Abstract
Background: A promoter variable number tandem repeat polymorphism (pVNTR) of CYP2C9 is described with three types of fragments: short (pVNTR-S), medium (pVNTR-M) and long (pVNTR-L). The pVNTR-S allele reduces the CYP2C9 mRNA level in the human liver, and it was found to be in high linkage disequilibrium (LD) with the CYP2C9*3 allele in a White American population. The aim of the present study is to determine the presence and frequency of CYP2C9 pVNTR in a Spanish population, as well as analyzing whether the pVNTR-S allele is in LD with the CYP2C9*3 allele in this population. Subjects and Methods: A total of 209 subjects from Spain participated in the study. The CYP2C9 promoter region was amplified and analyzed using capillary electrophoresis. Genotyping for CYP2C9*2 and *3 variants was performed using a fluorescence-based allele-specific TaqMan allelic discrimination assay. Results: The frequencies of CYP2C9 pVNTR-L, M and S variant alleles are 0.10, 0.82 and 0.08, respectively. A high LD between CYP2C9 pVNTR-S and CYP2C9*3 variant alleles is observed (D’ = 0.929, r2 = 0.884). Conclusion: The results from the present study show that both CYP2C9 pVNTR and CYP2C9*3 are in a high LD, which could help to better understand the lower metabolic activity exhibited by CYP2C9*3 allele carriers. These data might be relevant for implementation in the diverse clinical guidelines for the pharmacogenetic analysis of the CYP2C9 gene before treatment with different drugs, such as non-steroidal anti-inflammatory drugs, warfarin, phenytoin and statins.
1. Introduction
Cytochrome P450 2C9 (CYP2C9) is one of the four major isoforms of the CYP2C subfamily and is estimated to be involved in the metabolic clearance of 15–20% of all drugs with phase I metabolism [1,2]. The CYP2C9 protein is composed of 490 amino acids, with a size of approximately 55 KDa [3], being expressed mainly in the liver, where it comprises the second of the CYPs isoforms with a higher expression level [4].
CYP2C9 participates in the metabolism of drugs of great therapeutic importance, such as numerous non-steroidal anti-inflammatory drugs (celecoxib, diclofenac, ibuprofen, etc.), coumarin anticoagulants (warfarin and acenocoumarol), antidiabetic drugs (tolbutamide), antiepileptics (phenytoin) and antihypertensive drugs (losartan and irbesartan) [5,6,7]. Furthermore, CYP2C9 is involved in the metabolism of important endogenous compounds such as serotonin and, due to its epoxygenase activity, various polyunsaturated fatty acids such as arachidonic acid, converting them into different hydroxyeicosatetraenoic acids (HETEs) and epoxyeicosatrienoic acids (EETs) [5,8,9].
The CYP2C9 gene is located on chromosome 10, in the 10q23.33 cytogenetic location (chr10: 94938658-94990091, GRCh38), and is composed of 51434 bp with nine exons (GenBank accession: NG_008385.2; OMIM Entry: 601130). This gene is highly polymorphic, and, to date, 62 genetic variants have been described [10]. The most widely studied variants are CYP2C9*2 (p.R144C; rs1799853) and CYP2C9*3 (p. I359L; rs1057910), which seem to decrease the metabolic activity of this enzyme, mainly CYP2C9*3. This reduction in metabolic activity was confirmed in different in vivo studies, using drugs such as tolbutamide, phenytoin, losartan, diclofenac, and warfarin, where the individuals carrying the CYP2C9*2 and/or CYP2C9*3 variants showed either an increase in drug concentrations, elimination half-life, a reduction in drug clearance or required lower drug doses than subjects carrying CYP2C9*1/*1 variants [1,11,12,13,14,15]. In addition, adverse drug effects were reported in patients with CYP2C9*2 and/or CYP2C9*3 alleles treated with phenytoin [16,17,18,19], warfarin [20,21,22] or acenocoumarol [23]. Furthermore, members of the CYP2C subfamily, including CYP2C9, catalyze the oxidation of arachidonic acid to HETEs and EETs, promoting vasodilation and lowering blood pressure, which may play a role in the pathogenesis of chronic kidney disease (CKD) [24].
