A Systematic Review of Polygenic Models for Predicting Drug Outcomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Rationale and Scope of Review
2.2. Search Details
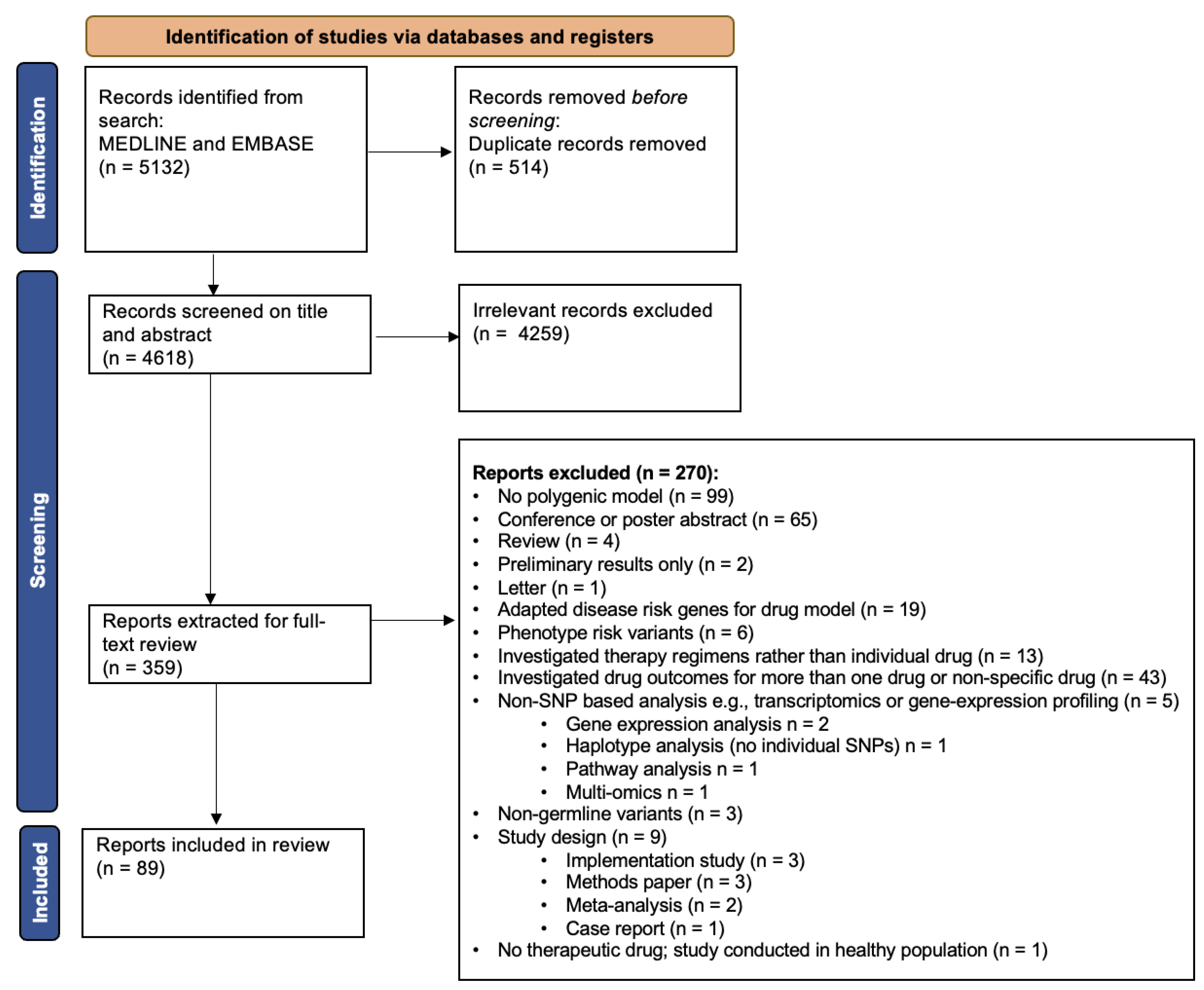
2.3. Study Selection
2.4. Data Extraction
2.5. Synthesis of Results
3. Results
3.1. Overview of Included Articles
3.2. Method of Gene-Selection for Developing a Polygenic Model Predicting Drug Outcomes
3.3. Overview of Methods Used to Develop Polygenic Predictions Models in Pharmacogenomics
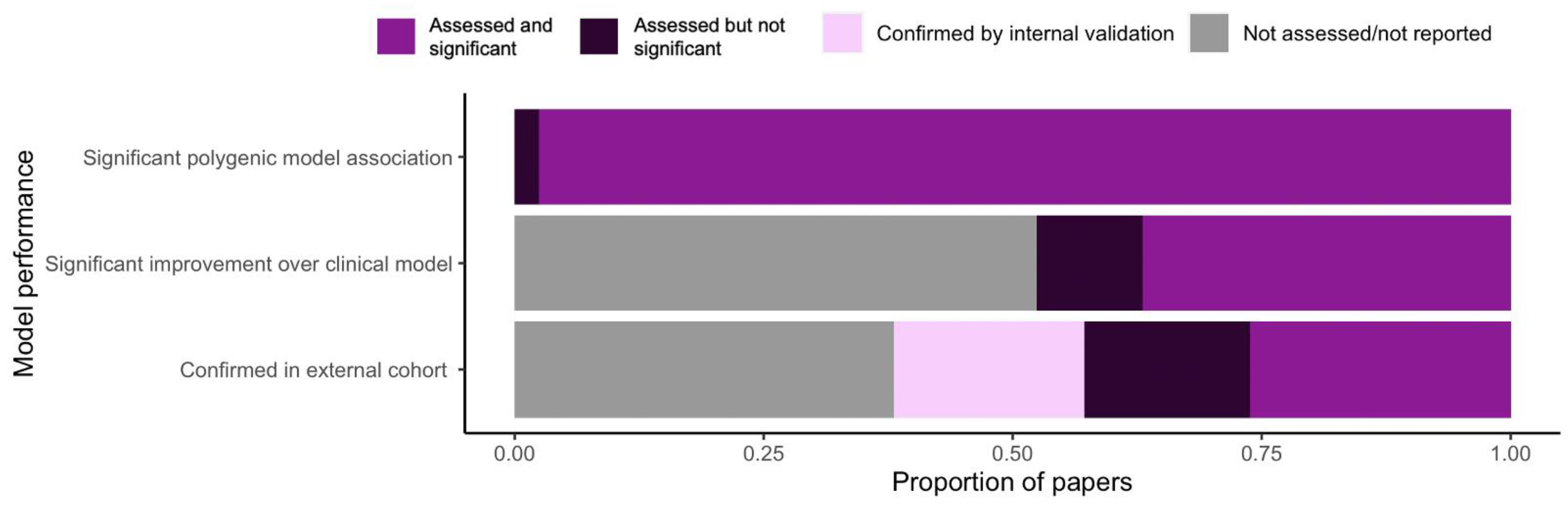
3.4. Performance of Polygenic Models in Pharmacogenomics Research
3.5. Validating the Performance of Polygenic Models
4. Discussion
4.1. Drug Outcomes Investigated
4.2. Methods for Polygenic Model Development
4.3. Model Performance
4.4. Model Validation
4.5. Study Limitations and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fisher, R.A. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 1918, 52, 399–433. [Google Scholar] [CrossRef]
- Zeggini, E.; Gloyn, A.L.; Barton, A.C.; Wain, L.V. Translational genomics and precision medicine: Moving from the lab to the clinic. Science 2019, 365, 1409–1413. [Google Scholar] [CrossRef]
- Wray, N.R.; Goddard, M.E.; Visscher, P.M. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007, 17, 1520–1528. [Google Scholar] [CrossRef] [PubMed]
- Pirmohamed, M.; Park, B.K. Genetic susceptibility to adverse drug reactions. Trends Pharmacol. Sci. 2001, 22, 298–305. [Google Scholar] [CrossRef]
- Alfirevic, A.; Pirmohamed, M. Genomics of Adverse Drug Reactions. Trends Pharmacol. Sci. 2017, 38, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Claussnitzer, M.; Cho, J.H.; Collins, R.; Cox, N.J.; Dermitzakis, E.T.; Hurles, M.E.; Kathiresan, S.; Kenny, E.E.; Lindgren, C.M.; Macarthur, D.G.; et al. A brief history of human disease genetics. Nature 2020, 577, 179–189. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Choi, S.W.; Mak, T.S.H.; O’Reilly, P.F. A guide to performing Polygenic Risk Score analyses. Nat. Protoc. 2020, 15, 2759–2772. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef]
- Guinan, K.; Beauchemin, C.; Tremblay, J.; Chalmers, J.; Woodward, M.; Tahir, M.R.; Hamet, P.; Lachaine, J. Economic Evaluation of a New Polygenic Risk Score to Predict Nephropathy in Adult Patients with Type 2 Diabetes. Can. J. Diabetes 2021, 45, 129–136. [Google Scholar] [CrossRef]
- Mavaddat, N.; Michailidou, K.; Dennis, J.; Lush, M.; Fachal, L.; Lee, A.; Tyrer, J.P.; Chen, T.H.; Wang, Q.; Bolla, M.K.; et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019, 104, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Forgetta, V.; Keller-Baruch, J.; Forest, M.; Durand, A.; Bhatnagar, S.; Kemp, J.P.; Nethander, M.; Evans, D.; Morris, J.A.; Kiel, D.P.; et al. Development of a polygenic risk score to improve screening for fracture risk: A genetic risk prediction study. PLoS Med. 2020, 17, e1003152. [Google Scholar] [CrossRef]
- Auwerx, C.; Sadler, M.C.; Reymond, A.; Kutalik, Z. From Pharmacogenetics to Pharmaco-Omics:Milestones and Future Directions. Hum. Genet. Genom. Adv. 2022, 3, 100100. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.K. Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 2010, 11, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.A.; Gong, L.; Whirl-Carrillo, M.; Gage, B.F.; Scott, S.A.; Stein, C.M.; Anderson, J.L.; Kimmel, S.E.; Lee, M.T.M.; Pirmohamed, M.; et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin. Pharmacol. Ther. 2011, 90, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M.; Johnson, J.A.; Langaee, T.Y.; Feng, H.; Stanaway, I.B.; Schwarz, U.I.; Ritchie, M.D.; Stein, C.M.; Roden, D.M.; Smith, J.D.; et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 2008, 112, 1022–1027. [Google Scholar] [CrossRef]
- Takeuchi, F.; McGinnis, R.; Bourgeois, S.; Barnes, C.; Eriksson, N.; Soranzo, N.; Whittaker, P.; Ranganath, V.; Kumanduri, V.; McLaren, W.; et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009, 5, e1000433. [Google Scholar] [CrossRef]
- The International Warfarin Pharmacogenetics Consortium. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. N. Engl. J. Med. 2009, 360, 753–764. [Google Scholar] [CrossRef]
- Johnson, D.; Wilke, M.A.P.; Lyle, S.M.; Kowalec, K.; Jorgensen, A.; Wright, G.E.B.; Drögemöller, B.I. A Systematic Review and Analysis of the Use of Polygenic Scores in Pharmacogenomics. Clin. Pharmacol. Ther. 2021, 111, 919–930. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yoshikawa, A.; Brennan, M.D.; Ramsey, T.L.; Meltzer, H.Y. Genetic predictors of antipsychotic response to lurasidone identified in a genome wide association study and by schizophrenia risk genes. Schizophr. Res. 2018, 192, 194–204. [Google Scholar] [CrossRef]
- Helmstaedter, C.; Mihov, Y.; Toliat, M.R.; Thiele, H.; Nuernberg, P.; Schoch, S.; Surges, R.; Elger, C.E.; Kunz, W.S.; Hurlemann, R. Genetic variation in dopaminergic activity is associated with the risk for psychiatric side effects of levetiracetam. Epilepsia 2013, 54, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Lexicomp. Available online: https://online-lexi-com.eu1.proxy.openathens.net/lco/action/home?siteid=2 (accessed on 17 May 2022).
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Shennan, M.; Reynolds, K.K.; Johnson, N.A.; Herrnberger, M.R.; Valdes, R. Estimation of warfarin maintenance dose based on VKORC1 (−1639 G > A) and CYP2C9 genotypes. Clin. Chem. 2007, 53, 1199–1205. [Google Scholar] [CrossRef]
- Sconce, E.A.; Khan, T.I.; Wynne, H.A.; Avery, P.; Monkhouse, L.; King, B.P.; Wood, P.; Kesteven, P.; Daly, A.K.; Kamali, F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: Proposal for a new dosing regimen. Blood 2005, 106, 2329–2333. [Google Scholar] [CrossRef]
- Gage, B.F.; Eby, C.; Milligan, P.E.; Banet, G.A.; Duncan, J.R. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb. Haemost. 2004, 91, 87–94. [Google Scholar] [CrossRef]
