Maximizing Small Biopsy Patient Samples: Unified RNA-Seq Platform Assessment of over 120,000 Patient Biopsies
Abstract
1. Introduction
2. Materials and Methods
2.1. Machine Learning-Based Classifiers
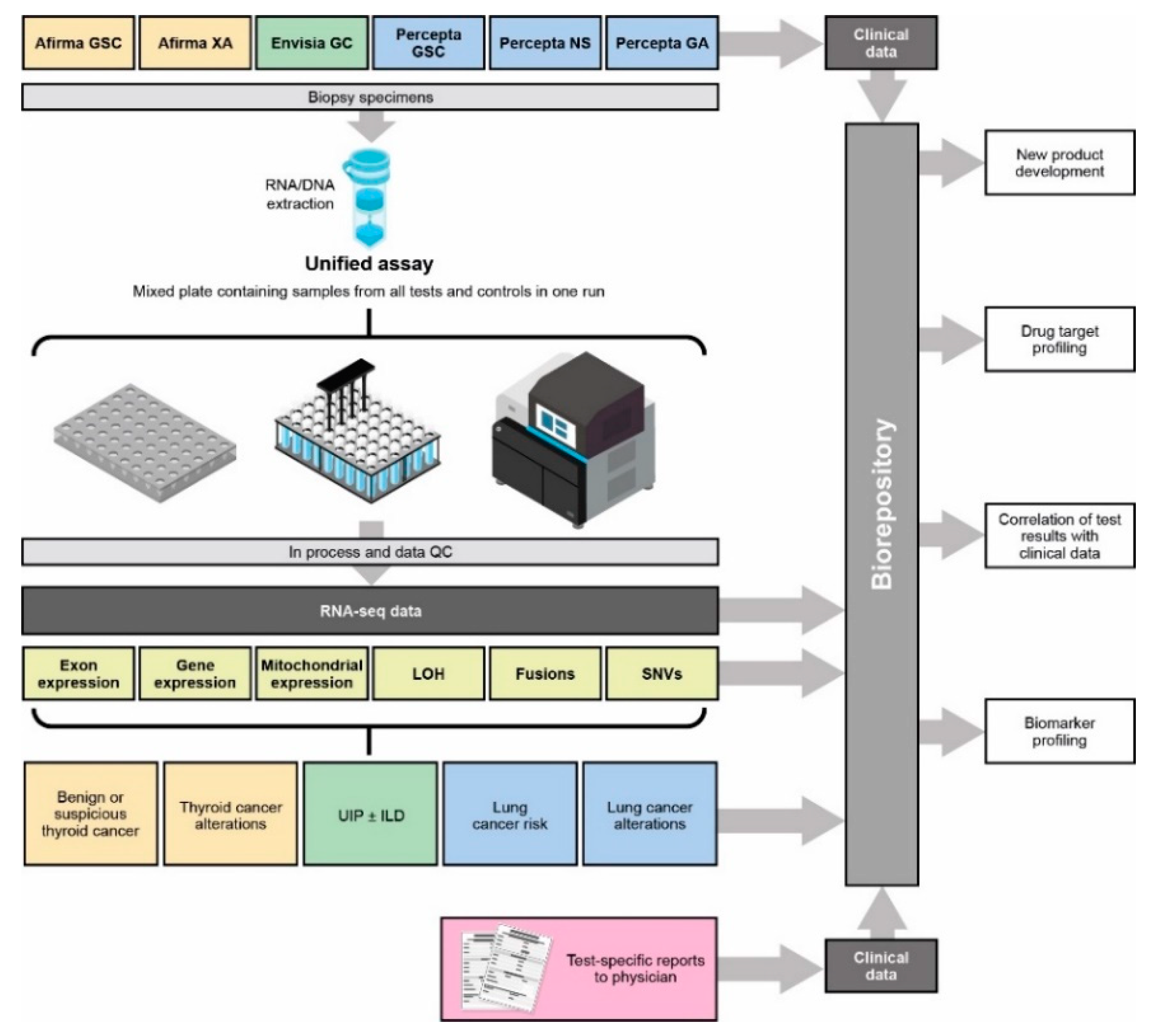
2.2. Sample Processing
2.3. RNA-Seq
2.4. Bioinformatics Pipeline
2.5. Downstream Analysis
2.6. Fusion Identification
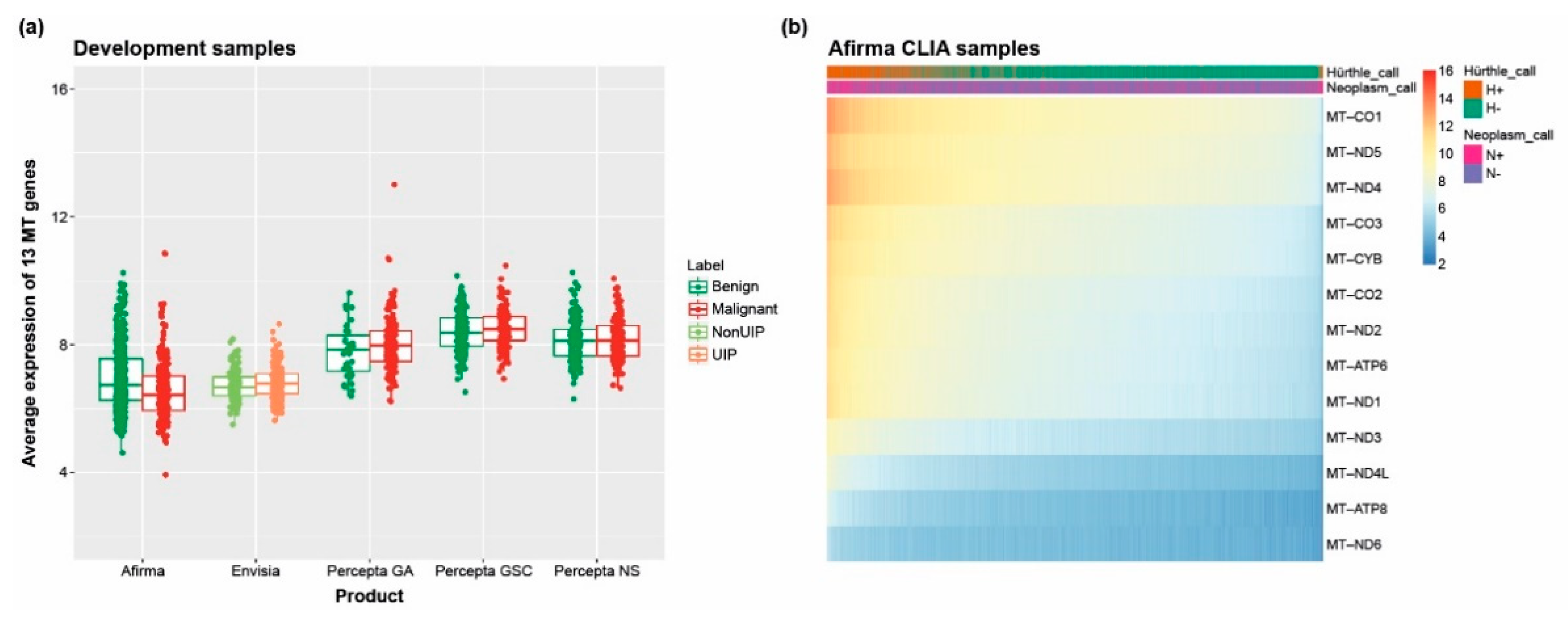
2.7. Mitochondrial Gene Expression and Hürthle Classification
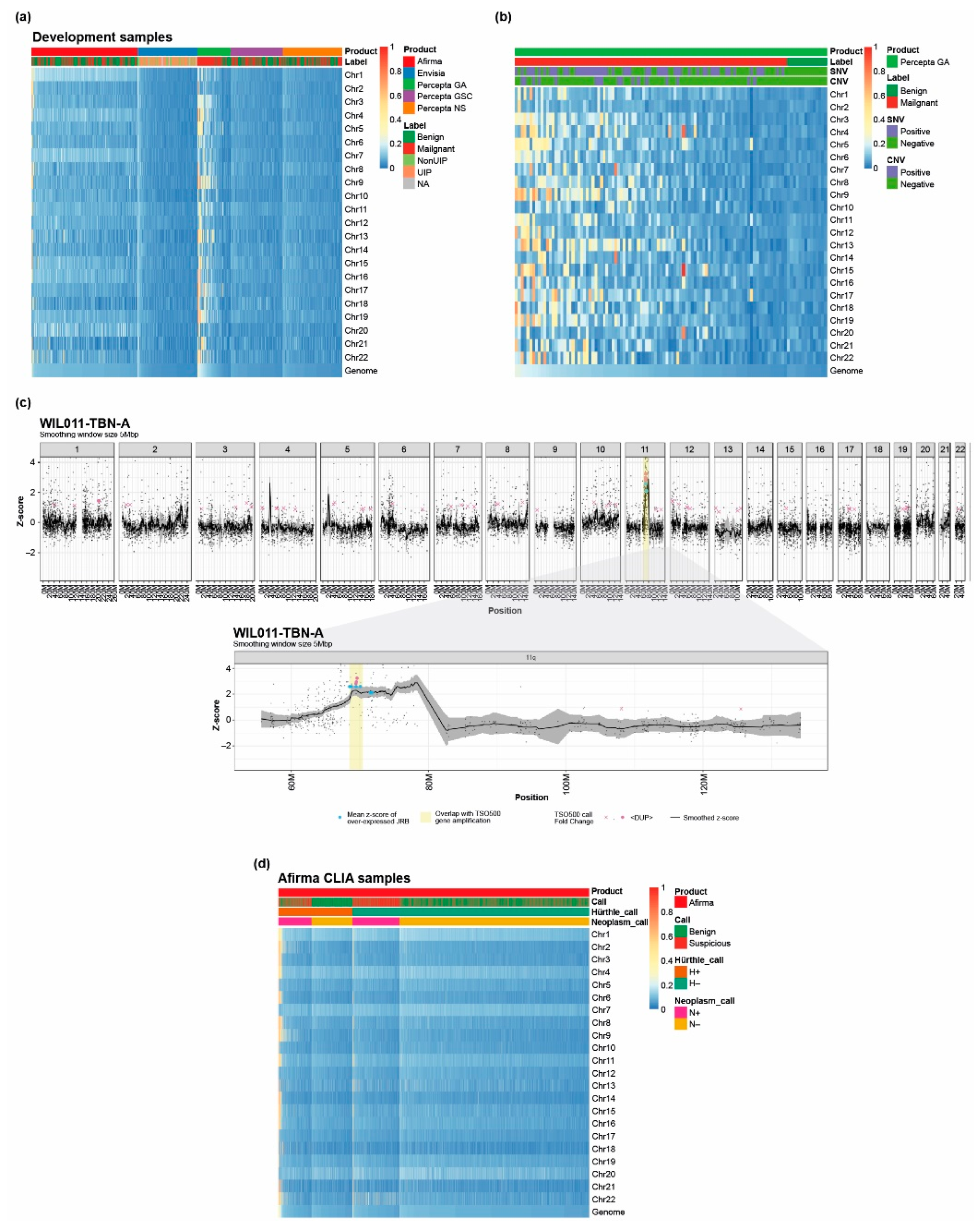
2.8. LOH Score Determination
2.9. Copy Number Variation (CNV) Identification
3. Results
3.1. Patient Samples
3.2. Use of an “Open” Platform and a Large Cohort Identifies Known and Novel Fusions, Including Rare and Unexpected Variant Combinations
3.3. Measuring Mitochondrial Gene Expression Identifies Challenging Subtypes
3.4. Loss of Heterozygosity Can Be Measured by the Unified Assay
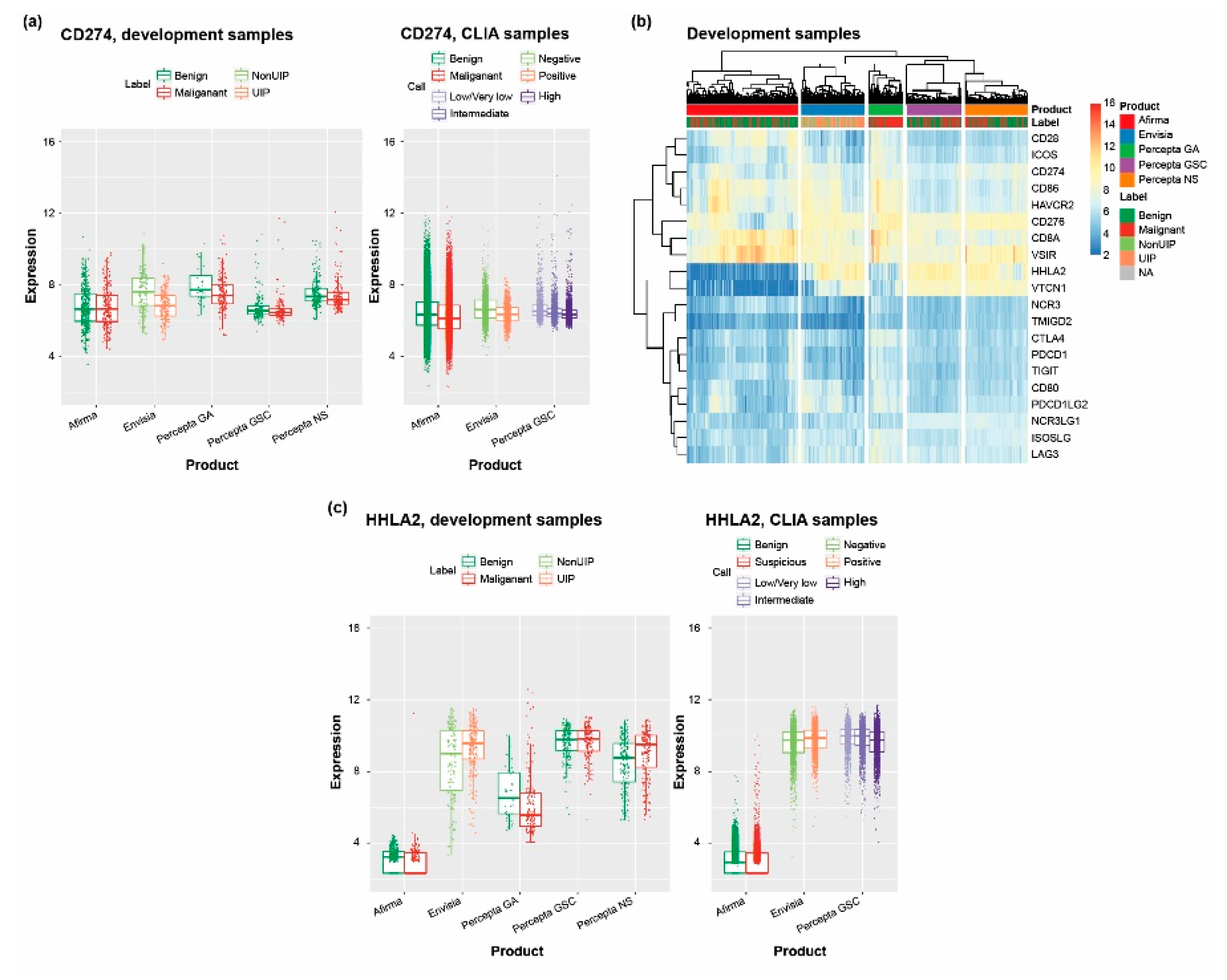
3.5. Scanning the Transcriptome of Clinical Samples for Drug Targets Reveals Potentially Useful Therapeutic Information
4. Discussion
4.1. Establishment of an Innovative Platform for Clinical Diagnostic Testing
4.2. Use of an Enrichment-Based Approach to Facilitate Diagnosis and Treatment Decisions
4.3. Identification of Disease-Related Variants by Comprehensive Testing
4.4. RNA-Seq–Based Identification of Potential Therapeutic Targets
4.5. Realizing the Potential of the Unified Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Smedley, D.; Smith, K.R.; Martin, A.; Thomas, E.A.; McDonagh, E.M.; Cipriani, V.; Ellingford, J.M.; Arno, G.; Tucci, A.; Vandrovcova, J.; et al. 100,000 genomes pilot on rare-disease diagnosis in health care—Preliminary report. N. Engl. J. Med. 2021, 385, 1868–1880. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef]
- Sokolenko, A.P.; Imyanitov, E.N. Molecular diagnostics in clinical oncology. Front. Mol. Biosci. 2018, 5, 76. [Google Scholar] [CrossRef] [PubMed]