Variable number tandem repeat polymorphisms (pVNTR) are DNA sequences in which a fragment (the size of which is higher than six base pairs) is consecutively repeated. The variation in the number of repeats, and not the repeated sequence, creates different alleles; these repeats usually have a high mutation rate, which makes them highly polymorphic [25]. Many microsatellites are found in non-coding DNA and are biologically silenced, while others are found in regulatory or even coding DNA; microsatellite dynamic mutations can lead to phenotypic changes and disease. Recent studies provide evidence that microsatellites can act as enhancers of disease-relevant regulatory genes [25]. When these VNTR polymorphisms are located in the promoter region, they can inhibit or promote gene expression in several ways by modifying transcription factors or other binding site proteins [25].
In a study by Wang et al. (2012), a pVNTR of CYP2C9 was identified [26]. This CYP2C9 pVNTR was located 4 kb upstream from the translation site (NC_000010.11; 94934570–94934705; GRCh38), affecting the expression of CYP2C9 mRNA in human liver [26]. In this region of CYP2C9, three types of fragments were found: short (pVNTR-S; 417–438 bp), medium (pVNTR-M; 446–488 bp) and long (pVNTR-L; 512–522 bp) alleles [26]. These three alleles have different lengths with diverse motif patterns [nTGnTAnTG(or CA)nTA(+/−CG)]. The pVNTR-L, pVNTR-M and pVNTR-M variants contain four, two and one motif copies, respectively [26].
In this study [26], the pVNTR-S allele was shown to reduce the promoter activity of the CYP2C9 enzyme in human liver. This decrease was associated with a 25% to 60% reduction in the CYP2C9 mRNA expression in human livers of pVNTR-S carriers compared to pVNTR-M and pVNTR-L [26].
Furthermore, it was observed that the pVNTR-S variant was present in high linkage disequilibrium (LD) with the CYP2C9*3 allele in a White American population [26], although this could not be observed in an African American [26] or an Egyptian [27] population.
Therefore, the aim of the present study was to determine the presence and frequency of CYP2C9 pVNTR in a Spanish population, as well as analyzing whether, for this population, the pVNTR-S allele is in LD with the CYP2C9*3 allele.
2. Materials and Methods
2.1. Subjects
The subjects included in the study (n = 209) were a group of 126 CKD patients (62.3% males; 68.2 ± 14.0 years, mean ± SD) recruited from the Nephrology Department of “Virgen del Puerto” Hospital (Plasencia, Spain) and a group of 83 subjects (74.7% females; 28.1 ± 11.3 years) from the University of Extremadura (Plasencia, Spain), mainly students and staff. The participants were part of a larger study that aimed to evaluate the relationship between different CYPs polymorphisms and the progression of chronic kidney disease. The inclusion criteria were being over the age of 18 years and having signed an informed consent form.
2.2. Determination of CYP2C9 pVNTR
A CYP2C9 fragment of 4216 bp upstream of the translation start site (NC_000010.11; 94934442–94934917; GRCh38) was PCR-amplified. This fragment contained CYP2C9 pVNTR (NC_000010.11; 94934570–94934705; GRCh38).
DNA was isolated and purified from blood samples (QIAmp Qiagen, Hilden, Germany), and the 10 μL PCR mixture consisted of 2 μL 5× MyTaq Reaction Buffer (Meridian Bioscience, Cincinnati, OH, USA), 0.2 μL of My Taq Polymerase (5 units/μL) (Meridian Bioscience, Cincinnati, OH, USA), 0.3 μL of pVNTR-forward primer (10 μM), 0.3 μL of pVNTR-reverse primer (10 μM) and 50–80 ng of DNA. Finally, nuclease-free water was added to a final volume of 10 μL.