- Nowak-Gottl, U.; Dietrich, K.; Schaffranek, D.; Eldin, N.S.; Yasui, Y.; Geisen, C.; Mitchell, L.G. In pediatric patients, age has more impact on dosing of vitamin K antagonists than VKORC1 or CYP2C9 genotypes. Blood 2010, 116, 6101–6105. [Google Scholar] [CrossRef]
- Lala, M.; Burckart, G.J.; Takao, C.M.; Pravica, V.; Momper, J.D.; Gobburu, J.V.S. Genetics-based pediatric warfarin dosage regimen derived using pharmacometric bridging. J. Pediatr. Pharmacol. Ther. 2013, 18, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.; Amstutz, U.; Hildebrand, C.; Rassekh, S.R.; Hosking, M.; Neville, K.; Leeder, J.S.; Hayden, M.R.; Ross, C.J.; Carleton, B.C. VKORC1 and CYP2C9 genotypes are predictors of warfarin-related outcomes in children. Pediatr. Blood Cancer 2014, 61, 1055–1062. [Google Scholar] [CrossRef]
- Vear, S.I.; Ayers, G.D.; van Driest, S.L.; Sidonio, R.F.; Stein, C.M.; Ho, R.H. The impact of age and CYP2C9 and VKORC1 variants on stable warfarin dose in the paediatric population. Br. J. Haematol. 2014, 165, 832–835. [Google Scholar] [CrossRef]
- Nguyen, N.; Anley, P.; Yu, M.Y.; Zhang, G.; Thompson, A.A.; Jennings, L.J. Genetic and clinical determinants influencing warfarin dosing in children with heart disease. Pediatr. Cardiol. 2013, 34, 984–990. [Google Scholar] [CrossRef]
- Moreau, C.; Bajolle, F.; Siguret, V.; Lasne, D.; Golmard, J.L.; Elie, C.; Beaune, P.; Cheurfi, R.; Bonnet, D.; Loriot, M.A. Vitamin K antagonists in children with heart disease: Height and VKORC1 genotype are the main determinants of the warfarin dose requirement. Blood 2012, 119, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Biss, T.T.; Avery, P.J.; Brandao, L.R.; Chalmers, E.A.; Williams, M.D.; Grainger, J.D.; Leathart, J.B.S.; Hanley, J.P.; Daly, A.K.; Kamali, F. VKORC1 and CYP2C9 genotype and patient characteristics explain a large proportion of the variability in warfarin dose requirement among children. Blood 2012, 119, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Wadelius, M.; Chen, L.Y.; Bumpstead, S.; Ghori, J.; Bentley, D.; McGinnis, R.; Deloukas, P.; Eriksson, N.; Wadelius, C. Association of warfarin dose with genes involved in its action and metabolism. Hum. Genet. 2007, 121, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Limdi, N.A.; Arnett, D.K.; McGwin, G.; Goldstein, J.A.; Beasley, T.M.; Adler, B.K.; Acton, R.T. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics 2008, 9, 511–526. [Google Scholar] [CrossRef]
- Spreafico, M.; Mannucci, P.M.; Peyvandi, F.; Lodigiani, C.; Rota, L.L.; van Leeuwen, Y.; Rosendaal, F.R.; Pizzotti, D. Effects of CYP2C9 and VKORC1 on INR variations and dose requirements during initial phase of anticoagulant therapy. Pharmacogenomics 2008, 9, 1237–1250. [Google Scholar] [CrossRef]
- Kurnik, D.; Qasem, H.; Sominski, S.; Halkin, H.; Gak, E.; Loebstein, R.; Li, C.; Stein, C. Effect of the VKORC1 ASP36TYR variant on warfarin dose requirement and pharmacogenetic dose prediction models. Clin. Pharmacol. Ther. 2012, 91 (Suppl. S1), S96–S135. [Google Scholar] [CrossRef]
- Teh, L.K.; Langmia, I.M.; Fazleen Haslinda, M.H.; Salleh, M.Z.; Ngow, H.A.; Roziah, M.J.; Harun, R.; Zakaria, Z.A. Clinical relevance of VKORC1 (G-1639A and C1173T) and CYP2C9*3 among patients on warfarin. J. Clin. Pharm. Ther. 2012, 37, 232–236. [Google Scholar] [CrossRef]
- Wang, D.; Wu, H.; Chong, J.; Lu, Y.; Yin, R.; Zhao, X.; Zhao, A.; Yang, J.; Chen, H.; Dai, D.P. Effects of rare CYP2C9 alleles on stable warfarin doses in Chinese Han patients with atrial fibrillation. Pharmacogenomics 2020, 21, 1021–1031. [Google Scholar] [CrossRef]
- Yoshida, K.; Nishizawa, D.; Ichinomiya, T.; Ichinohe, T.; Hayashida, M.; Fukuda K ichi Ikeda, K. Prediction formulas for individual opioid analgesic requirements based on genetic polymorphism analyses. PLoS ONE 2015, 10, e0116885. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Tsunedomi, R.; Fujita, Y.; Otori, T.; Ohba, M.; Kawai, Y.; Hirata, H.; Matsumoto, H.; Haginaka, J.; Suzuki, S.; et al. Pharmacogenetics-based area-under-curve model can predict efficacy and adverse events from axitinib in individual patients with advanced renal cell carcinoma. Oncotarget 2018, 9, 17160–17170. [Google Scholar] [CrossRef]
- Verhoef, T.I.; Redekop, W.K.; Buikema, M.M.; Schalekamp, T.; Van Der Meer, F.J.M.; LE Cessie, S.; Wessels, J.A.M.; Van Schie, R.M.F.; DE Boer, A.; Teichert, M.; et al. Long-term anticoagulant effects of the CYP2C9 and VKORC1 genotypes in acenocoumarol users. J. Thromb. Haemost. 2012, 10, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Chen, Y.J.; Fu, X.; Jiang, B.; Zhang, Y. Evolutionary Ensemble Learning Algorithm to Modeling of Warfarin Dose Prediction for Chinese. IEEE J. Biomed. Health Inform. 2019, 23, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Wenying, S.; Lingyan, C.; Xiaoye, H.; Meimei, Z.; Wensheng, C.; Lei, M.; Xiaoyan, L.; Jianing, H.; Tingyuan, P.; Jia, L.; et al. Cytochrome P450 Genetic Variations Can Predict mRNA Expression, Cyclophosphamide 4-Hydroxylation, and Treatment Outcomes in Chinese Patients with Non-Hodgkin’s Lymphoma. J. Clin. Pharmacol. 2017, 57, 886–898. [Google Scholar] [CrossRef]
- Shahabi, P.; Scheinfeldt, L.B.; Lynch, D.E.; Schmidlen, T.J.; Perreault, S.; Keller, M.A.; Kasper, R.; Wawak, L.; Jarvis, J.P.; Gerry, N.P.; et al. An expanded pharmacogenomics warfarin dosing table with utility in generalised dosing guidance. Thromb. Haemost. 2016, 116, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Perini, J.A.; Struchiner, C.J.; Silva-Assuncao, E.; Santana, I.S.C.; Rangel, F.; Ojopi, E.B.; Dias-Neto, E.; Suarez-Kurtz, G. Pharmacogenetics of warfarin: Development of a dosing algorithm for brazilian patients. Clin. Pharmacol. Ther. 2008, 84, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Marek, E.; Momper, J.D.; Hines, R.N.; Takao, C.M.; Gill, J.C.; Pravica, V.; Gaedigk, A.; Burckart, G.J.; Neville, K.A. Prediction of Warfarin Dose in Pediatric Patients: An Evaluation of the Predictive Performance of Several Models. J. Pediatr. Pharmacol. Ther. 2016, 21, 224–232. [Google Scholar] [CrossRef]
