Abstract
Relapsed and refractory (R/r) disease in paediatric acute leukaemia remains the first reason for treatment failure. Advances in molecular characterisation can ameliorate the identification of genetic biomarkers treatment strategies for this disease, especially in high-risk patients. The purpose of this study was to analyse a cohort of R/r children diagnosed with acute lymphoblastic (ALL) or myeloid (AML) leukaemia in order to offer them a targeted treatment if available. Advanced molecular characterisation of 26 patients diagnosed with R/r disease was performed using NGS, MLPA, and RT-qPCR. The clinical relevance of the identified alterations was discussed in a multidisciplinary molecular tumour board (MTB). A total of 18 (69.2%) patients were diagnosed with B-ALL, 4 (15.4%) with T-ALL, 3 (11.5%) with AML and 1 patient (3.8%) with a mixed-phenotype acute leukaemia (MPL). Most of the patients had relapsed disease (88%) at the time of sample collection. A total of 17 patients (65.4%) were found to be carriers of a druggable molecular alteration, 8 of whom (47%) received targeted therapy, 7 (87.5%) of them in addition to hematopoietic stem cell transplantation (HSCT). Treatment response and disease control were achieved in 4 patients (50%). In conclusion, advanced molecular characterisation and MTB can improve treatment and outcome in paediatric R/r acute leukaemias.
1. Introduction
Paediatric cancer is the main cause of disease-related mortality in children in developed countries [1]. Haematological malignancies are the most frequent cancer in this population, and within them, acute lymphoblastic leukaemia (ALL) is the predominant type, comprising 20% of total cancer cases occurring before the age of 19 [2,3].
The survival rates of paediatric leukaemia have dramatically increased over the last few decades; the current 5-year overall survival for ALL is 90% [4], constituting 65–70% for AML [5]. This achievement is due, among other reasons, to the establishment of international collaborative research platforms, the improvement of risk stratification and the introduction of multimodal innovative treatments, such as immunotherapy or chimeric antigen receptor (CAR) -T cell therapy [6,7].
Despite such improvements, R/r patients still represent a high-risk population with cure rates of less than 30% [8], due to disease progression or to the noxious effects, both in the short and long term, associated with the intensification of cytotoxic treatments [9]. Such poor prognosis is a powerful reason for the urgent need of novel, innovative therapies that are more specific and efficient in cases of no response.
On this basis, the concept and application of personalised medicine has led to a paradigm shift in the care of children with haematological malignancies. The exploration of genomics through the advent of next-generation sequencing (NGS) has enabled the identification of oncogenic tumour biomarkers and the progressive understanding of drug resistance mechanisms acquired by blasts at relapse. This constitutes a game-changing tool to potentially optimise molecular diagnosis, identify high-risk patients, find druggable mutations and support patient follow-up through time [10].
Within the possibilities offered by NGS, targeted panels are the most frequently used technique in clinical settings, due to their highly practical nature when compared to the whole genome or exome sequencing. Nevertheless, we must bear in mind a predominance of panels designed for adult cancer in the market [11]. The optimisation of panels by independent research groups to match paediatric molecular alterations has become the ultimate diagnostic approach; this is a cost-effective and flexible technique that allows for fast incorporation of the data that arises from clinical trials and publications in a timely manner [12].
Here, we report the experience of a paediatric haemato-oncology department from a tertiary hospital in Spain, performing molecular characterisation in a cohort of paediatric patients with acute leukaemia with the objective of potentially offering them targeted therapy.
2. Materials and Methods
2.1. Patients
Written informed consent to participate, approved by the hospital’s Ethics Committee, was obtained from the parents or legal guardians of the patients.
Isolated bone marrow disease was defined as ≥25% of blast cells in bone marrow aspirate (cytomorphology) and/or as presence of pathological clonality by flow cytometry similar to diagnosis. Central nervous system (CNS) involvement was defined as the presence of >5% of nucleated cells with bast evidence in cerebrospinal fluid (CSF). Isolated extramedullary relapse was defined by imaging evidence and biopsy confirmation of infiltration. Combined relapse was defined as the presence of ≥5% of blasts (cytomorphology) or presence of pathological clonality by flow cytometry, and at least one extramedullary involvement. Refractory disease was defined as the absence of complete remission (CR) after induction treatment.
A total of 26 patients diagnosed with relapsed or refractory acute leukaemia in a tertiary hospital between 2014 and 2021 were included in this study. CNS involvement was evaluated by lumbar puncture in all patients, and other extramedullary localisations were studied according to clinical suspicion. Diagnosis was confirmed by cytomorphology and immunophenotype, following the current guidelines and World Health Organization (WHO) criteria [13]. Cytogenetic data were obtained by cellular culture and karyotype analysis. Patients were classified depending on their karyotype in hypodiploid, hyperdiploid, complex or normal. For ALL, the rearrangements in MLL and ETV6-RUNX1, E2A-PBX1 and BCR-ABL translocations were analysed by FISH. For the patients with AML, MLL rearrangements were studied.
2.2. Samples
Bone marrow samples were collected by bone marrow aspirate either at diagnosis and/or at the moment of relapse/refractoriness. In the cases with isolated CNS relapse, blast cells were obtained from CSF. Tumour DNA and RNA from bone marrow or CSF samples were extracted using the Chemagic Automated DNA Separation System (Chemagen®, Baesweiler, Germany) and RNeasy Mini Kit (Qiagen®, Hilden, Germany), respectively, according to the manufacturer’s instructions. DNA and RNA concentration and integrity were determined using the TECAN spectrophotometer, Qubit fluorometer, and Agilent Bioanalyzer.