- Correia, L.; Magno, R.; Xavier, J.M.; de Almeida, B.P.; Duarte, I.; Esteves, F.; Ghezzo, M.; Eldridge, M.; Sun, C.; Bosma, A.; et al. Allelic expression imbalance of PIK3CA mutations is frequent in breast cancer and prognostically significant. NPJ Breast Cancer 2022, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Cummings, B.B.; Marshall, J.L.; Tukiainen, T.; Lek, M.; Donkervoort, S.; Foley, A.R.; Bolduc, V.; Waddell, L.B.; Sandaradura, S.A.; O’Grady, G.L.; et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017, 9, eaal5209. [Google Scholar] [CrossRef]
- Goovaerts, T.; Steyaert, S.; Vandenbussche, C.A.; Galle, J.; Thas, O.; Van Criekinge, W.; De Meyer, T. A comprehensive overview of genomic imprinting in breast and its deregulation in cancer. Nat. Commun. 2018, 9, 4120. [Google Scholar] [CrossRef]
- Grant, A.D.; Vail, P.; Padi, M.; Witkiewicz, A.K.; Knudsen, E.S. Interrogating mutant allele expression via customized reference genomes to define influential cancer mutations. Sci. Rep. 2019, 9, 12766. [Google Scholar] [CrossRef]
- Liu, Z.; Dong, X.; Li, Y. A genome-wide study of allele-specific expression in colorectal cancer. Front. Genet. 2018, 9, 570. [Google Scholar] [CrossRef]
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Wang, H.Y.; Deng, L.; Li, Y.Q.; Zhang, X.; Long, Y.K.; Zhang, X.; Feng, Y.F.; He, Y.; Tang, T.; Yang, X.H.; et al. Pan-cancer analysis of tumor mutational burden and homologous recombination DNA damage repair by targeted next-generation sequencing. Cancer Res. Treat. 2021, 53, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Angell, T.E.; Wirth, L.J.; Cabanillas, M.E.; Shindo, M.L.; Cibas, E.S.; Babiarz, J.E.; Hao, Y.; Kim, S.Y.; Walsh, P.S.; Huang, J.; et al. Analytical and clinical validation of expressed variants and fusions from the whole transcriptome of thyroid FNA samples. Front. Endocrinol. 2019, 10, 612. