Patient and Clinician Preferences for Genetic and Genomic Testing in Non-Small Cell Lung Cancer: A Discrete Choice Experiment
Abstract
:1. Introduction
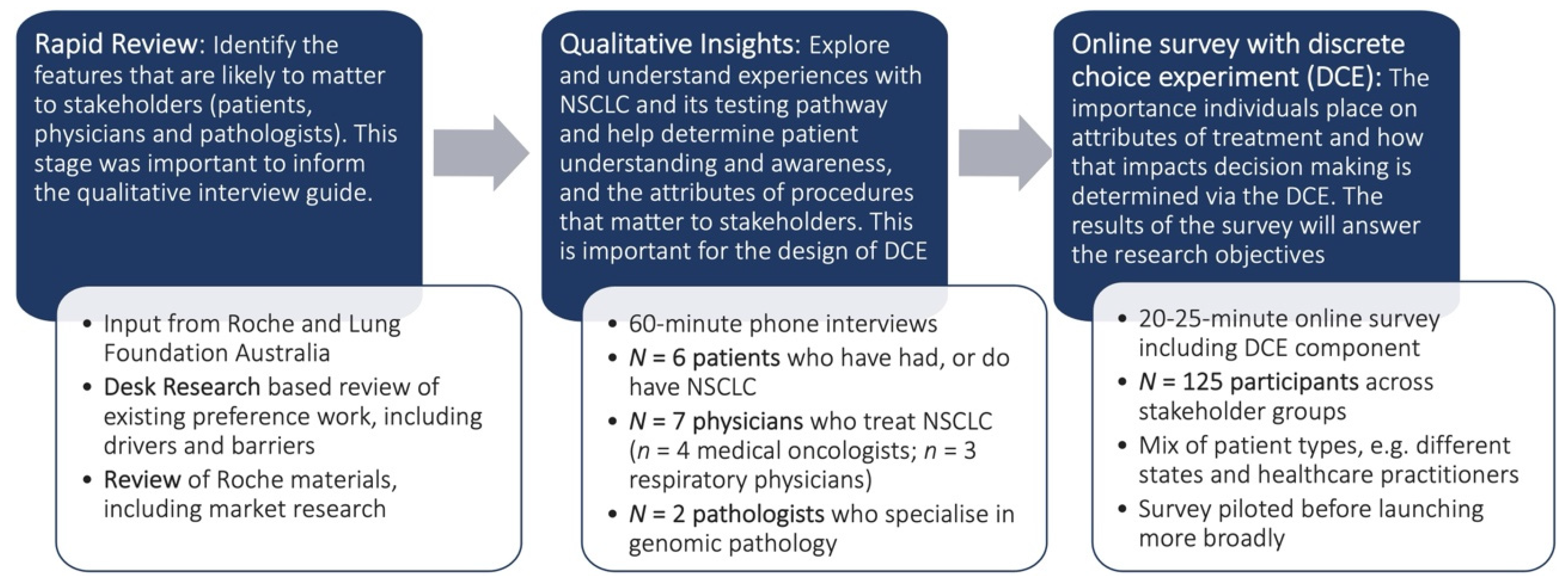
2. Materials and Methods
2.1. Interviews
2.1.1. Interview Participants
2.1.2. Interview Approach
2.1.3. Data Analysis
2.2. Discrete Choice Experiment
2.2.1. Survey Participants
2.2.2. Survey Instrument
2.3. Data Analysis
3. Results
3.1. Interviews
3.1.1. Participants
3.1.2. Patients
Patients cannot find opportunities to have their, you know, full genomic sequencing performed, pay for it if they wish, or just be clear with how much it costs, where to go and how long it is going to take. They’re the three things people want to know and yet to find that information is like finding a needle in a haystack, and if you don’t know … who’s who in the zoo then you miss out’. (Patient 4, Female, VIC)
You just offer so many more opportunities to patients to access treatments that are more personalised and targeted and you know provide them with really good quality of life’. (Patient 4, Female, VIC)
The more I know the better I equip myself with information and make better decisions. I don’t see any disadvantage in more knowledge yeah…the more you know the more information you know about your genes the better’. (Patient 3, Female, QLD)
I would go for the more detailed one, you would be able to have more of a chance of knowing what kind of cancer it mutates to which I don’t have, and then you’ll have the best chance of finding a treatment that suits you’. (Patient 6, Female, VIC)
It is only through genomic testing you know real sequencing of the cancer that has allowed them to take you know a combo of drugs. The patient demand is there for it, it is seen as a positive not a negative’. (Patient 2, Male, VIC)
3.1.3. Physicians
I think that’s what patients want because they want a road map of where to go from here rather than every so often saying, “Oh well we could do this other test and this might show this, and if we found that then we could do that’’. (Physician 2, Medical Oncologist, NSW)
3.1.4. Pathologists
Well, most molecular testing on cancer is one test for one gene for one drug, and you can have a patient centred approach where this is the patient they have a disease we test for all genes and if a gene has a mutation then they get a particular drug. The co-dependent methodology is fatally flawed in my opinion’ (Pathologist 1, Male, NSW)
3.1.5. Attributes Identified
3.2. Results of the DCE
3.2.1. Mixed Multinomial Logit Model Results for Patients
3.2.2. Mixed Multinomial Logit Model Results for Physicians