- Grossi, E.; Podda, G.M.; Pugliano, M.; Gabba, S.; Verri, A.; Carpani, G.; Buscema, M.; Casazza, G.; Cattaneo, M. Prediction of optimal warfarin maintenance dose using advanced artificial neural networks. Pharmacogenomics 2014, 15, 29–37. [Google Scholar] [CrossRef]
- Eriksson, N.; Wallentin, L.; Berglund, L.; Axelsson, T.; Connolly, S.; Eikelboom, J.; Ezekowitz, M.; Oldgren, J.; Pare, G.; Reilly, P.; et al. Genetic determinants of warfarin maintenance dose and time in therapeutic treatment range: A RE-LY genomics substudy. Pharmacogenomics 2016, 17, 1425–1439. [Google Scholar] [CrossRef]
- De Graan, A.J.M.; Elens, L.; Smid, M.; Martens, J.W.; Sparreboom, A.; Nieuweboer, A.J.M.; Friberg, L.E.; Elbouazzaoui, S.; Wiemer, E.A.C.; van der Holt, B.; et al. A pharmacogenetic predictive model for paclitaxel clearance based on the DMET platform. Clin. Cancer Res. 2013, 19, 5210–5217. [Google Scholar] [CrossRef]
- Altmann, V.; Schumacher-Schuh, A.F.; Rieck, M.; Callegari-Jacques, S.M.; Rieder, C.R.M.; Hutz, M.H. Influence of genetic, biological and pharmacological factors on levodopa dose in Parkinson’s disease. Pharmacogenomics 2016, 17, 481–488. [Google Scholar] [CrossRef]
- Kurnik, D.; Qasim, H.; Sominsky, S.; Lubetsky, A.; Markovits, N.; Li, C.; Stein, C.M.; Halkin, H.; Gak, E.; Loebstein, R. Effect of the VKORC1 D36Y variant on warfarin dose requirement and pharmacogenetic dose prediction. Thromb. Haemost. 2012, 108, 781–788. [Google Scholar] [PubMed]
- Finkelman, B.S.; French, B.; Bershaw, L.; Brensinger, C.M.; Streiff, M.B.; Epstein, A.E.; Kimmel, S.E. Predicting prolonged dose titration in patients starting warfarin. Pharmacoepidemiol. Drug Saf. 2016, 25, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Anton, A.I.; Cerezo-Manchado, J.J.; Padilla, J.; Perez-Andreu, V.; Corral, J.; Vicente, V.; Roldan, V.; Gonzalez-Conejero, R. Novel associations of VKORC1 variants with higher acenocoumarol requirements. PLoS ONE 2013, 8, e64469. [Google Scholar] [CrossRef]
- Dapia, I.; Garcia, I.; Martinez, C.J.; Arias, P.; Guerra, P.; Diaz, L.; Garcia, A.; Ochoa, D.; Tenorio, J.; Ramirez, E.; et al. Prediction models for voriconazole pharmacokinetics based on pharmacogenetics: AN exploratory study in a Spanish population. Int. J. Antimicrob. Agents 2019, 54, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Schalekamp, T.; Brassé, B.P.; Roijers, J.F.; Chahid, Y.; van Geest-Daalderop, J.H.; de Vries-Goldschmeding, H.; van Wijk, E.M.; Egberts, A.C.; de Boer, A. VKORC1 and CYP2C9 genotypes and acenocoumarol anticoagulation status: Interaction between both genotypes affects overanticoagulation. Clin. Pharmacol. Ther. 2006, 80, 13–22. [Google Scholar] [CrossRef]
- Hoskins, J.M.; Rosner, G.L.; Ratain, M.J.; McLeod, H.L.; Innocenti, F. Pharmacodynamic genes do not influence risk of neutropenia in cancer patients treated with moderately high-dose irinotecan. Pharmacogenomics 2009, 10, 1139–1146. [Google Scholar] [CrossRef]
- Chang, T.J.; Liu, P.H.; Liang, Y.C.; Chang, Y.C.; Jiang Y der Li, H.Y.; Lo, M.T.; Chen, H.S.; Chuang, L.M. Genetic predisposition and nongenetic risk factors of thiazolidinedione- related edema in patients with type 2 diabetes. Pharm. Genom. 2011, 21, 829–836. [Google Scholar] [CrossRef]
- Vispo, E.; Cevik, M.; Rockstroh, J.K.; Barreiro, P.; Nelson, M.; Scourfield, A.; Boesecke, C.; Wasmuth, J.C.; Soriano, V. Genetic determinants of idiopathic noncirrhotic portal hypertension in HIV-infected patients. Clin. Infect. Dis. 2013, 56, 1117–1122. [Google Scholar] [CrossRef]
- Visscher, H.; Ross, C.J.D.; Rassekh, S.R.; Sandor, G.S.S.; Caron, H.N.; van Dalen, E.C.; Kremer, L.C.; van der Pal, H.J.; Rogers, P.C.; Rieder, M.J.; et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatric Blood Cancer 2013, 60, 1375–1381. [Google Scholar] [CrossRef]
- Lima, A.; Bernardes, M.; Azevedo, R.; Monteiro, J.; Sousa, H.; Medeiros, R.; Seabra, V. SLC19A1, SLC46A1 and SLCO1B1 polymorphisms as predictors of methotrexate-related toxicity in Portuguese rheumatoid arthritis patients. Toxicol. Sci. 2014, 142, 196–209. [Google Scholar] [CrossRef]
- Custodio, A.; Moreno-Rubio, J.; Aparicio, J.; Gallego-Plazas, J.; Yaya, R.; Maurel, J.; Higuera, O.; Burgos, E.; Ramos, D.; Calatrava, A.; et al. Pharmacogenetic predictors of severe peripheral neuropathy in colon cancer patients treated with oxaliplatin-based adjuvant chemotherapy: A GEMCAD group study. Ann. Oncol. 2014, 25, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Ragia, G.; Tavridou, A.; Elens, L.; van Schaik, R.H.N.; Manolopoulos, V.G. CYP2C9*2 allele increases risk for hypoglycemia in POR*1/*1 type 2 diabetic patients treated with sulfonylureas. Exp. Clin. Endocrinol. Diabetes 2014, 122, 60–63. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, Q.; Fu, S.; Xu, M.; Zhang, T.; Xie, C.; Feng, J.; Chen, J.; Zang, A.; Cai, Y.; et al. A novel genetic score model of UGT1A1 and TGFB pathway as predictor of severe irinotecan-related diarrhea in metastatic colorectal cancer patients. J. Cancer Res. Clin. Oncol. 2016, 142, 1621–1628. [Google Scholar] [CrossRef] [PubMed]
- Chaix, M.A.; Parmar, N.; Kinnear, C.; Lafreniere-Roula, M.; Akinrinade, O.; Yao, R.; Miron, A.; Lam, E.; Meng, G.; Christie, A.; et al. Machine Learning Identifies Clinical and Genetic Factors Associated With Anthracycline Cardiotoxicity in Pediatric Cancer Survivors. JACC CardioOncology 2020, 2, 690–706. [Google Scholar] [CrossRef]
- Visscher, H.; Rassekh, S.R.; Sandor, G.S.; Caron, H.N.; van Dalen, E.C.; Kremer, L.C.; van der Pal, H.J.; Rogers, P.C.; Rieder, M.J.; Carleton, B.C.; et al. Genetic variants in SLC22A17 and SLC22A7 are associated with anthracycline-induced cardiotoxicity in children. Pharmacogenomics 2015, 16, 1065–1076. [Google Scholar] [CrossRef]