The sequences for the forward and reverse primers were 5′-TGTAGTCCCAGGTTGTCAAGAGG-FAM-3′ and 5′-CCAGTCTCTGTCTTTTCATCTCATTC-3′, respectively [26].
PCR was performed in a Veriti Thermal Cycler (Thermo Fisher Scientific, MA, USA). The PCR conditions were as follows: an initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 53 °C for 30 s and 72 °C for 1 min. A final cycle of 72 °C was applied for 10 min. PCR products were analyzed using capillary electrophoresis. Therefore, following PCR, the amplification products were diluted 1:10 with Hi-Di Formamide with 0.3% (v/v) of GeneScan™ 600 LIZ® Size Standard (Thermo Fisher Scientific, Waltham, MA, USA) for sizing DNA fragments in the 20–600 pb range. The samples were denatured at 95 °C for 5 min and then at 4 °C for 2 min. The denatured PCR products were electrophoresed through a 50 cm-long capillary by using POP-7 polymer (Thermo Fisher Scientific, MA, USA) in an Applied Biosystems Sanger Sequencing 3500 Series Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA). The parameters for capillary electrophoresis were dye set G5, an injection time of 8 s, an injection voltage of 1.6 Kv and an electrophoretic voltage of 19.5 Kv at a 60 °C temperature block. GeneScan Analysis v5.0 (Applied Biosystems, Thermo Fisher, Waltham, MA, USA) was used to automatically analyze and calculate the molecular size of the amplified alleles.
2.3. CYP2C9*2 and *3 Allele Analysis
Genotyping for CYP2C9*2 (rs1799853) and *3 (rs1057910) variants was performed using a fluorescence-based allele-specific TaqMan allelic discrimination assay. For each CYP2C9 single-nucleotide polymorphism for CYP2C9*2 and *3 allele identification, a pre-developed TaqMan assay reagent kit, containing one pair of PCR primers and one pair of fluorescent TaqMan probes, was purchased from Thermo Fisher Scientific (Waltham, MA, USA). PCR amplification for all single-nucleotide polymorphisms was performed in a PCR mixture consisting of 5 μL of 2× Ex Taq Premix, 0.2 μL of 50× Rox Reference Dye, 40× SNP Genotyping Assay Mix (Takara Bio Inc., Shiga, Japan) and 1 μL of DNA (0.25 ng/μL). Nuclease-free water was added to a final volume of 10 μL. Amplification was carried out in an ABI 7300 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The PCR conditions were as follows: an initial denaturation at 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 31 s. Two different types of fluorescence were measured at the end of the 60 °C segment of each cycle.
2.4. Statistical Analysis
Descriptive statistics were used, and results are presented as percentages and frequencies. The Hardy–Weinberg equilibrium was determined by comparing the genotype frequencies with the expected values using a contingency table χ2 statistic with Yates’s correction, and Fisher’s exact tests were used to compare differences in CYP2C9 pVNTR variant allele frequencies between different populations. p values of less than 0.05 were regarded as statistically significant.
Sample size was calculated by using two different calculators available online (Sample Size Calculator by the Australian Bureau of Statistics: https://www.abs.gov.au/websitedbs/d3310114.nsf/home/sample+size+calculator; accessed on 18 September 2021; Sample Size Calculator by Raosoft Inc.: http://www.raosoft.com/samplesize.html; accessed on 18 September 2021). The sample size ranged from 101 to 139 individuals, considering a percentage of 7% and 10%, respectively (expected frequency of the CYP2C9*3 allele in the Spanish population), with a confidence level of 95% and a margin of error of 5%.
Linkage disequilibrium statistics were obtained using SNPstats software (https://www.snpstats.net/; accessed on 18 September 2021). Statistical analyses were performed using GraphPad Prism 5.00 (GraphPad Software Inc., San Diego, CA, USA) and the SNPStats software (Catalan Institute of Oncology, Hospitalet de Llobregat, Spain).