3.3. Attribute Importance
3.4. Preference Share
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Australian Institute of Health and Welfare. Australian Burden of Disease Study: Impact and Causes of Illness and Death in Australia 2015; Australian Burden of Disease Series no. 19. Cat. no. BOD 22; AIHW: Canberra, Australia, 2019. [Google Scholar]
- Australian Institute of Health and Welfare. Burden of Cancer in Australia: Australian Burden of Disease Study 2011; AIHW: Canberra, Australia, 2017. [Google Scholar]
- National Cancer Control Indicators. Stage at Diagnosis by Type (Breast, Prostate, Colorectal, Lung). 2021. Available online: https://ncci.canceraustralia.gov.au/stage-diagnosis/stage-diagnosis-type (accessed on 20 December 2021).
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.; Chirieac, L.R.; D’Amico, T.A.; DeCamp, M.M.; Dilling, T.J.; Dobelbower, M.; et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2017, 15, 504–535. [Google Scholar] [CrossRef]
- Besse, B.; Adjei, A.; Baas, P.; Meldgaard, P.; Nicolson, M.; Paz-Ares, L.; Reck, M.; Smit, E.F.; Syrigos, K.; Stahel, R.; et al. 2nd ESMO Consensus Conference on Lung Cancer: Non-small-cell lung cancer first-line/second and further lines of treatment in advanced disease. Ann. Oncol. 2014, 25, 1475–1484. [Google Scholar] [CrossRef]
- Fumagalli, C.; Guerini-Rocco, E.; Barberis, M. Broad-based genomic sequencing in advanced non-small cell lung cancer in the dock. Transl. Lung Cancer Res. 2019, 8, S360–S363. [Google Scholar] [CrossRef] [PubMed]
- Cancer Council Australia Lung Cancer Guidelines Working Party. Clinical Practice Guidelines for the Treatment of Lung Cancer 2021. Available online: https://wiki.cancer.org.au/australia/Guidelines:Lung_cancer (accessed on 20 December 2021).
- Masciale, V.; Banchelli, F.; Grisendi, G.; D’Amico, R.; Maiorana, A.; Stefani, A.; Morandi, U.; Dominci, M.; Aramini, B. New Perspectives in Different Gene Expression Profiles for Early and Locally Advanced Non-Small Cell Lung Cancer Stem Cells. Front. Oncol. 2021, 11, 613198. [Google Scholar] [CrossRef]
- Therapeutic Goods Administration (TGA). 2022. Available online: https://www.tga.gov.au/ (accessed on 3 May 2022).
- P.B.S. Home. 2022. Available online: https://www.pbs.gov.au/pbs/home (accessed on 3 May 2022).
- Yuan, M.; Huang, L.-L.; Chen, J.-H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Australian Government Department of Health. MBS Online-Lung Cancer Tests 2022. Available online: http://www9.health.gov.au/mbs/search.cfm?q=lung+cancer+test&Submit=&sopt=S (accessed on 10 May 2022).