- Visscher, H.; Ross, C.J.D.; Rassekh, S.R.; Barhdadi, A.; Dubé, M.P.; Al-Saloos, H.; Sandor, G.S.; Caron, H.N.; van Dalen, E.C.; Kremer, L.C.; et al. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J. Clin. Oncol. 2012, 30, 1422–1428. [Google Scholar] [CrossRef]
- Vandell, A.G.; Walker, J.; Brown, K.S.; Zhang, G.; Lin, M.; Grosso, M.A.; Mercuri, M.F. Genetics and clinical response to warfarin and edoxaban in patients with venous thromboembolism. Heart 2017, 103, 1800–1805. [Google Scholar] [CrossRef]
- Vandell, A.G.; McDonough, C.W.; Gong, Y.; Langaee, T.Y.; Lucas, A.M.; Chapman, A.B.; Gums, J.G.; Beitelshees, A.L.; Bailey, K.R.; Johnson, R.J.; et al. Hydrochlorothiazide-induced hyperuricaemia in the pharmacogenomic evaluation of antihypertensive responses study. J. Intern. Med. 2014, 276, 486–497. [Google Scholar] [CrossRef]
- Suzuki, K.; Kakuta, Y.; Naito, T.; Takagawa, T.; Hanai, H.; Araki, H.; Sasaki, Y.; Sakuraba, H.; Sasaki, M.; Hisamatsu, T.; et al. Genetic Background of Mesalamine-induced Fever and Diarrhea in Japanese Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2021, 28, 21–31. [Google Scholar] [CrossRef]
- Serna, M.J.; Rivera-Caravaca, J.M.; Gonzalez-Conejero, R.; Esteve-Pastor, M.A.; Valdes, M.; Vicente, V.; Lip, G.Y.H.; Roldan, V.; Marin, F. Pharmacogenetics of vitamin K antagonists and bleeding risk prediction in atrial fibrillation. Eur. J. Clin. Investig. 2018, 48, e12929. [Google Scholar] [CrossRef]
- Redensek, S.; Bizjan, B.J.; Trost, M.; Dolzan, V. Clinical and Clinical-Pharmacogenetic Models for Prediction of the Most Common Psychiatric Complications Due to Dopaminergic Treatment in Parkinson’s Disease. Int. J. Neuropsychopharmacol. 2020, 23, 496–504. [Google Scholar] [CrossRef] [PubMed]
- Redensek, S.; Jenko Bizjan, B.; Trost, M.; Dolzan, V. Clinical-Pharmacogenetic Predictive Models for Time to Occurrence of Levodopa Related Motor Complications in Parkinson’s Disease. Front. Genet. 2019, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Park, M.W.; Her, S.H.; Kim, C.J.; SunCho, J.; Park, G.M.; Kim, T.S.; Choi, Y.S.; Park, C.S.; Koh, Y.S.; Park, H.J.; et al. Evaluation of the incremental prognostic value of the combination of CYP2C19 poor metabolizer status and ABCB1 3435 TT polymorphism over conventional risk factors for cardiovascular events after drug-eluting stent implantation in East Asians. Genet. Med. 2016, 18, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Palles, C.; Fotheringham, S.; Chegwidden, L.; Lucas, M.; Kerr, R.; Mozolowski, G.; Rosmarin, D.; Taylor, J.C.; Tomlinson, I.; Kerr, D. An Evaluation of the Diagnostic Accuracy of a Panel of Variants in DPYD and a Single Variant in ENOSF1 for Predicting Common Capecitabine Related Toxicities. Cancers 2021, 13, 1497. [Google Scholar] [CrossRef]
- Ooi, B.N.; Ying, A.F.; Koh, Y.Z.; Jin, Y.; Yee, S.W.; Lee, J.H.; Chong, S.S.; Tan, J.W.; Liu, J.; Lee, C.G.; et al. Robust Performance of Potentially Functional SNPs in Machine Learning Models for the Prediction of Atorvastatin-Induced Myalgia. Front. Pharmacol. 2021, 12, 605764. [Google Scholar] [CrossRef]
- Federico, N.; Stefania, F.F.; Rosalba, M.; Stefania, C.; Raffaella, G.; Giovanni, F.; Gabriele, I.; Antonia, M.; Carlotta, A.; Alfredo, F.; et al. Is a pharmacogenomic panel useful to estimate the risk of oxaliplatin-related neurotoxicity in colorectal cancer patients? Pharm. J. 2019, 19, 465–472. [Google Scholar] [CrossRef]
- Milosevic, G.; Kotur, N.; Krstovski, N.; Lazic, J.; Zukic, B.; Stankovic, B.; Janic, D.; Katsila, T.; Patrinos, G.P.; Pavlovic, S.; et al. Variants in TPMT, ITPA, ABCC4 and ABCB1 Genes as Predictors of 6-mercaptopurine Induced Toxicity in Children with Acute Lymphoblastic Leukemia. J. Med. Biochem. 2018, 37, 320–327. [Google Scholar] [CrossRef]
- Marcath, L.A.; Kidwell, K.M.; Vangipuram, K.; Gersch, C.L.; Rae, J.M.; Burness, M.L.; Griggs, J.J.; Van Poznak, C.; Hayes, D.F.; Smith, E.M.L.; et al. Genetic variation in EPHA contributes to sensitivity to paclitaxel-induced peripheral neuropathy. Br. J. Clin. Pharmacol. 2020, 86, 880–890. [Google Scholar] [CrossRef]
- Innocenti, F.; Kroetz, D.L.; Schuetz, E.; Dolan, M.E.; Ramirez, J.; Relling, M.; Chen, P.; Das, S.; Rosner, G.L.; Ratain, M.J. Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics. J. Clin. Oncol. 2009, 27, 2604–2614. [Google Scholar] [CrossRef]
- Cummins, N.W.; Neuhaus, J.; Chu, H.; Neaton, J.; Wyen, C.; Rockstroh, J.K.; Skiest, D.J.; Boyd, M.A.; Khoo, S.; Rotger, M.; et al. Investigation of Efavirenz Discontinuation in Multi-ethnic Populations of HIV-positive Individuals by Genetic Analysis. EBioMedicine 2015, 2, 706–712. [Google Scholar] [CrossRef]
- Chen, S.; Villeneuve, L.; Jonker, D.; Couture, F.; Laverdiere, I.; Cecchin, E.; Innocenti, F.; Toffoli, G.; Levesque, E.; Guillemette, C. ABCC5 and ABCG1 polymorphisms predict irinotecan-induced severe toxicity in metastatic colorectal cancer patients. Pharm. Genom. 2015, 25, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Chamorro, J.G.; Castagnino, J.P.; Aidar, O.; Musella, R.M.; Frias, A.; Visca, M.; Nogueras, M.; Costa, L.; Perez, A.; Caradonna, F.; et al. Effect of gene-gene and gene-environment interactions associated with antituberculosis drug-induced hepatotoxicity. Pharm. Genom. 2017, 27, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Biesiada, J.; Chidambaran, V.; Wagner, M.; Zhang, X.; Martin, L.J.; Meller, J.; Sadhasivam, S. Genetic risk signatures of opioid-induced respiratory depression following pediatric tonsillectomy. Pharmacogenomics 2014, 15, 1749–1762. Available online: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=medp&NEWS=N&AN=25493568NS- (accessed on 28 November 2021). [CrossRef] [PubMed]