3. Results
3.1. Determination of the Presence and Frequency of CYP2C9 pVNTR in a Spanish Population
The presence of CYP2C9 pVNTR variant was determined in all samples, as well as the CYP2C9*2 and CYP2C9*3 alleles, as described in the Material and Methods Section.
The microsatellite sequencing analysis showed three different fragment sizes: 419–431 bp, 446–487 bp and 510–517 bp; these fragments were grouped as pVNTR-S (short), pVNTR-M (medium) and pVNTR-L (long), respectively. Figure 1 shows the electropherograms of the six different diplotypes of CYP2C9 pVNTR found in the studied population.
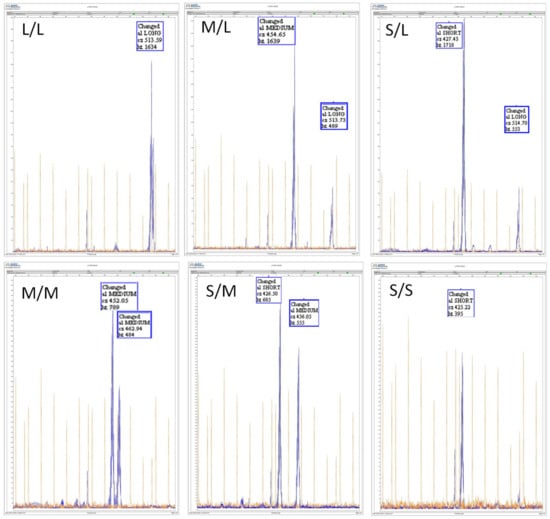
Figure 1.
Electropherograms of six different Variable number tandem repeat polymorphisms (pVNTR) CYP2C9 diplotypes found in the studied population (S: 419–431 bp; M: 446–487 bp; L: 510–517 bp).
There were no differences in the frequencies of CYP2C9 variants (*1, *2, *3 and pVNTR) between two sub-groups of participating subjects, so they were grouped into a single general population (Table 1).

Table 1.
Genotype and allelic frequency of CYP2C9 pVNTR *2 and *3 in a Spanish population (n = 209).
The presence of CYP2C9 pVNTR-L, M and S alleles could be observed, with a frequency of 0.10, 0.82 and 0.08, respectively. Furthermore, all alleles were in the Hardy–Weinberg equilibrium, both in the sub-groups and in the general population.
3.2. Comparison of the Frequency of CYP2C9 pVNTR between Different Populations
Concerning the frequencies of pVNTR from other previously studied populations (Table 2), it could be observed that the frequency of the pVNTR-S allele in the Spanish population was lower than in a Jordanian population (0.081 vs. 0.295; p < 0.0001) and did not differ from the rest of the studied populations. Regarding the pVNTR-L variant, it did not show significant differences from the rest of the populations, except with the frequency of a White American population, which was higher than in the Spanish subjects studied (0.103 vs. 0.152; p = 0.0094). In addition, the frequency of the pVNTR-M variant, which was the most studied in all populations, was similar to the rest of the populations investigated, except for the frequency of the Jordanian population, which was lower (0.816 vs. 0.627; p < 0.0001).

Table 2.
Minor allele frequency of CYP2C9 pVNTR alleles in different populations.
3.3. Analysis of LD between CYP2C9 pVNTR-S and CYP2C9*3 Alleles in a Spanish Population
Regarding the analysis of LD between CYP2C9 pVNTR-S and the CYP2C9*3 variant, it was observed that, in the Spanish population, these polymorphisms were in a high LD (D’ = 0.929, r2 = 0.884), similar to that observed in another population with Caucasian ancestry (White American population; Table 2). This LD between the *3 and pVNTR-S alleles of the CYP2C9 gene was not observed in either of the other two populations of non-Caucasian origin [26,27].