- Patel, Y.P.; Husereau, D.; Leighl, N.B.; Melosky, B.; Nam, J. Health and Budget Impact of Liquid-Biopsy-Based Comprehensive Genomic Profile (CGP) Testing in Tissue-Limited Advanced Non-Small Cell Lung Cancer (aNSCLC) Patients. Curr. Oncol. 2021, 28, 5278–5294. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; Tsao, M.S. Tumour tissue sampling for lung cancer management in the era of personalised therapy: What is good enough for molecular testing? Eur. Respir J. 2014, 44, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef]
- Smeltzer, M.P.; Wynes, M.W.; Lantuejoul, S.; Soo, R.; Ramalingam, S.S.; Varella-Garcia, M.; Taylor, M.M.; Richeimer, K.; Wood, K.; Howell, K.E.; et al. The International Association for the Study of Lung Cancer Global Survey on Molecular Testing in Lung Cancer. J. Thorac. Oncol. 2020, 15, 1434–1448. [Google Scholar] [CrossRef]
- Norris, S.; Belcher, A.; Howard, K.; Ward, R.L. Evaluating genetic and genomic tests for heritable conditions in Australia: Lessons learnt from health technology assessments. J. Community Genet. 2021, 9, 1–20. [Google Scholar]
- Parliment of the Commonwealth of Australia. The New Frontier–Delivering Better Health for All Australians. Inquiry into Approval Processes for New Drugs and Novel Medical Technologies in Australia; Parliament of the Commonwealth of Australia: Canberra, Australia, 2021. Available online: https://parlinfo.aph.gov.au/parlInfo/download/committees/reportrep/024755/toc_pdf/TheNewFrontier-DeliveringbetterhealthforallAustralians.pdf;fileType=application%2Fpdf (accessed on 5 December 2021).
- Australian Government Department of Health. MSAC Guideline Review. 2021. Available online: http://www.msac.gov.au/internet/msac/publishing.nsf/Content/guidelines-review (accessed on 5 December 2021).
- UK Government. New Implementation Plan to Deliver World-Leading Genomic Healthcare. 2021. Available online: https://www.gov.uk/government/news/new-implementation-plan-to-deliver-world-leading-genomic-healthcare (accessed on 5 December 2021).
- Public Policy Committee International Society of Pharmacoepidemiology. Guidelines for good pharmacoepidemiology practice (GPP). Pharmacoepidemiol. Drug Saf. 2016, 25, 2–10. [Google Scholar] [CrossRef]
- Thurstone, L.L. A law of comparative judgment. Psychol. Rev. 1927, 34, 273. [Google Scholar] [CrossRef]
- McFadden, D. Conditional logit analysis of qualitative choice behaviour. In Frontiers of Econometrics; Zarembka, P., Ed.; Academic Press: New York, NY, USA, 1974; pp. 105–142. [Google Scholar]
- Lancaster, K.J. A new approach to consumer theory. J. Polit Econ. 1966, 74, 132–157. [Google Scholar] [CrossRef]