- Anandi, P.; Dickson, A.L.; Feng, Q.; Wei, W.Q.; Dupont, W.D.; Plummer, D.; Liu, G.; Octaria, R.; Barker, K.A.; Kawai, V.K.; et al. Combining clinical and candidate gene data into a risk score for azathioprine-associated leukopenia in routine clinical practice. Pharm. J. 2020, 20, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Abaji, R.; Ceppi, F.; Patel, S.; Gagne, V.; Xu, C.J.; Spinella, J.F.; Colombini, A.; Parasole, R.; Buldini, B.; Basso, G.; et al. Genetic risk factors for VIPN in childhood acute lymphoblastic leukemia patients identified using whole-exome sequencing. Pharmacogenomics 2018, 19, 1181–1193. [Google Scholar] [CrossRef]
- Mega, J.L.; Walker, J.R.; Ruff, C.T.; Vandell, A.G.; Nordio, F.; Deenadayalu, N.; Murphy, S.A.; Lee, J.; Mercuri, M.F.; Giugliano, R.P.; et al. Genetics and the clinical response to warfarin and edoxaban: Findings from the randomised, double-blind ENGAGE AF-TIMI 48 trial. Lancet 2015, 385, 2280–2287. [Google Scholar] [CrossRef]
- Hamberg, A.K.; Friberg, L.E.; Hanseus, K.; Ekman-Joelsson, B.M.; Sunnegardh, J.; Jonzon, A.; Lundell, B.; Jonsson, E.N.; Wadelius, M. Warfarin dose prediction in children using pharmacometric bridging—Comparison with published pharmacogenetic dosing algorithms. Eur. J. Clin. Pharmacol. 2013, 69, 1275–1283. [Google Scholar] [CrossRef]
- Brugts, J.J.; Isaacs, A.; Boersma, E.; van Duijn, C.M.; Uitterlinden, A.G.; Remme, W.; Bertrand, M.; Ninomiya, T.; Ceconi, C.; Chalmers, J.; et al. Genetic determinants of treatment benefit of the angiotensin-convertingenzyme-inhibitor perindopril in patients with stable coronary arterydisease. Eur. Heart J. 2010, 31, 1854–1864. [Google Scholar] [CrossRef]
- Wu, X.; Lu, C.; Ye, Y.; Chang, J.; Yang, H.; Lin, J.; Gu, J.; Hong, W.K.; Stewart, D.; Spitz, M.R. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharm. Genom. 2008, 18, 955–965. [Google Scholar] [CrossRef]
- Oemrawsingh, R.M.; Akkerhuis, K.M.; van Vark, L.C.; Ken Redekop, W.; Rudez, G.; Remme, W.J.; Bertrand, M.E.; Fox, K.M.; Ferrari, R.; Jan Danser, A.H.; et al. Individualized angiotensin-converting enzyme (ACE)-inhibitor therapy in stable coronary artery disease based on clinical and pharmacogenetic determinants: The PERindopril GENEtic (PERGENE) risk model. J. Am. Heart Assoc. 2015, 5, e002688. [Google Scholar] [CrossRef]
- Sensorn, I.; Sukasem, C.; Sirachainan, E.; Chamnanphon, M.; Pasomsub, E.; Trachu, N.; Supavilai, P.; Pinthong, D.; Wongwaisayawan, S. ABCB1 and ABCC2 and the risk of distant metastasis in Thai breast cancer patients treated with tamoxifen. OncoTargets Ther. 2016, 9, 2121–2129. [Google Scholar] [CrossRef][Green Version]
- Lorés-Motta, L.; van Asten, F.; Muether, P.S.; Smailhodzic, D.; Groenewoud, J.M.; Omar, A.; Chen, J.; Koenekoop, R.K.; Fauser, S.; Hoyng, C.B.; et al. A genetic variant in NRP1 is associated with worse response to ranibizumab treatment in neovascular age-related macular degeneration. Pharm. Genom. 2016, 26, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Gagno, S.; D’Andrea, M.R.; Mansutti, M.; Zanusso, C.; Puglisi, F.; Dreussi, E.; Montico, M.; Biason, P.; Cecchin, E.; Iacono, D.; et al. A New Genetic Risk Score to Predict the Outcome of Locally Advanced or Metastatic Breast Cancer Patients Treated with First-Line Exemestane: Results From a Prospective Study. Clin. Breast Cancer 2019, 19, 137–145.e4. [Google Scholar] [CrossRef] [PubMed]
- Gui, H.; Zeld, N.; Williams, L.K.; Lanfear, D.E.; Sabbah, H.N.; Li, J.; She, R.; Luzum, J.A.; Donahue, M.P.; Kraus, W.E.; et al. Polygenic Score for beta-Blocker Survival Benefit in European Ancestry Patients with Reduced Ejection Fraction Heart Failure. Circ. Heart Fail. 2020, 13, e007012. [Google Scholar] [CrossRef]
- Grželj, J.; Mlinarič-Raščan, I.; Marko, P.B.; Marovt, M.; Gmeiner, T.; Šmid, A. Polymorphisms in GNMT and DNMT3b are associated with methotrexate treatment outcome in plaque psoriasis. Biomed. Pharmacother. 2021, 138, 111456. [Google Scholar] [CrossRef]
- Duconge, J.; Santiago, E.; Hernandez-Suarez, D.F.; Moneró, M.; López-Reyes, A.; Rosario, M.; Renta, J.Y.; González, P.; Ileana Fernández-Morales, L.; Antonio Vélez-Figueroa, L.; et al. Pharmacogenomic polygenic risk score for clopidogrel responsiveness among Caribbean Hispanics: A candidate gene approach. Clin. Transl. Sci. 2021, 14, 2254–2266. [Google Scholar] [CrossRef]
- Yin, J.Y.; Li, X.; Li, X.P.; Xiao, L.; Zheng, W.; Chen, J.; Mao, C.X.; Fang, C.; Cui, J.J.; Guo, C.X.; et al. Prediction models for platinum-based chemotherapy response and toxicity in advanced NSCLC patients. Cancer Lett. 2016, 377, 65–73. [Google Scholar] [CrossRef]
- Wessels, J.A.M.; van der Kooij, S.M.; le Cessie, S.; Kievit, W.; Barerra, P.; Allaart, C.F.; Huizinga, T.W.J.; Guchelaar, H.J. A clinical pharmacogenetic model to predict the efficacy of methotrexate monotherapy in recent-onset rheumatoid arthritis. Arthritis Rheum. 2007, 56, 1765–1775. Available online: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=med6&NEWS=N&AN=17530705 (accessed on 28 November 2021). [CrossRef]
- Wang, M.H.; Friton, J.J.; Raffals, L.E.; Leighton, J.A.; Pasha, S.F.; Picco, M.F.; Cushing, K.C.; Monroe, K.; Nix, B.D.; Newberry, R.D.; et al. Novel Genetic Risk Variants Can Predict Anti-TNF Agent Response in Patients With Inflammatory Bowel Disease. J. Crohn’s Colitis 2019, 13, 1036–1043. [Google Scholar] [CrossRef]
- Van der Leeuw, J.; Oemrawsingh, R.M.; van der Graaf, Y.; Brugts, J.J.; Deckers, J.W.; Bertrand, M.; Fox, K.; Ferrari, R.; Remme, W.J.; Simoons, M.L.; et al. Prediction of absolute risk reduction of cardiovascular events with perindopril for individual patients with stable coronary artery disease—Results from EUROPA. Int. J. Cardiol. 2015, 182, 194–199. [Google Scholar] [CrossRef]