2.3. Multiplex Ligand-Probe Amplification (MLPA)
Copy number alterations (CNAs) were analysed in B-ALL and T-ALL samples by MLPA. We used SALSA MLPA Probemix P335 ALL-IKZF1 and SALSA MLPA Probemix P383 T-ALL (MRC Holland®). The selection of the reference samples was performed according to the manufacturer’s instructions. In each experiment, we included at least 3 DNA samples obtained from peripheral blood of healthy individuals, with a normal copy number for the sequences detected by the target and reference probes (MRC-Holland®, www.mrcholland.com (accessed from July to December 2021)). CNAs were analysed using the ABI PRISM 3130 XL system and Coffalyser® NET Software (v.210604.1451) MRC-Holland® (Amsterdam, The Netherlands).
2.4. Quantitative Polymerase Chain Reaction (qPCR)
The quantification of the expression of the CRLF2 and WT1 genes was carried out by qPCR. The commercial TaqMan gene expression assays Hs00845692_m1 (Invitrogen®, Waltham, MA, USA) and the kit Ipsogen WT1 ProfileQuant (ELN) (Qiagen®) were used to target CRLF2 and WT1 genes, respectively.
WT1 expression was detected using the Ipsogen WT1 ProfileQuant kit following the manufacturer’s instructions. In the case of CRLF2, relative expression was calculated using the comparative Ct method and obtaining the relative fold-change value (2−ΔΔCt). At least three healthy control samples were included and a relative-fold change value in the patient sample above 2 was considered as CRLF2 overexpression [14].
2.5. Next-Generation Sequencing (NGS)
DNA samples were analysed using a customised panel including two different versions. First, version 1 (v.1), and a second and updated version 2 (v.2), where 91 and 182 genes, respectively, previously related to paediatric leukaemia and sensitivity to targeted therapies, were studied (Supplementary Table S1). The sequencing and the analysis of the raw data were carried out using the Illumina platform and a tailor-made pipeline, respectively. The resulting DNA sequence reads were mapped on the human reference sequence hg19 using Burrows–Wheeler Aligner (v0.7.17) [15]. PCR duplicates were removed using Picard (v2.18.25) [16], and recalibration of the reads was performed using the Genome Analysis Toolkit (GATK v4.1.4.9) [17]. SNPs and indels were detected using Pisces (v5.2.10.49) [18], Mutect2 (GATK v4.1.4.9) [19] and Lofreq (v2.1.5) [20]. Additionally, internal tandem duplications (ITD) in FLT3 gene were studied with the software Manta (v1.6.0) [21]. Variants were annotated in the Variant Call File (VCF) with gene names, predicted functional effect, protein positions and amino-acid changes, conservation scores, and population frequency data. The somatic variants identified were filtered and classified according to the American Association for Molecular Pathology (AMP) criteria [22] using VarSeq™ v2.2.0 (Golden Helix, Inc., Bozeman, MT, USA, www.goldenhelix.com (accessed from July to December 2021). Variant allele frequency (VAF) represents the fraction of reads containing a mutation divided by the total number of reads at a given locus and is a measure of mutational abundance. In our case, since bone marrow was used for DNA isolation and blast enrichment was not performed, the VAF refers to mutational abundance in bone marrow.
2.6. Molecular Tumour Board
Once the results were obtained and interpreted, the identified variants were considered for their diagnostic/prognostic and/or therapeutic potential.
A molecular tumour board (MTB) was constituted in order to create a multidisciplinary discussion platform that could optimise the decisions made in the final stage of this process. The main objective of the MTB was to facilitate discussions on the relevance and actionability of variants, accessibility and pursuit of clinical trials/drugs and treatment outcomes after prescription of targeted drugs.
This personalised medicine committee comprised the expertise of a wide range of professionals, including paediatric oncologists, pathologists, pharmacists, geneticists, bioinformatics, clinical trial experts and basic researchers. Figure 1 represents a schematisation of the work flow from the moment of sample extraction to the committee’s final decision and successive steps.
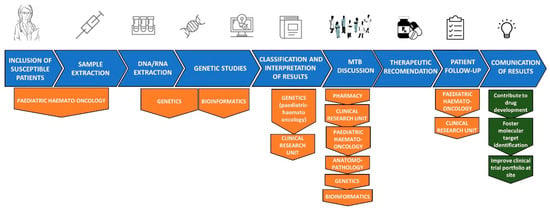
Figure 1.
Work flow from sample collection to MTB recommendation.
2.7. Statistical Analysis
For the quantitative variables, median and range were used. For the qualitative variants, absolute and relative frequencies were indicated.
3. Results
3.1. Clinical Data
Twenty-six patients were included in this study. The clinical data of the patients are summarised in Table 1. Detailed clinical data are provided in Supplementary Table S2.

Table 1.
Summary of clinical data.
The median age was 7.3 years (range 0.5–16.9) at diagnosis. The median time from diagnosis to relapse was 5 months (range 1–86). The median time from diagnosis to refractoriness was 6.8 months (range 1–19.1). A total of 65% of patients were male and the most frequent diagnosis was B-ALL (69.2%), followed by T-ALL (15.4%), AML (11.5%) and MPL (3.9%). Most patients had a normal karyotype (65.5%) and did not present CNS infiltration (88.5%). The median follow up was 49 months (range 6.1–154.6).
3.2. Molecular Findings
3.2.1. CRLF2 and WT1 Overexpression
The overexpression of the CRLF2 and WT1 genes has been described in acute lymphoblastic and myeloid leukaemia, respectively [23,24], and its assessment can be used, for instance, in addition to minimal residual disease, as a monitoring tool during follow-up in order to evaluate the persistence of disease and risk of relapse [25]. For that reason, the expression of CRLF2 was analysed by qPCR in the patients diagnosed with ALL, while WT1 was studied in AML and MPL patients. In this way, gene overexpression was studied in 21 patients (80.8%), including 18 diagnosed with ALL, 2 with AML and 1 with MPL. It was performed in a total of 24 samples, including 6 collected at the moment of diagnosis, 3 at refractoriness and 15 at relapse. Three patients (HRL 7, HRL 14 and HRL 25) were studied at two different time-points (diagnosis and relapse). CRLF2 and WT1 overexpression were detected in 4 patients diagnosed with ALL (15.4%, 2 with B-ALL and 2 with T-ALL) and in 3 patients diagnosed with AML (11.5%).