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Lu, J.; Hu, Z.; Pankratz, D.G.; Jiang, H.; Cao, M.; Marchisano, C.; Huiras, J.; Fedorowicz, G.; Wong, M.G.; et al. Analytical performance of Envisia: A genomic classifier for usual interstitial pneumonia. BMC Pulm. Med. 2017, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Qu, J.; Wu, S.; Hao, Y.; Zhang, J.; Ning, J.; Yang, X.; Lofaro, L.; Pankratz, D.G.; Babiarz, J.; et al. Improving lung cancer risk stratification leveraging whole transcriptome RNA sequencing and machine learning across multiple cohorts. BMC Med. Genomics 2020, 13, 151. [Google Scholar] [CrossRef]
- Grewal, J.K.; Tessier-Cloutier, B.; Jones, M.; Gakkhar, S.; Ma, Y.; Moore, R.; Mungall, A.J.; Zhao, Y.; Taylor, M.D.; Gelmon, K.; et al. Application of a neural network whole transcriptome-based pan-cancer method for diagnosis of primary and metastatic cancers. JAMA Netw Open 2019, 2, e192597. [Google Scholar] [CrossRef]
- Johnson, M.K.; Wu, S.; Pankratz, D.G.; Fedorowicz, G.; Anderson, J.; Ding, J.; Wong, M.; Cao, M.; Babiarz, J.; Lofaro, L.; et al. Analytical validation of the Percepta genomic sequencing classifier; an RNA next generation sequencing assay for the assessment of lung cancer risk of suspicious pulmonary nodules. BMC Cancer 2021, 21, 400. [Google Scholar] [CrossRef]
- Walter, W.; Shahswar, R.; Stengel, A.; Meggendorfer, M.; Kern, W.; Haferlach, T.; Haferlach, C. Clinical application of whole transcriptome sequencing for the classification of patients with acute lymphoblastic leukemia. BMC Cancer 2021, 21, 886. [Google Scholar] [CrossRef]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef]
- Piskol, R.; Ramaswami, G.; Li, J.B. Reliable identification of genomic variants from RNA-Seq data. Am. J. Hum. Genet. 2013, 93, 641–651. [Google Scholar] [CrossRef]
- Prodduturi, N.; Bhagwate, A.; Kocher, J.A.; Sun, Z. Indel sensitive and comprehensive variant/mutation detection from RNA sequencing data for precision medicine. BMC Med. Genom. 2018, 11, 67. [Google Scholar] [CrossRef]