- Sordillo, J.E.; Lutz, S.M.; McGeachie, M.J.; Lasky-Su, J.; Weiss, S.T.; Celedon, J.C.; Wu, A.C. Pharmacogenetic Polygenic Risk Score for Bronchodilator Response in Children and Adolescents with Asthma: Proof-of-Concept. J. Pers. Med. 2021, 11, 319. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.P.; Backman, J.D.; Reny, J.L.; Bergmeijer, T.O.; Mitchell, B.D.; Ritchie, M.D.; Dery, J.P.; Pakyz, R.E.; Gong, L.; Ryan, K.; et al. Pharmacogenomic polygenic response score predicts ischaemic events and cardiovascular mortality in clopidogrel-treated patients. Eur. Heart J. Cardiovasc. Pharmacother. 2020, 6, 203–210, Erratum in Eur. Heart J. Cardiovasc. Pharmacother. 2020, 6, 269. Available online: https://www.ncbi.nlm.nih.gov/pubmed/32386422 (accessed on 28 November 2021). [CrossRef]
- Leusink, M.; Maitland-van der Zee, A.H.; Ding, B.; Drenos, F.; van Iperen, E.P.; Warren, H.R.; Caulfield, M.J.; Cupples, L.A.; Cushman, M.; Hingorani, A.D.; et al. A genetic risk score is associated with statin-induced low-density lipoprotein cholesterol lowering. Pharmacogenomics 2016, 17, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Hlavaty, T.; Ferrante, M.; Henckaerts, L.; Pierik, M.; Rutgeerts, P.; Vermeire, S. Predictive model for the outcome of infliximab therapy in Crohn’s disease based on apoptotic pharmacogenetic index and clinical predictors. Inflamm. Bowel Dis. 2007, 13, 372–379. Available online: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=med6&NEWS=N&AN=17206723 (accessed on 28 November 2021). [CrossRef] [PubMed]
- Goricar, K.; Kovac, V.; Dolzan, V. Clinical-pharmacogenetic models for personalized cancer treatment: Application to malignant mesothelioma. Sci. Rep. 2017, 7, 46537. [Google Scholar] [CrossRef] [PubMed]
- Ciuculete, D.M.; Bandstein, M.; Benedict, C.; Waeber, G.; Vollenweider, P.; Lind, L.; Schioth, H.B.; Mwinyi, J. A genetic risk score is significantly associated with statin therapy response in the elderly population. Clin. Genet. 2017, 91, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.C.; Wong, H.S.C.; Chang, W.P.; Chen, B.K.; Wu, M.S.; Yang, K.D.; Hsieh, K.S.; Hsu, Y.W.; Liu, S.F.; Liu, X.; et al. Prediction for Intravenous Immunoglobulin Resistance by Using Weighted Genetic Risk Score Identified from Genome-Wide Association Study in Kawasaki Disease. Circ. Cardiovasc. Genet. 2017, 10, e001625. [Google Scholar] [CrossRef]
- Nelveg-Kristensen, K.E.; Busk Madsen, M.; Torp-Pedersen, C.; Kober, L.; Egfjord, M.; Berg Rasmussen, H.; Riis Hansen, P. Pharmacogenetic Risk Stratification in Angiotensin-Converting Enzyme Inhibitor-Treated Patients with Congestive Heart Failure: A Retrospective Cohort Study. PLoS ONE 2015, 10, e0144195. [Google Scholar] [CrossRef]
- Fransen, J.; Kooloos, W.M.; Wessels, J.A.M.; Huizinga, T.W.J.; Guchelaar, H.J.; van Riel, P.L.C.M.; Barrera, P. Clinical pharmacogenetic model to predict response of MTX monotherapy in patients with established rheumatoid arthritis after DMARD failure. Pharmacogenomics 2012, 13, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Eektimmerman, F.; Allaart, C.F.; Hazes, J.M.; Madhar, M.B.; den Broeder, A.A.; Fransen, J.; Swen, J.J.; Guchelaar, H.J. Validation of a clinical pharmacogenetic model to predict methotrexate nonresponse in rheumatoid arthritis patients. Pharmacogenomics 2019, 20, 85–93. [Google Scholar] [CrossRef]
- Amos, W.; Driscoll, E.; Hoffman, J.I. Candidate genes versus genome-wide associations: Which are better for detecting genetic susceptibility to infectious disease? Proc. R. Soc. B Biol. Sci. 2011, 278, 1183–1188. [Google Scholar] [CrossRef] [PubMed]
- McInnes, G.; Yee, S.W.; Pershad, Y.; Altman, R.B. Genomewide Association Studies in Pharmacogenomics. Clin. Pharmacol. Ther. 2021, 110, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, D.E.; Marsh, S.; McLeod, H.L. Caution with beta1-adrenergic receptor genotyping. Clin. Pharmacol. Ther. 2004, 76, 185–186. [Google Scholar] [CrossRef] [PubMed]
- De Villiers, C.B.; Kroese, M.; Moorthie, S. Understanding polygenic models, their development and the potential application of polygenic scores in healthcare. J. Med. Genet. 2020, 57, 725–732. [Google Scholar] [CrossRef]
- Curtis, M.J.; Abernethy, D.R. Replication—Why we need to publish our findings. Pharmacol. Res. Perspect. 2015, 3, e00164. [Google Scholar] [CrossRef]
- Agnelli, G.; Buller, H.R.; Cohen, A.; Curto, M.; Gallus, A.S.; Johnson, M.; Masiukiewicz, U.; Pak, R.; Thompson, J.; Raskob, G.E.; et al. Oral apixaban for the treatment of acute venous thromboembolism. N. Engl. J. Med. 2013, 369, 25–26. [Google Scholar] [CrossRef]
- Kirley, K.; Qato, D.M.; Kornfield, R.; Stafford, R.S.; Caleb Alexander, G. National Trends in Oral Anticoagulant Use in the United States, 2007–2011. Circ. Cardiovasc. Qual. Outcomes 2012, 5, 615–621. [Google Scholar] [CrossRef]
- Schulman, S.; Kearon, C.; Kakkar, A.K.; Mismetti, P.; Schellong, S.; Eriksson, H.; Baanstra, D.; Schnee, J.; Goldhaber, S.Z. Dabigatran versus warfarin in the treatment of acute venous thromboembolism. N. Engl. J. Med. 2009, 361, 2342–2352. [Google Scholar] [CrossRef]
- Budnitz, D.S.; Lovegrove, M.C.; Shehab, N.; Richards, C.L. Emergency hospitalizations for adverse drug events in older Americans. N. Engl. J. Med. 2011, 365, 2002–2012. [Google Scholar] [CrossRef]
- Johnson, J.A.; Cavallari, L.H. Warfarin Pharmacogenetics. Trends Cardiovasc. Med. 2015, 25, 33–41. [Google Scholar] [CrossRef]
- Daly, A.K. Pharmacogenomics of anticoagulants: Steps toward personal dosage. Genome Med. 2009, 1, 1–4. [Google Scholar] [CrossRef]
- Loebstein, R.; Dvoskin, I.; Halkin, H.; Vecsler, M.; Lubetsky, A.; Rechavi, G.; Amariglio, N.; Cohen, Y.; Ken-Dror, G.; Almog, S.; et al. A coding VKORC1 Asp36Tyr polymorphism predisposes to warfarin resistance. Blood 2007, 109, 2477–2480. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.M.; Stewart, K.; Dooley, M. The ten most common adverse drug reactions (ADRs) in oncology patients: Do they matter to you? Supportive Care Cancer 2004, 12, 626–633. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Cancer in Children in Canada (0–14 Years)—Canada.ca. Available online: https://www.canada.ca/en/public-health/services/chronic-diseases/cancer/cancer-children-canada-0-14-years.html (accessed on 18 May 2022).
