, J.; Tang, X.; Luo, R.; Yu, S.; Zhou, Y.; et al. Identifying HER2 from serum-derived exosomes in advanced gastric cancer as a promising biomarker for assessing tissue HER2 status and predicting the efficacy of trastuzumab-based therapy. Cancer Med. 2022. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Yu, G.; Sun, Z.; Wang, T.; Tian, X.; Duan, X.; Zhang, C. Exosomal miR-1246 and miR-155 as predictive and prognostic biomarkers for trastuzumab-based therapy resistance in HER2-positive breast cancer. Cancer Chemother. Pharmacol. 2020, 86, 761–772. [Google Scholar] [CrossRef]
- Onidani, K.; Shoji, H.; Kakizaki, T.; Yoshimoto, S.; Okaya, S.; Miura, N.; Sekikawa, S.; Furuta, K.; Lim, C.T.; Shibahara, T.; et al. Monitoring of cancer patients via next-generation sequencing of patient-derived circulating tumor cells and tumor DNA. Cancer Sci. 2019, 110, 2590–2599. [Google Scholar] [CrossRef]
- Qu, X.; Leung, T.C.N.; Ngai, S.-M.; Tsai, S.-N.; Thakur, A.; Li, W.-K.; Lee, Y.; Leung, L.; Ng, T.-H.; Yam, J.; et al. Proteomic Analysis of Circulating Extracellular Vesicles Identifies Potential Biomarkers for Lymph Node Metastasis in Oral Tongue Squamous Cell Carcinoma. Cells 2021, 10, 2179. [Google Scholar] [CrossRef]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef]
- Neal, R.D.; Johnson, P.; Clarke, C.A.; Hamilton, S.A.; Zhang, N.; Kumar, H.; Swanton, C.; Sasieni, P. Cell-Free DNA–Based Multi-Cancer Early Detection Test in an Asymptomatic Screening Population (NHS-Galleri): Design of a Pragmatic, Prospective Randomised Controlled Trial. Cancers 2022, 14, 4818. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J. Current and Future Perspectives of Cell-Free DNA in Liquid Biopsy. Curr. Issues Mol. Biol. 2022, 44, 2695–2709. [Google Scholar] [CrossRef]
| Molecular Target | Target Name | Body Fluid | Purpose | Sensitivity (%) | Specificity (%) | Reference |
|---|---|---|---|---|---|---|
| MicroRNAs | miR-24, miR-20a, miR-122, miR-150, miR-4419a, and miR-5100 | Blood (serum) | Cancer detection | 55.0 | 92.5 | [20] |
| miR-106-5p, miR-423-5p, and miR-193b-3p | Saliva | Cancer detection | 85.4 | 85.1 | [21] | |
| Extracellular vesicles (EVs) | miR-210 in EVs | Blood (plasma) | Cancer detection & prognostic marker | 92.3 | 86.6 | [22] |
| miR-24-3p in EVs | Saliva | Cancer detection | 64.4 | 80.0 | [23] | |
| miR-512-3p and miR-412-3p in EVs | Saliva | Cancer detection | - † | - † | [24] | |
| Alix in EVs | Blood (serum) | Cancer detection | 34.5 | 100.0 | [25] | |
| Alix in EVs | Saliva | Cancer detection | 34.8 | 100.0 | [25] | |
| miR-503-3p in EVs | Blood (serum) | Prognostic marker | - † | - † | [26] | |
| Cell-free DNAs | Human Arthobacter luteus (ALU) retrotransposon | Saliva | Cancer detection | 82.1 | 70.2 | [27] |
| beta-2-microglobulin | Saliva | Cancer detection | 71.4 | 81.4 | [28] | |
| Mitochondrial gene | Saliva | Cancer detection | 74.2 | 82.9 | [28] | |
| Proteins | CA19-9 | Blood (serum) | Cancer detection | - † | - † | [29] |
| CEA | Blood (serum) | Cancer detection | 66.27 | 61.21 | [29] | |
| SCC-Ag | Blood (serum) | Cancer detection | 73.37 | 68.10 | [29] | |
| IAP | Blood (serum) | Cancer detection | - † | - † | [30] | |
| Cyfra | Blood (serum) | Cancer detection | 60.36 | 81.03 | [29] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naito, Y.; Honda, K. Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges. J. Pers. Med. 2023, 13, 303. https://doi.org/10.3390/jpm13020303
Naito Y, Honda K. Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges. Journal of Personalized Medicine. 2023; 13(2):303. https://doi.org/10.3390/jpm13020303
Chicago/Turabian StyleNaito, Yutaka, and Kazufumi Honda. 2023. "Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges" Journal of Personalized Medicine 13, no. 2: 303. https://doi.org/10.3390/jpm13020303
APA StyleNaito, Y., & Honda, K. (2023). Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges. Journal of Personalized Medicine, 13(2), 303. https://doi.org/10.3390/jpm13020303