- Yuan, Y.; Ju, Y.S.; Kim, Y.; Li, J.; Wang, Y.; Yoon, C.J.; Yang, Y.; Martincorena, I.; Creighton, C.J.; Weinstein, J.N.; et al. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat. Genet. 2020, 52, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Duh, Q.Y.; Kloos, R.T.; Babiarz, J.; Harrell, R.M.; Traweek, S.T.; Kim, S.Y.; Fedorowicz, G.; Walsh, P.S.; Sadow, P.M.; et al. Identification of Hürthle cell cancers: Solving a clinical challenge with genomic sequencing and a trio of machine learning algorithms. BMC Syst. Biol. 2019, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Alexander, E.K.; Schorr, M.; Klopper, J.; Kim, C.; Sipos, J.; Nabhan, F.; Parker, C.; Steward, D.L.; Mandel, S.J.; Haugen, B.R. Multicenter clinical experience with the Afirma gene expression classifier. J. Clin. Endocrinol. Metab. 2014, 99, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Choi, Y.; Babiarz, J.E.; Kloos, R.T.; Kennedy, G.C.; Huang, J.; Walsh, P.S. Analytical verification performance of afirma genomic sequencing classifier in the diagnosis of cytologically indeterminate thyroid nodules. Front. Endocrinol. 2019, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.N.; Angell, T.E.; Babiarz, J.; Barth, N.M.; Blevins, T.; Duh, Q.Y.; Ghossein, R.A.; Harrell, R.M.; Huang, J.; Kennedy, G.C.; et al. Performance of a genomic sequencing classifier for the preoperative diagnosis of cytologically indeterminate thyroid nodules. JAMA Surg. 2018, 153, 817–824. [Google Scholar] [CrossRef]
- Hu, M.I.; Waguespack, S.G.; Dosiou, C.; Ladenson, P.W.; Livhits, M.J.; Wirth, L.J.; Sadow, P.M.; Krane, J.F.; Stack, B.C.; Zafereo, M.E.; et al. Afirma genomic sequencing classifier and Xpression Atlas molecular findings in consecutive Bethesda III-VI thyroid nodules. J. Clin. Endocrinol. Metab. 2021, 106, 2198–2207. [Google Scholar] [CrossRef]