3.2.2. Copy Number Alterations (CNAs)
The most frequent CNAs in childhood leukaemia were evaluated by MLPA, FISH and/or NGS. Principal deletions were found in CDKN2A (8 patients, 30.8%), CDKN2B (6 patients, 23%), IKZF1 (4 patients, 15.4%) and PAX5 (4 patients, 15.4%). Duplications were found in 6 patients (23%). The CNAs detected are shown in Supplementary Table S3.
3.2.3. Genetic Alterations Identified by NGS
NGS was performed in all patients. A total of 36 bone marrow samples were sequenced, 9 (25%) using v.1 and 27 (75%) using v.2 of the panel. Of them, 13 (36%) were collected at diagnosis, 3 (8.5%) at refractoriness and 20 (55.5%) at relapse. A total of 45 clinically relevant variants according to the AMP criteria [11] were found. Of them, 4 (8.9%) were classified as Tier I-diagnostic, 5 (11%) as Tier I-prognostic, 4 (8.9%) as Tier I-therapeutic; 12 (26.7%) as Tier II-diagnostic, 5 (11%) as Tier II-prognostic and 15 (33.3%) as Tier II-therapeutic. Tier I and Tier II variants were identified in 16 patients (61.5%). Among these 45 variants, 17 (37.8%) were found at diagnosis, 3 (6.7%) at refractoriness and 25 (55.5%) at relapse. Furthermore, 30 (66.7%) corresponded to single-nucleotide variants (SNV) and 15 (33.3%) to small insertions or deletions (indels). They were mostly missense mutations (n = 24; 53.3%), followed by frameshift mutations due to small indels (n = 12; 26.7%), non-sense mutations (n = 5; 11%), in-frame indels (n = 3; 6.7%) and splice-site variants (n = 1; 2.2%). The most frequently altered genes identified by NGS were FLT3, PTPN11, WT1, KRAS (3 patients, 12%) and PHF6, PTEN, NRAS, NT5C2 (2 patients; 8%). They mostly fall among key signaling pathways, such as the RAS/MAPK pathway, PI3K-AKT and JAK-STAT. Figure 2 shows the results of the advanced molecular characterisation of all the samples.
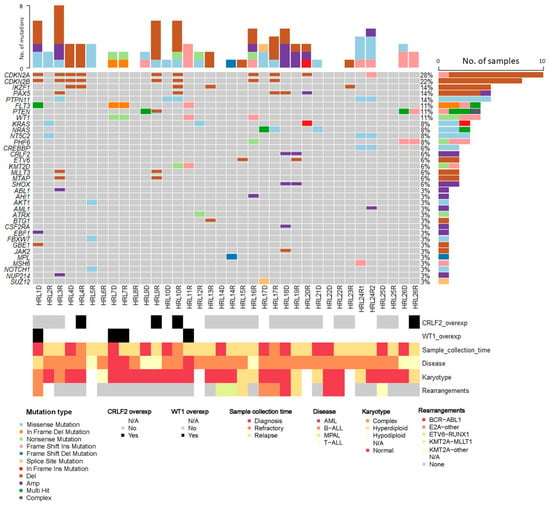
Figure 2.
Advanced molecular characterisation of the analysed samples.
When comparing the samples analysed by NGS at diagnosis with R/r, or in consecutive relapses (HRL 24), we found that patient HRL 7 presented the same oncogenic variants in both samples, whereas patients HRL 9, HRL10, HRL 17, HRL 24 and HRL 26 showed some differences (Supplementary Table S3). Nevertheless, in most cases, these differences were in variants with a low VAF and could be the result of differences in depth. As an exception, patient HRL 26 presented at diagnosis one mutation in PHF6 and two different mutations in PTEN, with a VAF between 0.3 and 0.4. At relapse, one of the PTEN mutation disappeared while the other doubled its proportion, becoming predominant.
3.3. Directed Therapy According to Genetic Characterisation
After performing a complete molecular characterisation, clinical and biological findings were discussed in the MTB in order to select high-risk patients or patients where a specific treatment could be offered. Of 26 patients, 16 (61.5%) were found to harbour a druggable mutation. Of them, 8 (50%) patients received personalised therapy after discussion by this committee. This kind of treatment was considered when no other standardised curable approach could be offered. The therapies were given inside a clinical trial whenever possible, and if not, within a compassionate use program. In addition, HSCT was performed in 7 (87.5%) patients. A total of 4 patients who received personalised treatment (50%) achieved a good response and control of their disease. Table 2 summarises this data.

Table 2.
Patients who received personalised treatment.
The remaining eight patients did not undergo any personalised treatment because of the preclinical status of the targeted drug (HRL 5 and HRL 26), the absence of potential clinical trial enrolment with denegation of a compassionate use program (HLR 12), adequate response to a second-line treatment after a first relapse (HRL 3 and HRL 20) or exitus before treatment could be started (HRL 1, HRL 9, and HRL 21).
4. Discussion
The outcomes in paediatric acute leukaemia have progressively improved over the last few decades [5,26]. However, survival rates in R/r disease are poor and constitute a group of patients that are challenging to treat [27]. Recent advances in molecular technologies, such as NGS, represent a very large promise to accelerate precision medicine for the care of children with cancer, and this is especially relevant in the case of high-risk patients. In this study, we showed that an NGS-based DNA panel, in combination with other molecular techniques, can be used to identify targetable genetic alterations in a high proportion of patients with R/r paediatric acute leukaemia. This molecular information, whenever possible, was used to select a personalised treatment that was able to control the disease in half of the cases.