In addition, 27/35 individuals in the Spanish population, carrying pVNTR-S and/or CYP2C9*3 variants, carried both polymorphisms, either in heterozygosity (77.1%) or in homozygosis (2.9%) (Figure 2). In contrast, only five individuals who presented with the pVNTR-S variant did not have the CYP2C9*3 variant (14.3%), as well as two CYP2C9*3 carrier individuals (5.7%) (Figure 2).
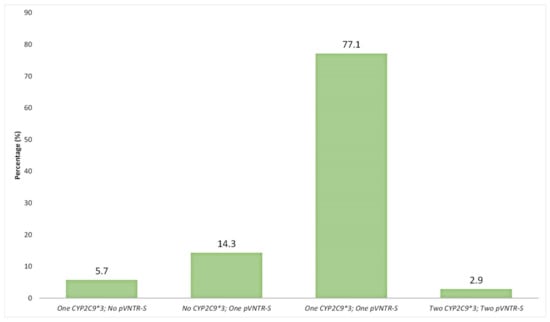
Figure 2.
Percentage (%) of individual carriers of zero, one or two pVNTR-S and/or CYP2C9*3 and (n = 35) among Spaniards (n = 209).
Therefore, according to the present results, 85% of the individuals in the Spanish population studied with pVNTR-S were also carriers of the CYP2C9*3 allele. Similarly, 93% of the carriers of the *3 allele also presented with the pVNTR-S variant.
4. Discussion
This is the first study where the presence and frequency of CYP2C9 pVNTR was analyzed in a European population, as well as the hypothetical association between pVNTR-S and CYP2C9*3 alleles in this population. Previously, two studies analyzed the frequency of CYP2C9 pVNTR in different populations: Jordanians [27], Egyptians and White and African Americans [26].
The CYP2C9 pVNTR-M variant was the most frequently studied in all populations; however, the frequency of this variant in the Spanish population was higher than in the Jordanian population [27]. Moreover, the frequencies of the pVNTR-S and pVNTR-L variants in the Spanish population were lower than in the Jordanian [27] and White American [26] populations, respectively.
Regarding the analytical methodology used, both previous studies [26,27] used different methods based on PCR technology to determine CYP2C9 pVNTR. In one method, a PCR with fluorescently labeled primers was first performed, and then the PCR products were sequenced [26]. In the other study, after amplifying the promoter region of CYP2C9 by PCR, the amplicons were visualized in polyacrylamide gels stained with ethidium bromide [27]. In our study, the CYP2C9 promoter region was PCR-amplified, but for the analysis of the amplicon products, these were separated by capillary electrophoresis; subsequently, the molecular size of the amplicons was calculated. The genetic analysis of microsatellites comprised a series of techniques in which DNA fragments were fluorescently labeled, separated by capillary electrophoresis, and the fragments were automatically sized. Sensitivity, simple preparation, and easy data analysis are some of advantages of this methodology because fragments differing by only one base pair are precisely sized, and no DNA cleanup (contrary to DNA sequencing) or genetic analysis software is required, which simplifies data analysis. Furthermore, a fragment analysis allows for the analysis of more than 20 loci in a single reaction, since alleles for overlapping loci are distinguished.
Concerning the analysis of LD between CYP2C9 pVNTR-S and CYP2C9*3 variants, it was observed that, in the Spanish population, these polymorphisms were in a high LD (D’ = 0.929, r2 = 0.884). This LD is similar to that observed in the other population of Caucasian origin: the White American population [26]. However, this LD between the *3 and pVNTR-S alleles of the CYP2C9 gene was not observed in either of the other two populations of non-Caucasian origin [26,27]. Notably, of the five individuals who were carriers of the CYP2C9 pVNTR-S variant and were not carriers of CYP2C9*3, four of the carriers were CYP2C9*1/*1, and the other carrier was *1/*2. No difference was found for ethnic origin, since all individuals were Spanish and from the same region.