- Lamb, C.; Rieger-Christ, K.; Reddy, C.; Ding, J.; Qu, J.; Wu, S.; Johnson, M.; Whitney, D.; Walsh, P.; Wilde, J.; et al. A nasal clinical-genomic classifier for assessing risk of malignancy in lung nodules demonstrates accurate performance independent of nodule size or cancer stage. Chest 2021, 160, A2518–A2519. [Google Scholar] [CrossRef]
- Babiarz, J.; Hao, Y.; Manqiu, C.; Bailey, G.; Wilson, D.S.; Krimsky, W.; Sarkar, S.; Bernstein, M.; Lee, H.; Wahidi, M.M.; et al. Detection of actionable molecular alterations through combined DNA/RNA molecular profiling of biopsies collected in early-stage lung cancer at time of diagnosis. J. Clin. Oncol. 2021, 39, e20546. [Google Scholar] [CrossRef]
- Choi, Y.; Liu, T.T.; Pankratz, D.G.; Colby, T.V.; Barth, N.M.; Lynch, D.A.; Walsh, P.S.; Raghu, G.; Kennedy, G.C.; Huang, J. Identification of usual interstitial pneumonia pattern using RNA-Seq and machine learning: Challenges and solutions. BMC Genom. 2018, 19, 101. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-Seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Haas, B.J.; Dobin, A.; Li, B.; Stransky, N.; Pochet, N.; Regev, A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 8 July 2022).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; RStudio. ggplot2. Available online: https://ggplot2.tidyverse.org/ (accessed on 30 June 2022).
- Allaire, J.J.; Ellis, P.; Gandrud, C.; Kuo, K.; Lewis, B.W.; Owen, J.; Russell, K.; Rogers, J.; Sese, C.; Yetman, C.J. D3 JavaScript Network Graphs from R. R Package Version 0.4. Available online: https://cran.r-project.org/web/packages/networkD3/networkD3.pdf (accessed on 30 June 2022).
- Kolde, R. pheatmap: Pretty Heatmaps. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 30 June 2022).
- Parsonage, H. Waterfalls. R package version 0.1.2. Available online: https://cran.r-project.org/web/packages/waterfalls/index.html (accessed on 30 June 2022).
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, D. Treemapify 2.5.3. Available online: http://packages.renjin.org/package/org.renjin.cran/treemapify (accessed on 30 June 2022).