We used a customised NGS panel in combination with MLPA and qPCR to detect the most frequent alterations in paediatric acute leukaemia. The most frequently altered genes in our cohort of patients (CDKN2A, CDKN2B, IKZF1, PAX5, PTNP11, FLT3, PTEN, CRLF2, WT1, KRAS, NRAS, PHF6, etc) have been previously reported in paediatric leukaemia [28,29,30,31,32] and are known to be involved in biological pathways related to leukemogenesis. We could also analyse, in some cases, samples at diagnosis and at relapse/refractoriness. Since some differences existed in the oncogenic variants identified in both samples, we highlight the importance of analysing the sample that is closest to the moment of treatment. When comparing samples from the same patient, collected at diagnosis and relapse, advanced molecular characterisation provides information about the clonal evolution of the disease, as has been previously reported [33].
Although a targeted approach has the inconvenience of being restricted to a limited number of genes, it also has some important advantages in clinical practice, such as the gain in efficiency both in terms of cost and time, and the more optimal coverage of targeted areas. In fact, our approach allowed the identification of oncogenic variants in 13 (50%) of the patients and the detection of actionable alterations in 17 (65.4%) of the patients. Unfortunately, only 8 (47%) of the patients with an actionable alteration were candidates to receive targeted treatment, in addition to hematopoietic stem cell transplantation that was performed in 7 (87.5%) of them. Among the patients who received personalised treatment, half of them achieved a good response.
In this study, the most frequently altered genes were CDKN2A and CDKN2B. Mutations in cell cycle regulators, such as deletions of CDKN2A/B gene (frequency: 5–20% in B-cell precursor and 60–80% in T-cell ALL), have a negative impact in the function of tumour suppressor genes such as TP53 and RB1 [28]. We found CDKN2A/B deletions in 2/4 (50%) T-ALL patients and in 6/18 (33.3%) patients with B-ALL. In addition, concomitant CDKN2A/B and PAX5 alterations were found in 4/8 patients (50%), all of them diagnosed with B-ALL, and a pathogenic somatic variant in CDKN2A was identified in one patient diagnosed with B-ALL. CDKN2A/B mutations can be potentially targeted with cyclin inhibitors, such as ribociclib or palbociclib, which are beginning to be introduced into clinical trials in children (NCT03740334) [28,34]. Ribociclib has been successfully employed in multiple malignancies, not only in ALL, but also in combination with other drugs [35,36,37]. It is an important drug to be considered, especially in T-ALL [38]. Nevertheless, resistance to cyclin inhibitors has been reported in patients with some concomitant mutations, such as RB1, p16 and especially TP53 [39,40,41,42,43]. This fact was studied before offering cyclin inhibitors. In our study, one patient that harboured a CDKN2A deletion and another one with a loss of function mutation in CDKN2A were treated with ribociclib. Response was maintained only in the first patient (50%).
Probably, the most common alterations in leukaemia are those found in the RAS/MAPK pathway (50% of relapsed B-ALL, 15% of T-ALL and 50% of AML), such as KRAS, NRAS and PTPN11 mutations [28,31]. We found 16 somatic variants (42%) in genes related to the RAS/MAPK pathway [44,45,46]. These variants, especially KRAS and NRAS ones, are localised in hot-spot regions. Since mutations in the RAS/MAPK pathway have been shown to be enriched at relapse, they are believed to constitute a factor for the development of drug resistance [28]. Although initially thought to be undruggable, there are ongoing research efforts to develop MEK or PIK3 inhibitors, such as trametinib [47,48], with controversial results. In this report, KRAS and NRAS mutations have been treated with trametinib, without response and fatal disease progression.
FLT3 mutations are frequently associated with AML in about one third of cases [49]. In this cohort, FLT3 was altered in 2/3 AML patients (67%). FLT3 main mutations can be classified in internal tandem duplicates (ITD), present in about 25% of patients, and somatic mutations in the tyrosine kinase domain (TKD), which occurs in approximately 5% of cases. There is a number of drugs being tested for these mutations, such as midostaurin, quizartinib or sorafenib [28]. Treatment with sunitinib (FLT3-TKD inhibitor) and quizartinib (FLT3-ITD inhibitor) was offered for compassionate use and in a clinical trial (NCT03793478), respectively, to two patients harbouring a FLT3 mutation, achieving CR of the disease with sunitinib. The results of the patient treated under the clinical trial are not discussed in this paper, as the trial is ongoing. The patient treated with sunitinib had a medullar relapse 4 months after its discontinuation (treated for 1 year). After NGS study of the relapse, the same somatic FLT3-TKD mutation present at diagnosis was found, and treatment with sunitinib was restarted after a second HSCT for an expected period of two years. This is an important example about the uncertain management of this kind of treatments and the controversy about how long to maintain them. Until now, there are no standardised guidelines nor recommendations about it.
It is known that 5–7% of patients with B-ALL show an overexpression of CRLF2 disrupting the JAK/STAT pathway, and this is commonly associated with activating JAK2 somatic mutations by rearrangements [12,43,50,51,52], rendering a possible therapeutic target for ruxolitinib, a JAK1/2 inhibitor. In this cohort, we discovered 4 cases with CRLF2 overexpression (15.4%). Treatment with JAK/SAT inhibitors, such as ruxolitinib, was given in 2 B-ALL patients (50%). All patients presented a response to this treatment.
In this cohort, all patients treated with these new drugs were candidates to receive HSCT. Treatment was sometimes started before transplantation in order to help to achieve CR or after the procedure, as a pre-emptive therapy. Although the performance of HSCT could act as a confounding factor when considering the effectiveness of targeted therapy, the case of the patient who relapsed after discontinuing sunitinib reinforces our hypothesis of its utility. Two patients died before HSCT could be performed due to disease progression.