- Impicciatore, P.; Choonara, I.; Clarkson, A.; Provasi, D.; Pandolfini, C.; Bonati, M. Incidence of adverse drug reactions in paediatric in/out-patients: A systematic review and meta-analysis of prospective studies. Br. J. Clin. Pharmacol. 2001, 52, 77–83. [Google Scholar] [CrossRef]
- Linskey, D.W.; Linskey, D.C.; McLeod, H.L.; Luzum, J.A. The need to shift pharmacogenetic research from candidate gene to genome-wide association studies. Pharmacogenomics 2021, 22, 1143–1150. [Google Scholar] [CrossRef]
- Maranville, J.C.; Cox, N.J. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharm. J. 2016, 16, 388–392. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Beesley, L.J.; Vandehaar, P.; Peng, R.B.; Salvatore, M.; Zawistowski, M.; Taliun, S.A.G.; Das, S.; Lefaive, J.; Kaleba, E.O.; et al. Exploring various polygenic risk scores for skin cancer in the phenomes of the Michigan genomics initiative and the UK Biobank with a visual catalog: PRSWeb. PLoS Genet. 2019, 15, e1008202. [Google Scholar] [CrossRef]
- Wagner, M.W.; Namdar, K.; Biswas, A.; Monah, S.; Khalvati, F.; Ertl-Wagner, B.B. Radiomics, machine learning, and artificial intelligence—What the neuroradiologist needs to know. Neuroradiology 2021, 63, 1957–1967. [Google Scholar] [CrossRef]
- Ngiam, K.Y.; Khor, I.W. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019, 20, e262–e273. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.; Telkar, N.; Reiker, T.; Walters, R.G.; Lin, K.; Eriksson, A.; Gurdasani, D.; Gilly, A.; Southam, L.; Tsafantakis, E.; et al. The transferability of lipid loci across African, Asian and European cohorts. Nat. Commun. 2019, 10, 4330. [Google Scholar] [CrossRef] [PubMed]
- Duncan, L.; Shen, H.; Gelaye, B.; Meijsen, J.; Ressler, K.; Feldman, M.; Peterson, R.; Domingue, B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019, 10, 3328. [Google Scholar] [CrossRef] [PubMed]
- Mostafavi, H.; Harpak, A.; Agarwal, I.; Conley, D.; Pritchard, J.K.; Przeworski, M. Variable prediction accuracy of polygenic scores within an ancestry group. ELife 2020, 9, e48376. [Google Scholar] [CrossRef] [PubMed]
- Van Driest, S.L.; McGregor, T.L. Pharmacogenetics in clinical pediatrics: Challenges and strategies. Pers. Med. 2013, 10, 661–671. [Google Scholar] [CrossRef]
- Brock, P.R.; Bellman, S.C.; Yeomans, E.C.; Pinkerton, C.R.; Pritchard, J. Cisplatin ototoxicity in children: A practical grading system. Med. Pediatr. Oncol. 1991, 19, 295–300. [Google Scholar] [CrossRef]
- Freyer, D.R.; Chen, L.; Krailo, M.D.; Knight, K.; Villaluna, D.; Bliss, B.; Pollock, B.H.; Ramdas, J.; Lange, B.; Van Hoff, D.; et al. Effects of sodium thiosulfate versus observation on development of cisplatin-induced hearing loss in children with cancer (ACCL0431): A multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2017, 18, 63–74. [Google Scholar] [CrossRef]
- Knight, K.R.G.; Kraemer, D.F.; Neuwelt, E.A. Ototoxicity in children receiving platinum chemotherapy: Underestimating a commonly occurring toxicity that may influence academic and social development. J. Clin. Oncol. 2005, 23, 8588–8596. [Google Scholar] [CrossRef]
- Drögemöller, B.I.; Wright, G.E.B.; Lo, C.; Le, T.; Brooks, B.; Bhavsar, A.P.; Rassekh, S.R.; Ross, C.J.D.; Carleton, B.C. Pharmacogenomics of Cisplatin-Induced Ototoxicity: Successes, Shortcomings, and Future Avenues of Research. Clin. Pharmacol. Ther. 2019, 106, 350–359. [Google Scholar] [CrossRef]
- King, K.A.; Brewer, C.C. Clinical trials, ototoxicity grading scales and the audiologist’s role in therapeutic decision making. Int. J. Audiol. 2018, 57 (Suppl. S4), S89–S98. [Google Scholar] [CrossRef]
- Schmidt, C.M.; Bartholomäus, E.; Deuster, D.; Heinecke, A.; Dinnesen, A.G. The “Muenster classification” of high frequency hearing loss following cisplatin chemotherapy. HNO 2007, 55, 299–306. [Google Scholar] [CrossRef]
- National Cancer Institute. Common Terminology Criteria for Adverse Events (CTCAE) Common Terminology Criteria for Adverse Events (CTCAE) v5.0. Published Online 2017. Available online: https://ctep.cancer.gov/protocoldevelopment/electronic_applications/docs/ctcae_v5_quick_reference_5x7.pdf (accessed on 14 January 2022).