- Soekhai, V.; de Bekker-Grob, E.W.; Ellis, A.R.; Vass, C.M. Discrete Choice Experiments in Health Economics: Past, Present and Future. Pharmacoeconomics 2019, 37, 201–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridges, J.F.; Hauber, A.B.; Marshall, D.; Lloyd, A.; Prosser, L.A.; Regier, D.A.; Johnson, F.R.; Mauskopf, J. Conjoint analysis applications in health--a checklist: A report of the ISPOR Good Research Practices for Conjoint Analysis Task Force. Value Health 2011, 14, 403–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, F.R.; Lancsar, E.; Marshall, D.; Kilambi, V.; Mühlbacher, A.; Regier, D.A.; Bresnahan, B.W.; Kanninen, B.; Bridges, J.F. Constructing experimental designs for discrete-choice experiments: Report of the ISPOR Conjoint Analysis Experimental Design Good Research Practices Task Force. Value Health 2013, 16, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Rose, J.M.; Bliemer, M.C. Constructing Efficient Stated Choice Experimental Designs. Transp. Rev. 2009, 29, 587–617. [Google Scholar] [CrossRef]
- Gonzalez, J.M. A Guide to Measuring and Interpreting Attribute Importance. Patient 2019, 12, 287–295. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Sholl, L.M.; Aisner, D.L.; Varella-Garcia, M.; Berry, L.D.; Dias-Santagata, D.; Wistuba, I.I.; Chen, H.; Fujimoto, J.; Kugler, K.; Franklin, W.A.; et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2015, 10, 768–777. [Google Scholar] [CrossRef] [Green Version]
- Prabhakar, C. Next Generation Sequencing in Lung Cancer Using Small Volume Biopsies; The University of Sydney: Sydney, Australia, 2020. [Google Scholar]
- Zhao, S.; Zhang, Z.; Zhan, J.; Zhao, X.; Chen, X.; Xiao, L.; Wu, K.; Ma, Y.; Li, M.; Yang, Y.; et al. Utility of comprehensive genomic profiling in directing treatment and improving patient outcomes in advanced non-small cell lung cancer. BMC Med. 2021, 19, 223. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Kim, D.W.; Nakagawa, K.; Seto, T.; Crinó, L.; Ahn, M.J.; De Pas, T.; Besse, B.; Solomon, B.J.; Blackhall, F.; et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 2013, 368, 2385–2394. [Google Scholar] [CrossRef] [Green Version]
- Lazzari, C.; Bulotta, A.; Cangi, M.G.; Bucci, G.; Pecciarini, L.; Bonfiglio, S.; Lorusso, V.; Ippati, S.; Arrigoni, G.; Grassini, G.; et al. Next Generation Sequencing in Non-Small Cell Lung Cancer: Pitfalls and Opportunities. Diagnostics 2020, 10, 1092. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; OHaire, S.; Khuong-Quang, D.A.; Markman, B.; Gan, H.K.; Ekert, P.G.; O’Byrne, K.J.; Millward, M.; Solomon, B.J.; Tran, B.; et al. Evaluating barriers to uptake of comprehensive genomic profiling (CGP) in advanced cancer patients (pts). Am. Soc. Clin. Oncol. 2020, 38, 2033. [Google Scholar] [CrossRef]
- Erdmann, A.; Rehmann-Sutter, C.; Bozzaro, C. Patients’ and professionals’ views related to ethical issues in precision medicine: A mixed research synthesis. BMC Med. Ethics 2021, 22, 116. [Google Scholar] [CrossRef]
- Pharmaceutical Benefits Scheme. Esbriet (Pirfenidone) PBS Criteria. 2019. Available online: https://www.pbs.gov.au/pbs/search?term=PIRFENIDONE&analyse=false&search-type=medicines (accessed on 10 May 2022).