- Seo, J.S.; Ju, Y.S.; Lee, W.C.; Shin, J.Y.; Lee, J.K.; Bleazard, T.; Lee, J.; Jung, Y.J.; Kim, J.O.; Shin, J.Y.; et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012, 22, 2109–2119. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.Z.; Siperstein, A.; Sadow, P.M.; Golding, A.C.; Kennedy, G.C.; Kloos, R.T.; Ladenson, P.W. Extending expressed RNA genomics from surgical decision making for cytologically indeterminate thyroid nodules to targeting therapies for metastatic thyroid cancer. Cancer Cytopathol. 2019, 127, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Raghu, G.; Flaherty, K.R.; Lederer, D.J.; Lynch, D.A.; Colby, T.V.; Myers, J.L.; Groshong, S.D.; Larsen, B.T.; Chung, J.H.; Steele, M.P.; et al. Use of a molecular classifier to identify usual interstitial pneumonia in conventional transbronchial lung biopsy samples: A prospective validation study. Lancet Respir. Med. 2019, 7, 487–496. [Google Scholar] [CrossRef]
- Richeldi, L.; Scholand, M.B.; Lynch, D.A.; Colby, T.V.; Myers, J.L.; Groshong, S.D.; Chung, J.H.; Benzaquen, S.; Nathan, S.D.; Davis, J.R.; et al. Utility of a molecular classifier as a complement to high-resolution computed tomography to identify usual interstitial pneumonia. Am. J. Respir. Crit. Care Med. 2021, 203, 211–220. [Google Scholar] [CrossRef]
- Alexander, E.K.; Kennedy, G.C.; Baloch, Z.W.; Cibas, E.S.; Chudova, D.; Diggans, J.; Friedman, L.; Kloos, R.T.; LiVolsi, V.A.; Mandel, S.J.; et al. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. N. Engl. J. Med. 2012, 367, 705–715. [Google Scholar] [CrossRef]
- Bhorade, S.; Bernstein, M.; Dotson, D.L.; Feller-Kopman, D.; Lee, H.; Choi, Y.; Lofaro, L.; Huang, J.; Whitney, D.; Stevenson, C.; et al. Accuracy of the next generation Percepta Genomic Sequencing Classifier (GSC) for the diagnosis of suspicious intermediate pulmonary nodules. In Proceedings of the American Association for Bronchology and Interventional Pulmonology, Denver, CO, USA, 15–17 August 2019. [Google Scholar]
- Silvestri, G.A.; Vachani, A.; Whitney, D.; Elashoff, M.; Porta Smith, K.; Ferguson, J.S.; Parsons, E.; Mitra, N.; Brody, J.; Lenburg, M.E.; et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. N. Engl. J. Med. 2015, 373, 243–251. [Google Scholar] [CrossRef]
- Perez-Rogers, J.F.; Gerrein, J.; Anderlind, C.; Liu, G.; Zhang, S.; Alekseyev, Y.; Porta Smith, K.; Whitney, D.; Johnson, W.E.; Elashoff, D.A.; et al. Shared gene expression alterations in nasal and bronchial epithelium for lung cancer detection. J. Natl. Cancer Inst. 2017, 109, djw327. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Wang, Q.; Torres-Garcia, W.; Zheng, S.; Vegesna, R.; Kim, H.; Verhaak, R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 2015, 34, 4845–4854. [Google Scholar] [CrossRef] [PubMed]
- Chase, A.; Ernst, T.; Fiebig, A.; Collins, A.; Grand, F.; Erben, P.; Reiter, A.; Schreiber, S.; Cross, N.C. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica 2010, 95, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Vellichirammal, N.N.; Albahrani, A.; Banwait, J.K.; Mishra, N.K.; Li, Y.; Roychoudhury, S.; Kling, M.J.; Mirza, S.; Bhakat, K.K.; Band, V.; et al. Pan-cancer analysis reveals the diverse landscape of novel sense and antisense fusion transcripts. Mol. Ther. Nucleic Acids 2020, 19, 1379–1398. [Google Scholar] [CrossRef]
- Park, H.J.; Yang, M.J.; Oh, J.H.; Yang, Y.S.; Kwon, M.S.; Song, C.W.; Yoon, S. Genome-wide transcriptional response during the development of bleomycin-induced pulmonary fibrosis in Sprague-Dawley rats. Toxicol. Res. 2010, 26, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Hescheler, D.A.; Riemann, B.; Hartmann, M.J.M.; Michel, M.; Faust, M.; Bruns, C.J.; Alakus, H.; Chiapponi, C. Targeted therapy of papillary thyroid cancer: A comprehensive genomic analysis. Front. Endocrinol. 2021, 12, 748941. [Google Scholar] [CrossRef]
- Yakushina, V.D.; Lerner, L.V.; Lavrov, A.V. Gene fusions in thyroid cancer. Thyroid 2018, 28, 158–167. [Google Scholar] [CrossRef]
- Botton, T.; Talevich, E.; Mishra, V.K.; Zhang, T.; Shain, A.H.; Berquet, C.; Gagnon, A.; Judson, R.L.; Ballotti, R.; Ribas, A.; et al. Genetic heterogeneity of BRAF fusion kinases in melanoma affects drug responses. Cell Rep. 2019, 29, 573–588. [Google Scholar] [CrossRef]