In conclusion, this is the first study where the presence and frequency of CYP2C9 pVNTR were analyzed in a European population, and the results of the present study show that the CYP2C9 pVNTR and CYP2C9*3 variants are in LD, which can help to better understand the lower metabolic activity exhibited by CYP2C9*3 allele carriers. Furthermore, the genetic analysis of microsatellites used in the present study to determine the CYP2C9 pVNTR showed advantages compared with other previous methodologies [26,27] because fragments differing by only one base pair were precisely sized without DNA cleanup (contrary to DNA sequencing), and the use of genetic analysis software was useful for simplifying data analysis.
Our data could be implemented in diverse clinical guidelines for the pharmacogenetic analysis of the CYP2C9 gene before treatment with different drugs, such as non-steroidal/anti-inflammatory drugs, warfarin, phenytoin and statins [28,29,30,31].
Nevertheless, larger clinical studies are needed to define whether pVNTR-S has an effect in vivo, or whether the low activity attributed to the CYP2C9*3 allele is really a combination of the effects on CYP2C9 expression caused by the presence of pVNTR-S, along with effects on catalytic activity from the CYP2C9*3 variant. However, further studies should be performed to evaluate the potential relationship of this pVNTR with other CYP2C9 variants, such as CYP2C9*5, *6, *8 and *11, which are more frequent in other non-Caucasian populations, for example, populations with African ancestry [32].
Author Contributions
Conceptualization, P.D. and G.S.-D.; methodology, Y.G.-M., P.D. and G.S.-D.; formal analysis, P.D., Y.G.-M.; investigation, P.D., G.S.-D., Y.G.-M. and M.Á.S.-S.; writing—original draft preparation, P.D.; writing—review and editing, P.D., G.S.-D., Y.G.-M. and M.Á.S.-S.; funding acquisition, P.D. and M.Á.S.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Junta de Extremadura and European Regional Development Fund (FEDER) grant (IB16138; V Plan Regional de I + D + i).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Clinical Research Ethics Committee of Cáceres (Extremadura Health Service; reference: MASR/2016), and by the Bioethics and Biosecurity Committee (University of Extremadura; reference: 64/2016).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available in deidentified form on request from the corresponding author. The data are not publicly available due to privacy restrictions.
Acknowledgments
The authors thank all patients who kindly participated in the study, as well as Vanesa Garcia-Bernalt Funes for his support and help with kidney patients, Raquel Mayordomo, Ana Pérez-Pico and Esther Mingorance for their support with volunteers’ recruitment, and the technical and human support provided by the Facility of Bioscience Applied Techniques of SAIUEx (financed by UEX, Junta de Extremadura, MICINN, FEDER and FSE). In addition, the priceless help of Marisol López-López for the English edition of this manuscript is also acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lee, C.R.; Goldstein, J.A.; Pieper, J.A. Cytochrome P450 2C9 polymorphisms: A comprehensive review of the in-vitro and human data. Pharmacogenetics 2002, 12, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.K.; Rettie, A.E.; Fowler, D.M.; Miners, J.O. Pharmacogenomics of CYP2C9: Functional and Clinical Considerations. J. Pers. Med. 2017, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, J.A.; de Morais, S.M. Biochemistry and molecular biologyofthe human CYP2C subfamily. Pharmacogenetics 1994, 4, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Soars, M.G.; Gelboin, H.V.; Krausz, K.W.; Riley, R.J. A comparison of relative abundance; activity factor and inhibitory monoclonal antibody approaches in the characterization of human CYP enzymology. Br. J. Clin. Pharmacol. 2003, 55, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Rettie, A.E.; Jones, J.P. Clinical and toxicological relevance of CYP2C9: Drug-druginteractions and pharmacogenetics. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 477–494. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.F.; Zhou, Z.W.; Huang, M. Polymorphisms of human cytochrome P450 2C9 and the functional relevance. Toxicology 2010, 278, 165–188. [Google Scholar] [CrossRef]
- Wang, B.; Wang, J.; Huang, S.Q.; Su, H.H.; Zhou, S.F. Genetic polymorphism of the human cytochrome P450 2C9 gene and its clinical significance. Curr. Drug Metab. 2009, 10, 781–834. [Google Scholar] [CrossRef]
- Westphal, C.; Konkel, A.; Schunck, W.H. CYP-eicosanoids—A new link between omega-3 fatty acids and cardiac disease? Prostaglandins Other Lipid Mediat. 2011, 96, 99–108. [Google Scholar] [CrossRef]
- Spector, A.A.; Kim, H.Y. Cytochrome P450 epoxygenase pathway of polyunsaturated fatty acid metabolism. Biochim. Biophys. Acta 2015, 1851, 356–365. [Google Scholar] [CrossRef] [Green Version]
- The Pharmacogene Variation (PharmVar) Consortium. Available online: https://www.pharmvar.org/gene/CYP2C9 (accessed on 18 February 2022).