- Best, S.; Stark, Z.; Phillips, P.; Wu, Y.; Long, J.C.; Taylor, N.; Braithwaite, J.; Christodoulou, J.; Goranitis, I. Clinical genomic testing: What matters to key stakeholders? Eur. J. Hum. Genet. 2020, 28, 866–873. [Google Scholar] [CrossRef]
- Dall’Olio, F.G.; Conci, N.; Rossi, G.; Fiorentino, M.; De Giglio, A.; Grilli, G.; Altimari, A.; Gruppioni, E.; Filippini, D.M.; Di Federico, A.; et al. Comparison of Sequential Testing and Next Generation Sequencing in advanced Lung Adenocarcinoma patients—A single centre experience. Lung Cancer 2020, 149, 5–9. [Google Scholar] [CrossRef]
- Pennell, N.A.; Mutebi, A.; Zhou, Z.Y.; Ricculli, M.L.; Tang, W.; Wang, H.; Guerin, A.; Arnhart, T.; Dalal, A.; Sasane, M.; et al. Economic Impact of Next-Generation Sequencing Versus Single-Gene Testing to Detect Genomic Alterations in Metastatic Non–Small-Cell Lung Cancer Using a Decision Analytic Model. JCO Precis. Oncol. 2019, 3, 1–9. [Google Scholar] [CrossRef]
- Thavaneswaran, S.; Ballinger, M.; Butow, P.; Meiser, B.; Goldstein, D.; Lin, F.; Napier, C.; Thomas, D.; Best, M. The experiences and needs of Australian medical oncologists in integrating comprehensive genomic profiling into clinical care: A nation-wide survey. Oncotarget 2021, 12, 2169–2176. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.L.; Klein, W.M.; Erby, L.A.; Smith, C.H.; Roter, D.L. The impact of the number of tests presented and a provider recommendation on decisions about genetic testing for cancer risk. Patient Educ. Couns. 2021, 104, 265–275. [Google Scholar] [CrossRef]
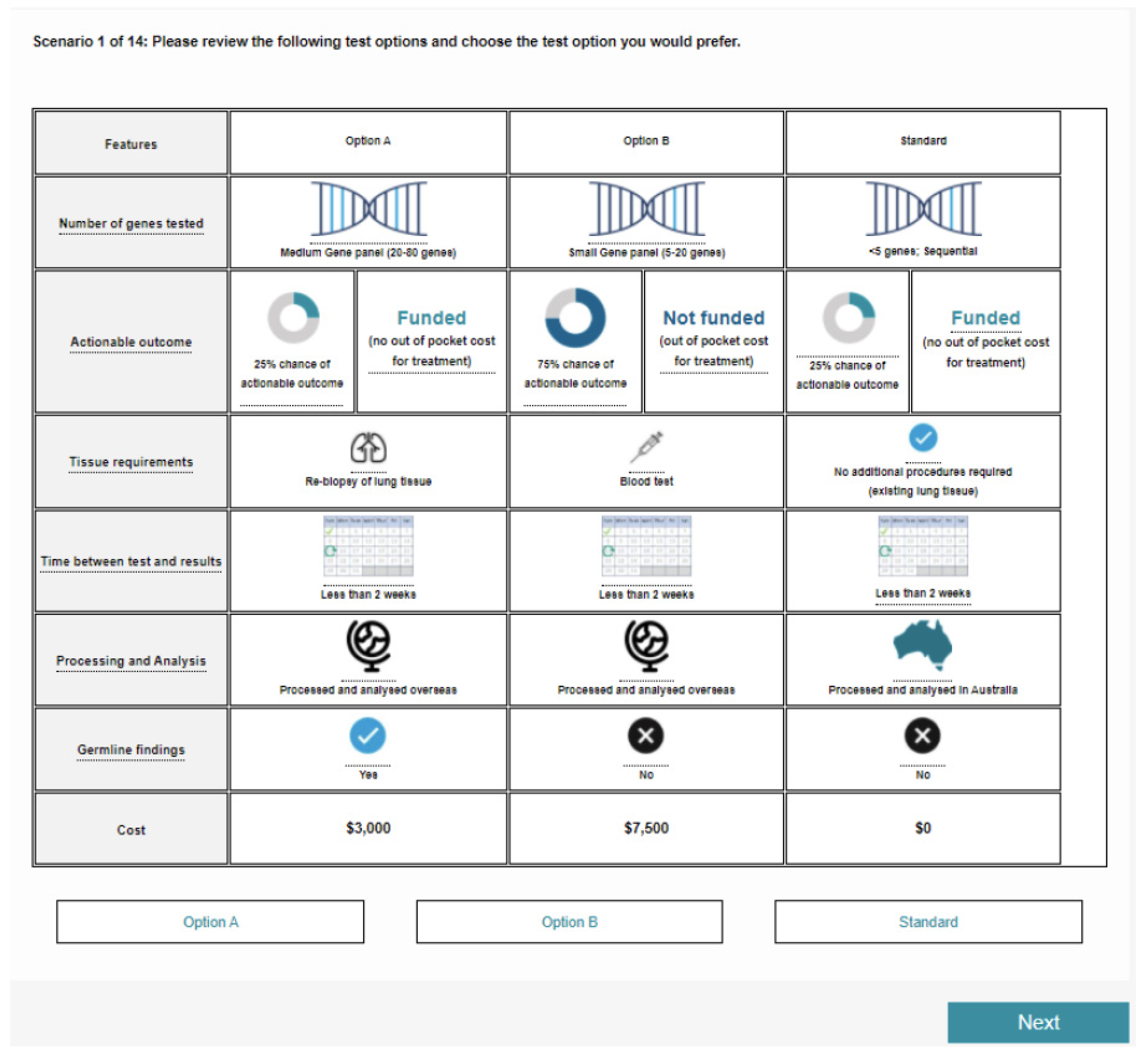

| Attribute Title Description in Patient Survey | Level Further Information (if Applicable) |
|---|---|
| Number of genes tested Number of genetic mutations tested for on the panel, some of which may have a potentially actionable outcome | Small gene panel (5–20 genes) 5–20 genetic mutations |
| Medium gene panel (20–80 genes) 20–80 genetic mutations | |
| Comprehensive Gene panel (or CGP) (80–500 genes) 80–500 genetic mutations | |
| Whole Exome/Whole Genome sequencing (>500) More than 500 genetic mutations | |
| Chance of actionable outcome Likelihood that the test will result in an actionable outcome for your treatment. e.g., targeted therapy; change/cessation of ineffective treatment; dosing requirements/adjustments | <1% chance <1% chance that the test may impact treatment decisions |
| 25% chance ** 25% chance that the test may impact treatment decisions | |
| 50% chance 50% chance that the test may impact treatment decisions | |
| 75% chance 75% chance that the test may impact treatment decisions | |
| Funding for treatment Whether the actionable outcome for your treatment is funded (no out of pocket cost to you) or not funded (may be an out of pocket cost to you) | Funded ** No out of pocket costs for treatment. e.g., targeted therapy is covered by the PBS; clinical trial |
| Not funded Potential out of pocket costs for treatment e.g., overseas clinical trials; medication not covered on PBS | |