An important fact to take into account is the difficulty in obtaining new drugs that have not been approved for paediatric R/r acute leukaemia. All new therapies should ideally be administered within the context of clinical trials [53]. However, trials are not always available for patients, and several other limitations, such as eligibility criteria, must be considered [54]. Due to this limitation, its use depends, on many occasions, upon compassionate, off-label or extended use prescriptions, with the consequent safety and efficacy concerns. The future existence of robust evidence in this field relies on the strengthening and implementation of precision medicine strategies that promote oncogenic variant identification and clinical trial enrolment [55].
In addition, previously described variants associated with prognosis have been found. For example, variants related to drug resistance, such as NT5C2 and MSH6, in ALL have been linked to thiopurines resistance [56,57]. In the same way, variants are also related to a worse prognosis with higher risk of refractoriness and relapse in AML, such as WT1 [58,59,60], or with a favourable outcome related to a better treatment response, such FBXW7 and NOTCH1 in T-ALL [61,62,63].
It is becoming increasingly clear that advanced molecular characterisation is an important tool in paediatric oncology. It has been employed in tertiary hospitals in the past few years, not only in acute leukaemia, but in all paediatric tumours, mostly solid tumours [64]. Understanding the molecular landscape of acute leukaemia can lead us to the possibility of better risk stratification and the pathway of personalised medicine [65]. Accordingly, at the present time, the management of the paediatric oncological patient requires a multidisciplinary approach, where different and important points of view must be integrated. In this line, in our centre, a molecular tumour board has been recently created. Paediatric haemato-oncologists, oncological geneticists and pharmacists are key pieces in its integration.
This paper summarises the recent experience in the creation of a molecular leukaemia tumour board in a Spanish tertiary hospital. Although the reported data are promising, this study has points that should be improved. For example, it does not focus on the new cytogenetic alterations that have been shown to be associated with an unfavourable prognosis. In addition, the heterogeneity and small sample size, the absence of bone marrow samples at diagnosis from all the patients and the different follow-up times of this study are limitations. New studies with a larger number of patients, in which not only the advanced molecular characterisation is taken into account but also the detailed cytogenetic study, would provide more information on this type of disease.
5. Conclusions
Advanced molecular characterisation, especially NGS, represents a paradigm shift in oncology. A better understanding of the disease is essential in order to improve not only diagnostic strategies, but it may also be an important therapeutic tool.
In reference to paediatric R/r acute leukaemia, the information given by molecular characterisation is a very important strategy that should be implemented as part of standard clinical practice, both at diagnosis, and more importantly, in refractory or relapsed disease.
It also should be taken into account in conventional guidelines at the moment of diagnosis for risk stratification, although information on prognostic significance of some variants is still lacking.
Personalised medicine and the implementation of new therapies are nowadays the present of contemporary oncology, and represent the pathway to the achievement of the total cure.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/2075-4426/12/6/881/s1. Table S1: Summary of the genes included in the panel; Table S2: Summary of clinical data of the patients; Table S3: Summary of the advanced molecular characterisation of the probes analysed.
Author Contributions
Conceptualisation: A.E.-L. and A.P.-M.; methodology: A.E.-L., N.M., A.P.-S. and V.G.-G.; software: B.R.-C.; validation and formal analysis: B.R.-C., A.P.-S., A.E.-L., N.M. and V.G.-G.; investigation: all the authors; data curation: B.R.-C., A.E.-L., N.M. and V.G.-G.; writing—original draft preparation: V.G.-G., N.M. and P.V.-S.; writing—review and editing: V.G.-G.; N.M., B.O.-F., P.G.-G., J.M., B.G., I.M.-R., S.S.R.-P., P.E.-M., A.I.-N. and A.E.-L.; supervision and project administration: A.E.-L. and A.P.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of La Paz University Hospital.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Acknowledgments
Our gratitude to Fondos FEDER (FIS) PI18/01301, as well as Instituto de Salud Carlos III (ISCIII) and especially to the Cris contra el Cáncer Foundation (http://criscancer.org).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Pui, C.-H.; Gajjar, A.J.; Kane, J.R.; Qaddoumi, I.A.; Pappo, A.S. Challenging Issues in Pediatric Oncology. Nat. Rev. Clin. Oncol. 2011, 8, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Downing, J.R.; Wilson, R.K.; Zhang, J.; Mardis, E.R.; Pui, C.-H.; Ding, L.; Ley, T.J.; Evans, W.E. The Pediatric Cancer Genome Project. Nat. Genet. 2012, 44, 619–622. [Google Scholar] [CrossRef]