- Yasar, U.; Forslund-Bergengren, C.; Tybring, G.; Dorado, P.; Llerena, A.; Sjöqvist, F.; Eliasson, E.; Dahl, M.L. Pharmacokinetics of losartan and its metabolite E-3174 in relation to the CYP2C9 genotype. Clin. Pharmacol. Ther. 2002, 71, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Dorado, P.; Berecz, R.; Norberto, M.J.; Yasar, U.; Dahl, M.L.; LLerena, A. CYP2C9 genotypes and diclofenac metabolism in Spanish healthy volunteers. Eur. J. Clin. Pharmacol. 2003, 59, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Dorado, P.; Beltrán, L.J.; Machín, E.; Peñas-Lledó, E.M.; Terán, E.; Llerena, A. Losartan hydroxylation phenotype in an Ecuadorian population: Influence of CYP2C9 genetic polymorphism, habits and gender. Pharmacogenomics 2012, 13, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Kirchheiner, J.; Brockmöller, J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin. Pharmacol. Ther. 2005, 77, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Vázquez, A.; Dorado, P.; Fricke-Galindo, I.; Jung-Cook, H.; Monroy-Jaramillo, N.; Martínez-Juárez, I.E.; Familiar-López, I.; Peñas-Lledó, E.; LLerena, A.; López-López, M. CYP2C9; CYP2C19; ABCB1 genetic polymorphisms and phenytoin plasma concentrations in Mexican-Mestizo patients with epilepsy. Pharmacogenom. J. 2016, 16, 286–292. [Google Scholar] [CrossRef]
- Ninomiya, H.; Mamiya, K.; Matsuo, S.; Ieiri, I.; Higuchi, S.; Tashiro, N. Genetic polymorphism of the CYP2C subfamily and excessive serum phenytoin concentration with central nervous system intoxication. Ther. Drug Monit. 2000, 22, 230–232. [Google Scholar] [CrossRef]
- Brandolese, R.; Scordo, M.G.; Spina, E.; Gusella, M.; Padrini, R. Severe phenytoin intoxication in a subject homozygous for CYP2C9*3. Clin. Pharmacol. Ther. 2001, 70, 391–394. [Google Scholar] [CrossRef]
- Kidd, R.S.; Curry, T.B.; Gallagher, S.; Edeki, T.; Blaisdell, J.; Goldstein, J.A. Identification of a null allele of CYP2C9 in an African-American exhibiting toxicity to phenytoin. Pharmacogenetics 2001, 11, 803–808. [Google Scholar] [CrossRef]
- Dorado, P.; López-Torres, E.; Peñas-Lledó, E.M.; Martínez-Antón, J.; Llerena, A. Neurological toxicity after phenytoin infusion in a pediatric patient with epilepsy: Influence of CYP2C9; CYP2C19 and ABCB1 genetic polymorphisms. Pharmacogenom. J. 2013, 13, 359–361. [Google Scholar] [CrossRef]
- Aithal, G.P.; Day, C.P.; Kesteven, P.J.; Daly, A.K. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 1999, 353, 717–719. [Google Scholar] [CrossRef]
- Margaglione, M.; Colaizzo, D.; D’Andrea, G.; Brancaccio, V.; Ciampa, A.; Grandone, E.; Di Minno, G. Genetic modulation of oral anticoagulation with warfarin. Thromb. Haemost. 2000, 84, 775–778. [Google Scholar]