| Tissue requirements What kind of tissue is needed for the genetic mutation test, and whether other procedures are required | No additional procedures required ** No additional procedures required (existing lung tissue) |
| Blood test | |
| Re-biopsy of lung tissue | |
| Time between test and results Test turnaround: time to receive results that may impact clinical decisions | Less than 2 weeks ** |
| 2–4 weeks | |
| 4–6 weeks | |
| 6–12 weeks | |
| Location of processing and analysis Where the sample is processed and analysed. This includes tissue transport and storage, data analysis and interpretation of results for delivery | Australia ** |
| Overseas | |
| Interpretation and reporting * Information included in the report that is returned, and what degree of interpretation it provides for assisting in clinical decisions. A full list of genetic mutations tested for, even when no alterations are found, will be delivered as an appendix in all tests | Reports on genetic mutation status only ** |
| Reports on genetic mutation status and eligibility for established targeted therapies | |
| Reports on genetic mutation status and eligibility for targeted therapies that are established or under investigation | |
| Reports on genetic mutation status and eligibility for targeted therapies that are established or under investigation. Access to local Decision Support Service *** | |
| Germline findings Whether there is a 15% chance of detecting additional hereditary or inherited mutations. These findings do not guide treatment decisions, though may impact yours and your family decisions in other ways | Yes |
| No ** | |
| Cost of test Out of pocket cost for the test | $0 ** |
| $1500 | |
| $3000 | |
| $4500 | |
| $6000 | |
| $7500 |
| Demographic | Patient Respondents (n = 45) | Clinician Respondents (n = 44) |
|---|---|---|
| n (%) | n (%) | |
| Gender identity | ||
| Female | 27 (60%) | 9 (20%) |
| Male | 18 (40%) | 34 (77%) |
| Not answered | 1 (2%) | |
| Age | ||
| 31–40 | 10 (22%) | 14 (32%) |
| 41–50 | 12 (27%) | 17 (39%) |
| 51–60 | 7 (16%) | 9 (20%) |
| 61–70 | 12 (27%) | 3 (7%) |
| 71–80 | 4 (9%) | 1 (2%) |
| Area of residence/practice | ||
| Metropolitan/City | 31 (69%) | |
| Regional | 8 (18%) | |
| Rural | 6 (13%) | |
| Stage at diagnosis | - | |
| Early stage (I or II) | 16 (36%) | |
| Advanced stage (III or IV) | 26 (58%) | |
| Unknown/Not staged | 3 (7%) | |
| Current stage | - | |
| Early stage (I or II) | 3 (7%) | |
| Advanced stage (III or IV) | 36 (80%) | |
| Unknown/Not staged | 6 (13%) | |
| Current line of therapy * | - | |
| unknown | 2 (4%) | |
| 1 | 11 (24%) | |
| 2 | 13 (29%) | |
| 3 | 14 (31%) | |
| ≥4 | 5 (11%) | |
| Length of time treating patients with lung cancer | ||
| 3–4 years | 4 (9%) | |
| 5–6 years | 5 (11%) | |
| 7–8 years | 6 (14%) | |
| 9–10 years | 3 (7%) | |
| >10 years | 26 (59%) | |
| Specialty type | ||
| Medical oncologist | 23 (52%) | |
| Respiratory physician | 21 (48%) | |
| Proportion of patients for whom genetic mutation testing for NSCLC has been ordered, mean (SD) | 59% (36%) |