- Chang, K.W.; Chinosornvatana, N. Practical grading system for evaluating cisplatin ototoxicity in children. J. Clin. Oncol. 2010, 28, 1788–1795. [Google Scholar] [CrossRef]
- Clemens, E.; Brooks, B.; De Vries, A.C.H.; van Grotel, M.; van den Heuvel-Eibrink, M.M.; Carleton, B. A comparison of the Muenster, SIOP Boston, Brock, Chang and CTCAEv4.03 ototoxicity grading scales applied to 3799 audiograms of childhood cancer patients treated with platinum-based chemotherapy. PLoS ONE 2019, 14, e0210646. [Google Scholar] [CrossRef] [PubMed]
- Jarrett, D.; Stride, E.; Vallis, K.; Gooding, M.J. Applications and limitations of machine learning in radiation oncology. Br. J. Radiol. 2019, 92, 20190001. [Google Scholar] [CrossRef] [PubMed]
- Phang-Lyn, S.; Llerena, V.A. Biochemistry, Biotransformation; StatPearls: Tampa, FL, USA, 2021. [Google Scholar]
- Joober, R.; Schmitz, N.; Annable, L.; Boksa, P. Publication bias: What are the challenges and can they be overcome? J. Psychiatry Neurosci. 2012, 37, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Bhagat, A.; Kleinerman, E.S. Anthracycline-Induced Cardiotoxicity: Causes, Mechanisms, and Prevention. Adv. Exp. Med. Biol. 2020, 1257, 181–192. [Google Scholar] [CrossRef]
- Callejo, A.; Sedó-Cabezón, L.; Juan, I.D.; Llorens, J. Cisplatin-induced ototoxicity: Effects, mechanisms and protection strategies. Toxics 2015, 3, 268–293. [Google Scholar] [CrossRef]
- Triarico, S.; Romano, A.; Attinà, G.; Capozza, M.A.; Maurizi, P.; Mastrangelo, S.; Ruggiero, A. Vincristine-Induced Peripheral Neuropathy (VIPN) in Pediatric Tumors: Mechanisms, Risk Factors, Strategies of Prevention and Treatment. Int. J. Mol. Sci. 2021, 22, 4112. [Google Scholar] [CrossRef]
- Tellor, K.B.; Nguyen, S.N.; Bultas, A.C.; Armbruster, A.L.; Greenwald, N.A.; Yancey, A.M. Evaluation of the impact of body mass index on warfarin requirements in hospitalized patients. Ther. Adv. Cardiovasc. Dis. 2018, 12, 207–216. [Google Scholar] [CrossRef]
- Johnson, J.A. Pharmacogenetics in clinical practice: How far have we come and where are we going? Pharmacogenomics 2013, 14, 835–843. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Haidar, C.E.; Wilkinson, M.R.; Crews, K.R.; Baker, D.K.; Kornegay, N.M.; Yang, W.; Pui, C.H.; Reiss, U.M.; Gaur, A.H.; et al. PG4KDS: A model for the clinical implementation of pre-emptive pharmacogenetics. Am. J. Med. Genet. C Semin. Med. Genet. 2014, 166, 45–55. [Google Scholar] [CrossRef]
- Danahey, K.; Borden, B.A.; Furner, B.; Yukman, P.; Hussain, S.; Saner, D.; Volchenboum, S.L.; Ratain, M.J.; O’Donnell, P.H. Simplifying the use of pharmacogenomics in clinical practice: Building the genomic prescribing system. J. Biomed. Inform. 2017, 75, 110–121. [Google Scholar] [CrossRef]
- Eadon, M.T.; Desta, Z.; Levy, K.D.; Decker, B.S.; Pierson, R.C.; Pratt, V.M.; Callaghan, J.T.; Rosenman, M.B.; Carpenter, J.S.; Holmes, A.M.; et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clin. Pharmacol. Ther. 2016, 100, 63–66. [Google Scholar] [CrossRef]
- Lip, G.Y.H.; Frison, L.; Halperin, J.L.; Lane, D.A. Comparative validation of a novel risk score for predicting bleeding risk in anticoagulated patients with atrial fibrillation: The HAS-BLED (hypertension, abnormal renal/liver function, stroke, bleeding history or predisposition, labile INR, elderly, drugs/alcohol concomitantly) score. J. Am. Coll. Cardiol. 2011, 57, 173–180. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Ann. Intern. Med. 2015, 162, 55–63. [Google Scholar] [CrossRef]
- Ware, M.P.H.E.B.; Schmitz, L.L.; Faul, M.P.H.J.; Gard, A.M.; Mitchell, C.; Smith, M.P.H.J.A.; Zhao, W.; Weir, D.; Kardia, S.L. Heterogeneity in polygenic scores for common human traits. BioRxiv 2017, 106062. [Google Scholar] [CrossRef]
- Wand, H.; Lambert, S.A.; Tamburro, C.; Iacocca, M.A.; O’Sullivan, J.W.; Sillari, C.; Kullo, I.J.; Rowley, R.; Dron, J.S.; Brockman, D.; et al. Improving reporting standards for polygenic scores in risk prediction studies. Nature 2021, 591, 211–219. [Google Scholar] [CrossRef]
- Moons, K.G.M.; Kengne, A.P.; Woodward, M.; Royston, P.; Vergouwe, Y.; Altman, D.G.; Grobbee, D.E. Risk prediction models: I. Development, internal validation, and assessing the incremental value of a new (bio)marker. Heart 2012, 98, 683–690. [Google Scholar] [CrossRef]
- Moons, K.G.M.; Kengne, A.P.; Grobbee, D.E.; Royston, P.; Vergouwe, Y.; Altman, D.G.; Woodward, M. Risk prediction models: II. External validation, model updating, and impact assessment. Heart 2012, 98, 691–698. [Google Scholar] [CrossRef]
- Moons, K.G.M. Criteria for Scientific Evaluation of Novel Markers: A Perspective. Clin. Chem. 2010, 56, 537–541. [Google Scholar] [CrossRef][Green Version]
- Ivanov, J.; Tu, J.V.; Naylor, C.D. Ready-Made, Recalibrated, or Remodeled? Circulation 1999, 99, 2098–2104. [Google Scholar] [CrossRef]
- What Do We Mean by Validating a Prognostic Model?—Altman—2000—Statistics in Medicine—Wiley Online Library. Available online: https://onlinelibrary.wiley.com/doi/pdf/10.1002/ (accessed on 10 May 2022).
- Steyerberg, E.W.; Borsboom, G.J.J.M.; van Houwelingen, H.C.; Eijkemans, M.J.C.; Habbema, J.D.F. Validation and updating of predictive logistic regression models: A study on sample size and shrinkage. Stat. Med. 2004, 23, 2567–2586. [Google Scholar] [CrossRef]
- Moons, K.G.M.; Altman, D.G.; Vergouwe, Y.; Royston, P. Prognosis and prognostic research: Application and impact of prognostic models in clinical practice. BMJ 2009, 338, 1487–1490. [Google Scholar] [CrossRef]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef]
- Martin, A.R.; Kanai, M.; Kamatani, Y.; Okada, Y.; Neale, B.M.; Daly, M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019, 51, 584–591. [Google Scholar] [CrossRef]
- Kerminen, S.; Martin, A.R.; Koskela, J.; Ruotsalainen, S.E.; Havulinna, A.S.; Surakka, I.; Palotie, A.; Perola, M.; Salomaa, V.; Daly, M.J.; et al. Geographic Variation and Bias in the Polygenic Scores of Complex Diseases and Traits in Finland. Am. J. Hum. Genet. 2019, 104, 1169–1181. [Google Scholar] [CrossRef]
- Kim, M.S.; Patel, K.P.; Teng, A.K.; Berens, A.J.; Lachance, J. Genetic disease risks can be misestimated across global populations 06 Biological Sciences 0604 Genetics. Genome Biol. 2018, 19, 179. [Google Scholar] [CrossRef]
- Slunecka, J.L.; van der Zee, M.D.; Beck, J.J.; Johnson, B.N.; Finnicum, C.T.; Pool, R.; Hottenga, J.J.; de Geus, E.J.; Ehli, E.A. Implementation and implications for polygenic risk scores in healthcare. Hum. Genom. 2021, 15, 46. [Google Scholar] [CrossRef]
- Abdullah-Koolmees, H.; van Keulen, A.M.; Nijenhuis, M.; Deneer, V.H.M. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and RNPGx Guidelines. Front. Pharmacol. 2021, 11, 25. [Google Scholar] [CrossRef]

| n = 89 | |
|---|---|
| Development cohort size (n) | |
| Median (range) | 269 (37.0, 8726) |
| Validation cohort size (n) | |
| Median (range) | 187 (16.0, 14,348) |
| Method of SNP-selection for inclusion in polygenic model | |
| Candidate-gene | 74 (83.1%) |
| Genome-wide association | 11 (12.4%) |
| Validation of existing polygenic model | 4 (4.5%) |
| Method for model development | |
| Machine Learning | 11 (12.4%) |
| Regression-based method | 68 (76.4%) |
| Other | 10 (11.2%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siemens, A.; Anderson, S.J.; Rassekh, S.R.; Ross, C.J.D.; Carleton, B.C. A Systematic Review of Polygenic Models for Predicting Drug Outcomes. J. Pers. Med. 2022, 12, 1394. https://doi.org/10.3390/jpm12091394
Siemens A, Anderson SJ, Rassekh SR, Ross CJD, Carleton BC. A Systematic Review of Polygenic Models for Predicting Drug Outcomes. Journal of Personalized Medicine. 2022; 12(9):1394. https://doi.org/10.3390/jpm12091394
Chicago/Turabian StyleSiemens, Angela, Spencer J. Anderson, S. Rod Rassekh, Colin J. D. Ross, and Bruce C. Carleton. 2022. "A Systematic Review of Polygenic Models for Predicting Drug Outcomes" Journal of Personalized Medicine 12, no. 9: 1394. https://doi.org/10.3390/jpm12091394
APA StyleSiemens, A., Anderson, S. J., Rassekh, S. R., Ross, C. J. D., & Carleton, B. C. (2022). A Systematic Review of Polygenic Models for Predicting Drug Outcomes. Journal of Personalized Medicine, 12(9), 1394. https://doi.org/10.3390/jpm12091394