- Onciu, M.; Pui, C.-H. Diagnosis and Classification. In Childhood Leukemias; Pui, C.-H., Ed.; Cambridge University Press: Cambridge, UK, 2012; pp. 21–48. ISBN 978-0-511-97763-3. [Google Scholar]
- Inaba, H.; Mullighan, C.G. Pediatric Acute Lymphoblastic Leukemia. Haematology 2020, 105, 2524–2539. [Google Scholar] [CrossRef] [PubMed]
- de Morais, R.V.; de Souza, M.V.; de Silva, K.A.S.; Santiago, P.; Lorenzoni, M.C.; Lorea, C.F.; de Junior, C.G.C.; Taniguchi, A.N.R.; Scherer, F.F.; Michalowski, M.B.; et al. Epidemiological Evaluation and Survival of Children with Acute Myeloid Leukemia. J. Pediatr. 2021, 97, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.M.; Neglia, J.P.; Woods, W.G.; Sandlund, J.T.; Pui, C.-H.; Kun, L.E.; Robison, L.L.; Green, D.M. Lessons from the Past: Opportunities to Improve Childhood Cancer Survivor Care through Outcomes Investigations of Historical Therapeutic Approaches for Pediatric Hematological Malignancies. Pediatr. Blood Cancer 2012, 58, 334–343. [Google Scholar] [CrossRef]
- Hucks, G.; Rheingold, S.R. The Journey to CAR T Cell Therapy: The Pediatric and Young Adult Experience with Relapsed or Refractory B-ALL. Blood Cancer J. 2019, 9, 10. [Google Scholar] [CrossRef]
- von Stackelberg, A.; Völzke, E.; Kühl, J.-S.; Seeger, K.; Schrauder, A.; Escherich, G.; Henze, G.; Tallen, G. ALL-REZ BFM Study Group Outcome of Children and Adolescents with Relapsed Acute Lymphoblastic Leukaemia and Non-Response to Salvage Protocol Therapy: A Retrospective Analysis of the ALL-REZ BFM Study Group. Eur. J. Cancer 2011, 47, 90–97. [Google Scholar] [CrossRef]
- Kopp, L.M.; Gupta, P.; Pelayo-Katsanis, L.; Wittman, B.; Katsanis, E. Late Effects in Adult Survivors of Pediatric Cancer: A Guide for the Primary Care Physician. Am. J. Med. 2012, 125, 636–641. [Google Scholar] [CrossRef]
- Tran, T.H.; Shah, A.T.; Loh, M.L. Precision Medicine in Pediatric Oncology: Translating Genomic Discoveries into Optimized Therapies. Clin. Cancer Res. 2017, 23, 5329–5338. [Google Scholar] [CrossRef][Green Version]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef]
- Hiemenz, M.C.; Ostrow, D.G.; Busse, T.M.; Buckley, J.; Maglinte, D.T.; Bootwalla, M.; Done, J.; Ji, J.; Raca, G.; Ryutov, A.; et al. OncoKids: A Comprehensive Next-Generation Sequencing Panel for Pediatric Malignancies. J. Mol. Diagn. 2018, 20, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Beau, M.M.L.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 Revision to the World Health Organization Classification of Myeloid Neoplasms and Acute Leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Renneville, A.; Hermitte, F.; Hills, R.K.; Daly, S.; Jovanovic, J.V.; Gottardi, E.; Fava, M.; Schnittger, S.; Weiss, T.; et al. Real-Time Quantitative Polymerase Chain Reaction Detection of Minimal Residual Disease by Standardized WT1 Assay to Enhance Risk Stratification in Acute Myeloid Leukemia: A European LeukemiaNet Study. J. Clin. Oncol. 2009, 27, 5195–5201. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Picard Tools—By Broad Institute. Available online: https://broadinstitute.github.io/picard/ (accessed on 13 March 2022).
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Dunn, T.; Berry, G.; Emig-Agius, D.; Jiang, Y.; Lei, S.; Iyer, A.; Udar, N.; Chuang, H.-Y.; Hegarty, J.; Dickover, M.; et al. Pisces: An Accurate and Versatile Variant Caller for Somatic and Germline next-Generation Sequencing Data. Bioinformatics 2019, 35, 1579–1581. [Google Scholar] [CrossRef]
- Benjamin, D.; Sato, T.; Cibulskis, K.; Getz, G.; Stewart, C.; Lichtenstein, L. Calling Somatic SNVs and Indels with Mutect2. BioRxiv 2019, 861054. [Google Scholar] [CrossRef]
- Wilm, A.; Aw, P.P.K.; Bertrand, D.; Yeo, G.H.T.; Ong, S.H.; Wong, C.H.; Khor, C.C.; Petric, R.; Hibberd, M.L.; Nagarajan, N. LoFreq: A Sequence-Quality Aware, Ultra-Sensitive Variant Caller for Uncovering Cell-Population Heterogeneity from High-Throughput Sequencing Datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.-L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Kim, H.-J.; Shin, S.-H.; Yahng, S.-A.; Lee, S.-E.; Cho, B.-S.; Eom, K.-S.; Kim, Y.-J.; Lee, S.; Min, C.-K.; et al. Serial Measurement of WT1 Expression and Decrement Ratio until Hematopoietic Cell Transplantation as a Marker of Residual Disease in Patients with Cytogenetically Normal Acute Myelogenous Leukemia. Biol. Blood Marrow. Transplant. 2013, 19, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Gottardi, E.; Saglio, G. WT1 Overexpression in Acute Myeloid Leukemia and Myelodysplastic Syndromes. Methods Mol. Med. 2006, 125, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.-H.; Yang, J.J.; Hunger, S.P.; Pieters, R.; Schrappe, M.; Biondi, A.; Vora, A.; Baruchel, A.; Silverman, L.B.; Schmiegelow, K.; et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J. Clin. Oncol. 2015, 33, 2938–2948. [Google Scholar] [CrossRef] [PubMed]