| Parameter | Symbol | Parameter | SE | T-Ratio | |
|---|---|---|---|---|---|
| Random | |||||
| Number of genes tested | |||||
| Medium gene panel (20–80 genes) | 𝛽𝐺𝑀 | 0.1311 | 0.183 | 0.71 | |
| Comprehensive gene panel (80–500 genes) | 𝛽𝐺𝐶 | 0.2499 | 0.175 | 1.43 | |
| Whole exome/whole genome sequencing (More than 500 genes) | 𝛽𝐺𝑊 | 0.2198 | 0.203 | 1.08 | |
| RC: Small Gene panel (5–20 genes) | |||||
| Tissue requirements | |||||
| Blood test required | 𝛽𝑇𝐵 | 0.0089 | 0.128 | 0.07 | |
| Existing lung tissue (no procedures required) | 𝛽𝑇𝑁 | 0.5523 | *** | 0.18 | 3.08 |
| RC: Re-biopsy of lung tissue | |||||
| Time between test and results | |||||
| Less than 2 weeks | 𝛽𝑇𝑀𝐸2 | 0.8012 | *** | 0.225 | 3.57 |
| 2–4 weeks | 𝛽𝑇𝑀𝐸4 | 0.1570 | 0.185 | 0.85 | |
| 4–6 weeks | 𝛽𝑇𝑀𝐸6 | 0.1693 | 0.176 | 0.96 | |
| RC: 6–12 weeks | |||||
| Cost of test | |||||
| Cost | 𝛽𝐶𝑂𝑆𝑇 | −0.0004 | *** | 0 | −5.66 |
| Chance of actionable outcome and funding | |||||
| Actionable outcome: <1%, Funded | 𝛽𝐴𝐶1𝐹 | −2.3698 | *** | 0.481 | −4.93 |
| Actionable outcome: 25%, Not funded | 𝛽𝐴𝐶25𝑁𝐹 | −1.1763 | *** | 0.326 | −3.61 |
| Actionable outcome: 25%, Funded | 𝛽𝐴𝐶25𝐹 | −0.3719 | 0.263 | −1.41 | |
| Actionable outcome: 50%, Not funded | 𝛽𝐴𝐶50𝑁𝐹 | 1.6214 | *** | 0.296 | 5.48 |
| Actionable outcome: 50%, Funded | 𝛽𝐴𝐶50𝐹 | 1.5471 | *** | 0.299 | 5.17 |
| Actionable outcome: 75%, Not funded | 𝛽𝐴𝐶75𝑁𝐹 | 2.3385 | *** | 0.424 | 5.51 |
| Actionable outcome: 75%, Funded | 𝛽𝐴𝐶75𝐹 | 3.7469 | *** | 0.695 | 5.39 |
| RC: Actionable outcome: <1%, Not funded | |||||
| Location of processing and analysis | |||||
| Australia | 𝛽𝐿𝑂𝐶 | 0.4814 | *** | 0.13 | 3.72 |
| RC: Overseas | |||||
| Germline findings | |||||
| Yes | 𝛽𝐺𝐸𝑅𝑀 | 0.4626 | *** | 0.118 | 3.93 |
| RC: No | |||||
| Non-random | |||||
| Constant | |||||
| Current testing scheme | CUR | −1.2161 | *** | 0.448 | −2.71 |
| Parameters | Symbol | Parameter | SE | T-ratio | |
|---|---|---|---|---|---|
| Random | |||||
| Tissue requirements | |||||
| Blood test required | 𝛽𝑇𝐵 | 0.1808 | 0.1382 | 1.31 | |
| Existing lung tissue (no procedures required) | 𝛽𝑇𝑁 | 0.857 | *** | 0.2353 | 3.64 |
| RC: Re-biopsy of lung tissue | |||||
| Time between test and results | |||||
| Less than 2 weeks | 𝛽𝑇𝑀𝐸2 | 1.0958 | *** | 0.309 | 3.55 |
| 2–4 weeks | 𝛽𝑇𝑀𝐸4 | 0.2182 | 0.1936 | 1.13 | |
| 4–6 weeks | 𝛽𝑇𝑀𝐸6 | −0.0401 | 0.2067 | −0.19 | |
| RC: 6–12 weeks | |||||
| Cost of test | |||||
| Cost | 𝛽𝐶𝑂𝑆𝑇 | −0.0006 | *** | 0.0001 | −5.28 |
| Chance of actionable outcome and funding | |||||
| Actionable outcome: <1%, Funded | 𝛽𝐴𝐶1𝐹 | −2.9017 | *** | 0.6291 | −4.61 |
| Actionable outcome: 25%, Not funded | 𝛽𝐴𝐶25𝑁𝐹 | −1.777 | *** | 0.4391 | −4.05 |
| Actionable outcome: 25%, Funded | 𝛽𝐴𝐶25𝐹 | −0.5473 | * | 0.3163 | −1.73 |
| Actionable outcome: 50%, Not funded | 𝛽𝐴𝐶50𝑁𝐹 | 0.5834 | ** | 0.2607 | 2.24 |
| Actionable outcome: 50%, Funded | 𝛽𝐴𝐶50𝐹 | 2.9653 | *** | 0.476 | 6.23 |