- Ross, J.S.; Wang, K.; Chmielecki, J.; Gay, L.; Johnson, A.; Chudnovsky, J.; Yelensky, R.; Lipson, D.; Ali, S.M.; Elvin, J.A.; et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int. J. Cancer 2016, 138, 881–890. [Google Scholar] [CrossRef]
- Kumari, S.; Adewale, R.; Klubo-Gwiezdzinska, J. The molecular landscape of Hürthle cell thyroid cancer is associated with altered mitochondrial function-a comprehensive review. Cells 2020, 9, 1570. [Google Scholar] [CrossRef] [PubMed]
- Ganly, I.; Makarov, V.; Deraje, S.; Dong, Y.; Reznik, E.; Seshan, V.; Nanjangud, G.; Eng, S.; Bose, P.; Kuo, F.; et al. Integrated genomic analysis of Hürthle cell cancer reveals oncogenic drivers, recurrent mitochondrial mutations, and unique chromosomal landscapes. Cancer Cell 2018, 34, 256–270. [Google Scholar] [CrossRef] [PubMed]
- Gopal, R.K.; Kübler, K.; Calvo, S.E.; Polak, P.; Livitz, D.; Rosebrock, D.; Sadow, P.M.; Campbell, B.; Donovan, S.E.; Amin, S.; et al. Widespread chromosomal losses and mitochondrial DNA alterations as genetic drivers in Hürthle cell carcinoma. Cancer Cell 2018, 34, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef]
- Thodou, E.; Canberk, S.; Schmitt, F. Challenges in cytology specimens with Hürthle cells. Front. Endocrinol. 2021, 12, 701877. [Google Scholar] [CrossRef]
- Naccache, J.M.; Gibiot, Q.; Monnet, I.; Antoine, M.; Wislez, M.; Chouaid, C.; Cadranel, J. Lung cancer and interstitial lung disease: A literature review. J. Thorac. Dis. 2018, 10, 3829–3844. [Google Scholar] [CrossRef]
- Govindan, R.; Ding, L.; Griffith, M.; Subramanian, J.; Dees, N.D.; Kanchi, K.L.; Maher, C.A.; Fulton, R.; Fulton, L.; Wallis, J.; et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012, 150, 1121–1134. [Google Scholar] [CrossRef]
- Rabizadeh, S.; Garner, C.; Sanborn, J.Z.; Benz, S.C.; Reddy, S.; Soon-Shiong, P. Comprehensive genomic transcriptomic tumor-normal gene panel analysis for enhanced precision in patients with lung cancer. Oncotarget 2018, 9, 19223–19232. [Google Scholar] [CrossRef]
- Rhee, J.K.; Lee, S.; Park, W.Y.; Kim, Y.H.; Kim, T.M. Allelic imbalance of somatic mutations in cancer genomes and transcriptomes. Sci. Rep. 2017, 7, 1653. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pan-cancer genome and transcriptome analyses of 1699 paediatric leukaemias and solid tumours. Nature 2018, 555, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Markman, B.; Javier Ramos, F.; Capdevila, J.; Tabernero, J. EGFR and KRAS in colorectal cancer. Adv. Clin. Chem. 2010, 51, 71–119. [Google Scholar] [CrossRef]
- Kremer, L.S.; Bader, D.M.; Mertes, C.; Kopajtich, R.; Pichler, G.; Iuso, A.; Haack, T.B.; Graf, E.; Schwarzmayr, T.; Terrile, C.; et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017, 8, 15824. [Google Scholar] [CrossRef]
- Iyama, K.; Matsuse, M.; Mitsutake, N.; Rogounovitch, T.; Saenko, V.; Suzuki, K.; Ashizawa, M.; Ookouchi, C.; Suzuki, S.; Mizunuma, H.; et al. Identification of three novel fusion oncogenes, SQSTM1/NTRK3, AFAP1L2/RET, and PPFIBP2/RET, in thyroid cancers of young patients in Fukushima. Thyroid 2017, 27, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Hamatani, K.; Eguchi, H.; Koyama, K.; Mukai, M.; Nakachi, K.; Kusunoki, Y. A novel RET rearrangement (ACBD5/RET) by pericentric inversion, inv(10)(p12.1;q11.2), in papillary thyroid cancer from an atomic bomb survivor exposed to high-dose radiation. Oncol. Rep. 2014, 32, 1809–1814. [Google Scholar] [CrossRef][Green Version]
- Skálová, A.; Banečkova, M.; Thompson, L.D.R.; Ptáková, N.; Stevens, T.M.; Brcic, L.; Hyrcza, M.; Michal, M., Jr.; Simpson, R.H.W.; Santana, T.; et al. Expanding the molecular spectrum of secretory carcinoma of salivary glands with a novel VIM-RET fusion. Am. J. Surg. Pathol. 2020, 44, 1295–1307. [Google Scholar] [CrossRef]
- Davis, A.A.; Patel, V.G. The role of PD-L1 expression as a predictive biomarker: An analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer 2019, 7, 278. [Google Scholar] [CrossRef]
- Cheng, H.; Borczuk, A.; Janakiram, M.; Ren, X.; Lin, J.; Assal, A.; Halmos, B.; Perez-Soler, R.; Zang, X. Wide expression and significance of alternative immune checkpoint molecules, B7x and HHLA2, in PD-L1 negative human lung cancers. Clin. Cancer Res. 2018, 24, 1954–1964. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, M.; Cadranel, J.; Lusque, A.; Meyer, N.; Gounant, V.; Moro-Sibilot, D.; Michot, J.M.; Raimbourg, J.; Girard, N.; Guisier, F.; et al. Immune-checkpoint inhibitors associated with interstitial lung disease in cancer patients. Eur Respir. J. 2017, 50, 1700050. [Google Scholar] [CrossRef]