- Oskarsson, T.; Söderhäll, S.; Arvidson, J.; Forestier, E.; Montgomery, S.; Bottai, M.; Lausen, B.; Carlsen, N.; Hellebostad, M.; Lähteenmäki, P.; et al. Relapsed Childhood Acute Lymphoblastic Leukemia in the Nordic Countries: Prognostic Factors, Treatment and Outcome. Haematologica 2016, 101, 68–76. [Google Scholar] [CrossRef]
- Rau, R.E.; Loh, M.L. Using Genomics to Define Pediatric Blood Cancers and Inform Practice. Hematol. Am. Soc. Hematol. Educ. Program 2018, 2018, 286–300. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pan-Cancer Genome and Transcriptome Analyses of 1,699 Paediatric Leukaemias and Solid Tumours. Nature 2018, 555, 371–376. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Wang, B.-Y.; Zhang, W.-N.; Huang, J.-Y.; Li, B.-S.; Zhang, M.; Jiang, L.; Li, J.-F.; Wang, M.-J.; Dai, Y.-J.; et al. Genomic Profiling of Adult and Pediatric B-Cell Acute Lymphoblastic Leukemia. EBioMedicine 2016, 8, 173–183. [Google Scholar] [CrossRef]
- Liu, Y.; Easton, J.; Shao, Y.; Maciaszek, J.; Wang, Z.; Wilkinson, M.R.; McCastlain, K.; Edmonson, M.; Pounds, S.B.; Shi, L.; et al. The Genomic Landscape of Pediatric and Young Adult T-Lineage Acute Lymphoblastic Leukemia. Nat. Genet. 2017, 49, 1211–1218. [Google Scholar] [CrossRef]
- Mercher, T.; Schwaller, J. Pediatric Acute Myeloid Leukemia (AML): From Genes to Models Toward Targeted Therapeutic Intervention. Front. Pediatr. 2019, 7, 401. [Google Scholar] [CrossRef]
- Inaba, H.; Greaves, M.; Mullighan, C.G. Acute Lymphoblastic Leukaemia. Lancet 2013, 381, 1943–1955. [Google Scholar] [CrossRef]
- Zhang, W.; Kuang, P.; Liu, T. Prognostic Significance of CDKN2A/B Deletions in Acute Lymphoblastic Leukaemia: A Meta-Analysis. Ann. Med. 2019, 51, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Bortolozzi, R.; Mattiuzzo, E.; Trentin, L.; Accordi, B.; Basso, G.; Viola, G. Ribociclib, a Cdk4/Cdk6 Kinase Inhibitor, Enhances Glucocorticoid Sensitivity in B-Acute Lymphoblastic Leukemia (B-All). Biochem. Pharmacol. 2018, 153, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Bride, K.L.; Hu, H.; Tikhonova, A.; Fuller, T.J.; Vincent, T.L.; Shraim, R.; Li, M.M.; Carroll, W.L.; Raetz, E.A.; Aifantis, I.; et al. Rational Drug Combinations with CDK4/6 Inhibitors in Acute Lymphoblastic Leukemia. Haematologica 2021. [Google Scholar] [CrossRef]
- Bautista, F.; Paoletti, X.; Rubino, J.; Brard, C.; Rezai, K.; Nebchi, S.; Andre, N.; Aerts, I.; De Carli, E.; van Eijkelenburg, N.; et al. Phase I or II Study of Ribociclib in Combination With Topotecan-Temozolomide or Everolimus in Children With Advanced Malignancies: Arms A and B of the AcSé-ESMART Trial. J. Clin. Oncol. 2021, 39, 3546–3560. [Google Scholar] [CrossRef]
- Pikman, Y.; Alexe, G.; Roti, G.; Conway, A.S.; Furman, A.; Lee, E.S.; Place, A.E.; Kim, S.; Saran, C.; Modiste, R.; et al. Synergistic Drug Combinations with a CDK4/6 Inhibitor in T-Cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2017, 23, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Konecny, G.E.; Winterhoff, B.; Kolarova, T.; Qi, J.; Manivong, K.; Dering, J.; Yang, G.; Chalukya, M.; Wang, H.-J.; Anderson, L.; et al. Expression of P16 and Retinoblastoma Determines Response to CDK4/6 Inhibition in Ovarian Cancer. Clin. Cancer Res. 2011, 17, 1591–1602. [Google Scholar] [CrossRef]
- Hamilton, E.; Infante, J.R. Targeting CDK4/6 in Patients with Cancer. Cancer Treat. Rev. 2016, 45, 129–138. [Google Scholar] [CrossRef]
- Roskoski, R. Cyclin-Dependent Protein Serine/Threonine Kinase Inhibitors as Anticancer Drugs. Pharmacol. Res. 2019, 139, 471–488. [Google Scholar] [CrossRef]
- Witkiewicz, A.K.; Knudsen, K.E.; Dicker, A.P.; Knudsen, E.S. The Meaning of P16(Ink4a) Expression in Tumors: Functional Significance, Clinical Associations and Future Developments. Cell Cycle 2011, 10, 2497–2503. [Google Scholar] [CrossRef]
- Du, Q.; Guo, X.; Wang, M.; Li, Y.; Sun, X.; Li, Q. The Application and Prospect of CDK4/6 Inhibitors in Malignant Solid Tumors. J. Hematol. Oncol. 2020, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Antić, Ž.; Yu, J.; Van Reijmersdal, S.V.; Van Dijk, A.; Dekker, L.; Segerink, W.H.; Sonneveld, E.; Fiocco, M.; Pieters, R.; Hoogerbrugge, P.M.; et al. Multiclonal Complexity of Pediatric Acute Lymphoblastic Leukemia and the Prognostic Relevance of Subclonal Mutations. Haematologica 2021, 106, 3046–3055. [Google Scholar] [CrossRef] [PubMed]
- Malinowska-Ozdowy, K.; Frech, C.; Schönegger, A.; Eckert, C.; Cazzaniga, G.; Stanulla, M.; zur Stadt, U.; Mecklenbräuker, A.; Schuster, M.; Kneidinger, D.; et al. KRAS and CREBBP Mutations: A Relapse-Linked Malicious Liaison in Childhood High Hyperdiploid Acute Lymphoblastic Leukemia. Leukemia 2015, 29, 1656–1667. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-H.; Wang, H.-S.; Qian, X.-W.; Zhu, X.-H.; Miao, H.; Yu, Y.; Meng, J.-H.; Le, J.; Jiang, J.-Y.; Cao, P.; et al. Ras Pathway Mutation Feature in the Same Individuals at Diagnosis and Relapse of Childhood Acute Lymphoblastic Leukemia. Transl. Pediatr. 2020, 9, 4–12. [Google Scholar] [CrossRef]