| Actionable outcome: 75%, Not funded | 𝛽𝐴𝐶75𝑁𝐹 | 2.5349 | *** | 0.4698 | 5.40 |
| Actionable outcome: 75%, Funded | 𝛽𝐴𝐶75𝐹 | 5.1731 | *** | 0.8243 | 6.28 |
| RC: Actionable outcome: <1%, Not funded | |||||
| Non-random | |||||
| Number of genes tested | |||||
| Medium gene panel (20–80 genes) | 𝛽𝐺𝑀 | 0.7027 | ** | 0.3316 | 2.12 |
| Comprehensive gene panel (80–500 genes) | 𝛽𝐺𝐶 | 0.6578 | * | 0.3837 | 1.71 |
| Whole exome/whole genome sequencing (More than 500 genes) | 𝛽𝐺𝑊 | 0.9801 | ** | 0.4289 | 2.29 |
| RC: Small Gene panel (5–20 genes) | |||||
| Location of processing and analysis | |||||
| Australia | 𝛽𝐿𝑂𝐶 | 0.1712 | 0.1377 | 1.24 | |
| RC: Overseas | |||||
| Germline findings | |||||
| Yes | 𝛽𝐺𝐸𝑅𝑀 | 0.2758 | ** | 0.1333 | 2.07 |
| RC: No | |||||
| Interpretation and reporting | |||||
| Reports on genetic mutation status and eligibility for established targeted therapies | 𝛽𝑅𝐸𝑃𝑇𝑇 | −0.2217 | 0.207 | −1.07 | |
| Reports on genetic mutation status and eligibility for targeted therapies that are established or under investigation | 𝛽𝑅𝐸𝑃𝐼𝑁𝑉 | 0.0613 | 0.213 | 0.29 | |
| Reports on genetic mutation status and eligibility for targeted therapies that are established or under investigation. Access to local Decision Support Service | 𝛽𝑅𝐸𝑃𝐷𝑆𝑆 | 0.6043 | ** | 0.281 | 2.15 |
| RC: Reports of genetic mutation status only | |||||
| Constant | |||||
| Current testing scheme | CUR | −1.1812 | ** | 0.5729 | −2.06 |
| WES/WGS | CGP | |||||||
|---|---|---|---|---|---|---|---|---|
| Funding of treatment | Funded | Not funded | Funded | Not funded | ||||
| Physician | Patient | Physician | Patient | Physician | Patient | Physician | Patient | |
| TOTAL UPTAKE If all re-tests are: | ||||||||
| Tumour re-biopsy | 94.17% | 87.17% | 61.47% | 64.10% | 94.69% | 87.11% | 68.22% | 87.90% |
| Blood test | 96.98% | 89.68% | 70.26% | 68.34% | 97.26% | 89.64% | 77.04% | 90.31% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fifer, S.; Ordman, R.; Briggs, L.; Cowley, A. Patient and Clinician Preferences for Genetic and Genomic Testing in Non-Small Cell Lung Cancer: A Discrete Choice Experiment. J. Pers. Med. 2022, 12, 879. https://doi.org/10.3390/jpm12060879
Fifer S, Ordman R, Briggs L, Cowley A. Patient and Clinician Preferences for Genetic and Genomic Testing in Non-Small Cell Lung Cancer: A Discrete Choice Experiment. Journal of Personalized Medicine. 2022; 12(6):879. https://doi.org/10.3390/jpm12060879
Chicago/Turabian StyleFifer, Simon, Robyn Ordman, Lisa Briggs, and Andrea Cowley. 2022. "Patient and Clinician Preferences for Genetic and Genomic Testing in Non-Small Cell Lung Cancer: A Discrete Choice Experiment" Journal of Personalized Medicine 12, no. 6: 879. https://doi.org/10.3390/jpm12060879
APA StyleFifer, S., Ordman, R., Briggs, L., & Cowley, A. (2022). Patient and Clinician Preferences for Genetic and Genomic Testing in Non-Small Cell Lung Cancer: A Discrete Choice Experiment. Journal of Personalized Medicine, 12(6), 879. https://doi.org/10.3390/jpm12060879