- Taube, J.; Halsall, D.; Baglin, T. Influence of cytochrome P-450 CYP2C9 polymorphisms on warfarin sensitivity and risk of over-anticoagulation in patients on long-term treatment. Blood 2000, 96, 1816–1819. [Google Scholar] [CrossRef] [PubMed]
- Verstuyft, C.; Morin, S.; Robert, A.; Loriot, M.A.; Beaune, P.; Jaillon, P.; Becquemont, L. Early acenocoumarol overanticoagulation among cytochrome P450 2C9 poor metabolizers. Pharmacogenetics 2001, 11, 735–737. [Google Scholar] [CrossRef] [PubMed]
- Dreisbach, A.W.; Smith, S.V.; Kyle, P.B.; Ramaiah, M.; Amenuke, M.; Garrett, M.R.; Lirette, S.T.; Griswold, M.E.; Roman, R.J. Urinary CYP eicosanoid excretion correlates with glomerular filtration in African-Americans with chronic kidney disease. Prostaglandins Other Lipid Mediat. 2014, 113–115, 45–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakhtiari, M.; Park, J.; Ding, Y.C.; Shleizer-Burko, S.; Neuhausen, S.L.; Halldórsson, B.V.; Stefánsson, K.; Gymrek, M.; Bafna, V. Variable number tandem repeats mediate the expression of proximal genes. Nat. Commun. 2021, 12, 2075. [Google Scholar] [CrossRef]
- Wang, D.; Sun, X.; Gong, Y.; Gawronski, B.E.; Langaee, T.Y.; Shahin, M.H.; Khalifa, S.I.; Johnson, J.A. CYP2C9 promoter variable number tandem repeat polymorphism regulates mRNA expression in human livers. Drug Metab. Dispos. 2012, 40, 884–891. [Google Scholar] [CrossRef] [Green Version]
- Al-Eitan, L.N.; Almasri, A.Y.; Al-Habahbeh, S.O. Impact of a variable number tandem repeat in the CYP2C9 promoter on warfarin sensitivity and responsiveness in Jordanians with cardiovascular disease. Pharmgenom. Pers. Med. 2019, 21, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Theken, K.N.; Lee, C.R.; Gong, L.; Caudle, K.E.; Formea, C.M.; Gaedigk, A.; Klein, T.E.; Agúndez, J.A.G.; Grosser, T. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin. Pharmacol. Ther. 2020, 108, 191–200. [Google Scholar] [CrossRef]
- Johnson, J.A.; Caudle, K.E.; Gong, L.; Whirl-Carrillo, M.; Stein, C.M.; Scott, S.A.; Lee, M.T.; Gage, B.F.; Kimmel, S.E.; Perera, M.A.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin. Pharmacol. Ther. 2017, 102, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Karnes, J.H.; Rettie, A.E.; Somogyi, A.A.; Huddart, R.; Fohner, A.E.; Formea, C.M.; Ta Michael Lee, M.; Llerena, A.; Whirl-Carrillo, M.; Klein, T.E.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update. Clin. Pharmacol. Ther. 2021, 109, 302–309. [Google Scholar] [CrossRef]
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for SLCO1B1; ABCG2; and CYP2C9 and statin-associated musculoskeletal symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef]
- PharmGKB. CYP2C9 Frequency Table. Available online: https://api.pharmgkb.org/v1/download/file/attachment/CYP2C9_frequency_table.xlsx (accessed on 18 April 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).