- Jerchel, I.S.; Hoogkamer, A.Q.; Ariës, I.M.; Steeghs, E.M.P.; Boer, J.M.; Besselink, N.J.M.; Boeree, A.; van de Ven, C.; de Groot-Kruseman, H.A.; de Haas, V.; et al. RAS Pathway Mutations as a Predictive Biomarker for Treatment Adaptation in Pediatric B-Cell Precursor Acute Lymphoblastic Leukemia. Leukemia 2018, 32, 931–940. [Google Scholar] [CrossRef]
- Kerstjens, M.; Driessen, E.M.C.; Willekes, M.; Pinhanços, S.S.; Schneider, P.; Pieters, R.; Stam, R.W. MEK Inhibition Is a Promising Therapeutic Strategy for MLL-Rearranged Infant Acute Lymphoblastic Leukemia Patients Carrying RAS Mutations. Oncotarget 2017, 8, 14835–14846. [Google Scholar] [CrossRef]
- Kennedy, V.E.; Smith, C.C. FLT3 Mutations in Acute Myeloid Leukemia: Key Concepts and Emerging Controversies. Front. Oncol. 2020, 10, 612880. [Google Scholar] [CrossRef]
- Harvey, R.C.; Tasian, S.K. Clinical Diagnostics and Treatment Strategies for Philadelphia Chromosome-like Acute Lymphoblastic Leukemia. Blood Adv. 2020, 4, 218–228. [Google Scholar] [CrossRef]
- Palmi, C.; Vendramini, E.; Silvestri, D.; Longinotti, G.; Frison, D.; Cario, G.; Shochat, C.; Stanulla, M.; Rossi, V.; Di Meglio, A.M.; et al. Poor Prognosis for P2RY8-CRLF2 Fusion but Not for CRLF2 over-Expression in Children with Intermediate Risk B-Cell Precursor Acute Lymphoblastic Leukemia. Leukemia 2012, 26, 2245–2253. [Google Scholar] [CrossRef]
- Yamashita, Y.; Shimada, A.; Yamada, T.; Yamaji, K.; Hori, T.; Tsurusawa, M.; Watanabe, A.; Kikuta, A.; Asami, K.; Saito, A.M.; et al. IKZF1 and CRLF2 Gene Alterations Correlate with Poor Prognosis in Japanese BCR-ABL1-Negative High-Risk B-Cell Precursor Acute Lymphoblastic Leukemia. Pediatr. Blood Cancer 2013, 60, 1587–1592. [Google Scholar] [CrossRef]
- Joseph, P.D.; Craig, J.C.; Caldwell, P.H.Y. Clinical Trials in Children. Br. J. Clin. Pharmacol. 2015, 79, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Kyr, M.; Svobodnik, A.; Stepanova, R.; Hejnova, R. N-of-1 Trials in Pediatric Oncology: From a Population-Based Approach to Personalized Medicine—A Review. Cancers 2021, 13, 5428. [Google Scholar] [CrossRef] [PubMed]
- Moerdler, S.; Zhang, L.; Gerasimov, E.; Zhu, C.; Wolinsky, T.; Roth, M.; Goodman, N.; Weiser, D.A. Physician Perspectives on Compassionate Use in Pediatric Oncology. Pediatr. Blood Cancer 2019, 66, e27545. [Google Scholar] [CrossRef] [PubMed]
- Dieck, C.L.; Ferrando, A. Genetics and Mechanisms of NT5C2-Driven Chemotherapy Resistance in Relapsed ALL. Blood 2019, 133, 2263–2268. [Google Scholar] [CrossRef]
- Barz, M.J.; Hof, J.; Groeneveld-Krentz, S.; Loh, J.W.; Szymansky, A.; Astrahantseff, K.; von Stackelberg, A.; Khiabanian, H.; Ferrando, A.A.; Eckert, C.; et al. Subclonal NT5C2 Mutations Are Associated with Poor Outcomes after Relapse of Pediatric Acute Lymphoblastic Leukemia. Blood 2020, 135, 921–933. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Hu, J.; Ren, Y.; Wang, H. Clinical Features and Prognosis of Normal Karyotype Acute Myeloid Leukemia Pediatric Patients with WT1 Mutations: An Analysis Based on TCGA Database. Hematology 2020, 25, 79–84. [Google Scholar] [CrossRef]
- Zidan, M.A.A.; Kamal Shaaban, H.M.; Elghannam, D.M. Prognostic Impact of Wilms Tumor Gene Mutations in Egyptian Patients with Acute Myeloid Leukemia with Normal Karyotype. Hematology 2014, 19, 267–274. [Google Scholar] [CrossRef]
- Owen, C.; Fitzgibbon, J.; Paschka, P. The Clinical Relevance of Wilms Tumour 1 (WT1) Gene Mutations in Acute Leukaemia. Hematol. Oncol. 2010, 28, 13–19. [Google Scholar] [CrossRef]
- Valliyammai, N.; Nancy, N.K.; Sagar, T.G.; Rajkumar, T. Study of NOTCH1 and FBXW7 Mutations and Its Prognostic Significance in South Indian T-Cell Acute Lymphoblastic Leukemia. J. Pediatr. Hematol. Oncol. 2018, 40, e1–e8. [Google Scholar] [CrossRef]
- Fogelstrand, L.; Staffas, A.; Wasslavik, C.; Sjögren, H.; Söderhäll, S.; Frost, B.-M.; Forestier, E.; Degerman, S.; Behrendtz, M.; Heldrup, J.; et al. Prognostic Implications of Mutations in NOTCH1 and FBXW7 in Childhood T-ALL Treated According to the NOPHO ALL-1992 and ALL-2000 Protocols. Pediatr. Blood Cancer 2014, 61, 424–430. [Google Scholar] [CrossRef]
- Park, M.-J.; Taki, T.; Oda, M.; Watanabe, T.; Yumura-Yagi, K.; Kobayashi, R.; Suzuki, N.; Hara, J.; Horibe, K.; Hayashi, Y. FBXW7 and NOTCH1 Mutations in Childhood T Cell Acute Lymphoblastic Leukaemia and T Cell Non-Hodgkin Lymphoma. Br. J. Haematol. 2009, 145, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Inaba, H.; Azzato, E.M.; Mullighan, C.G. Integration of Next-Generation Sequencing to Treat Acute Lymphoblastic Leukemia with Targetable Lesions: The St. Jude Children’s Research Hospital Approach. Front. Pediatr. 2017, 5, 258. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.-H. Precision Medicine in Acute Lymphoblastic Leukemia. Front. Med. 2020, 14, 689–700. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).