- Kim, J.; Myers, A.C.; Chen, L.; Pardoll, D.M.; Truong-Tran, Q.A.; Lane, A.P.; McDyer, J.F.; Fortuno, L.; Schleimer, R.P. Constitutive and inducible expression of b7 family of ligands by human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2005, 33, 280–289. [Google Scholar] [CrossRef]
- Wan, H.; Huang, X.; Cong, P.; He, M.; Chen, A.; Wu, T.; Dai, D.; Li, W.; Gao, X.; Tian, L.; et al. Identification of hub genes associated with COVID-19 and idiopathic pulmonary fibrosis by integrated bioinformatics analysis. Front. Mol. Biosci 2021, 8, 711239. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhu, G.; Xu, H.; Yao, S.; Zhou, G.; Zhu, Y.; Tamada, K.; Huang, L.; Flies, A.D.; Broadwater, M.; et al. B7-H3 promotes pathogenesis of autoimmune disease and inflammation by regulating the activity of different T cell subsets. PLoS ONE 2015, 10, e0130126. [Google Scholar] [CrossRef] [PubMed]
- Tsuyuki, S.; Tsuyuki, J.; Einsle, K.; Kopf, M.; Coyle, A.J. Costimulation through B7-2 (CD86) is required for the induction of a lung mucosal T helper cell 2 (TH2) immune response and altered airway responsiveness. J. Exp. Med. 1997, 185, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Duitman, J.; van den Ende, T.; Spek, C.A. Immune checkpoints as promising targets for the treatment of idiopathic pulmonary fibrosis? J. Clin. Med. 2019, 8, 1547. [Google Scholar] [CrossRef]
- US Food & Drug Administration. Design Control Guidance for Medical Device Manufacturers. Guidance for Industry. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/design-control-guidance-medical-device-manufacturers (accessed on 30 June 2022).

| Indication | Development Samples, n | Classifier (CLIA) Samples, n |
|---|---|---|
| Thyroid (Afirma GSC) | 634 | 109,912 |
| ILD (Envisia GC) | 359 | 3025 |
| Lung (Percepta GSC) | 311 | 5521 |
| Lung (Percepta NS) | 356 | |
| Lung (Percepta GA) | 194 |
| Properties | Benefits over Current Diagnostic Tools |
|---|---|
| Open platform |
|
| Enrichment-based approach |
|
| Wider dynamic range than microarrays and RT-qPCR for detecting expression differences |
|
| Use of the same analy tically validated clinical assay for both research and CLIA samples |
|
| Detection and quantitation of both nuclear and mitochondrial transcripts |
|
| Creation of complex RNA signatures for diagnosis as well as providing prognostic and/or predictive information using all transcripts as features in machine learning |
|
| Detection and quantitation of known and novel translocations/fusions |
|
| Chromosome and genome-level LOH measurements, and identification of specific CNVs |
|
| Collection of full transcriptome data on every patient sample, creating a large biorepository for Pharma to mine |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walsh, P.S.; Hao, Y.; Ding, J.; Qu, J.; Wilde, J.; Jiang, R.; Kloos, R.T.; Huang, J.; Kennedy, G.C. Maximizing Small Biopsy Patient Samples: Unified RNA-Seq Platform Assessment of over 120,000 Patient Biopsies. J. Pers. Med. 2023, 13, 24. https://doi.org/10.3390/jpm13010024
Walsh PS, Hao Y, Ding J, Qu J, Wilde J, Jiang R, Kloos RT, Huang J, Kennedy GC. Maximizing Small Biopsy Patient Samples: Unified RNA-Seq Platform Assessment of over 120,000 Patient Biopsies. Journal of Personalized Medicine. 2023; 13(1):24. https://doi.org/10.3390/jpm13010024
Chicago/Turabian StyleWalsh, P. Sean, Yangyang Hao, Jie Ding, Jianghan Qu, Jonathan Wilde, Ruochen Jiang, Richard T. Kloos, Jing Huang, and Giulia C. Kennedy. 2023. "Maximizing Small Biopsy Patient Samples: Unified RNA-Seq Platform Assessment of over 120,000 Patient Biopsies" Journal of Personalized Medicine 13, no. 1: 24. https://doi.org/10.3390/jpm13010024
APA StyleWalsh, P. S., Hao, Y., Ding, J., Qu, J., Wilde, J., Jiang, R., Kloos, R. T., Huang, J., & Kennedy, G. C. (2023). Maximizing Small Biopsy Patient Samples: Unified RNA-Seq Platform Assessment of over 120,000 Patient Biopsies. Journal of Personalized Medicine, 13(1), 24. https://doi.org/10.3390/jpm13010024






