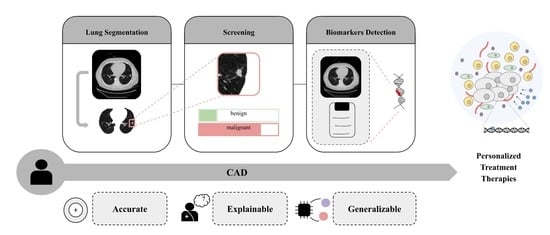
Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges
Abstract
1. Introduction
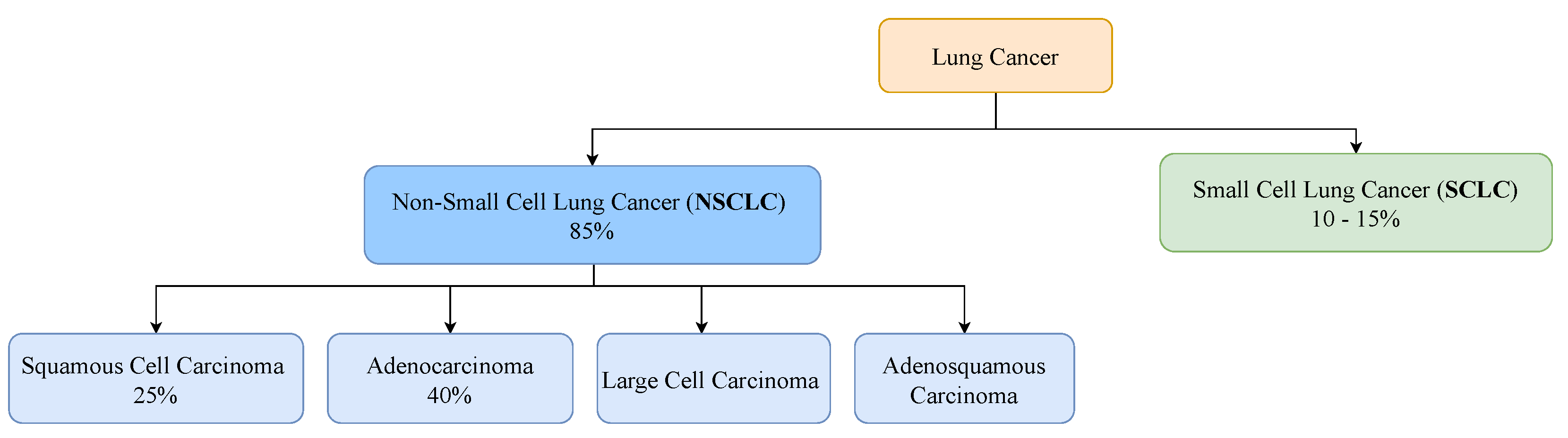
2. Clinical Pathway for Lung Cancer
2.1. Screening
2.2. Diagnosis
2.2.1. Initial Evaluation
2.2.2. Tissue Biopsy
2.2.3. Liquid Biopsy
2.2.4. Staging
2.3. Treatment Plan
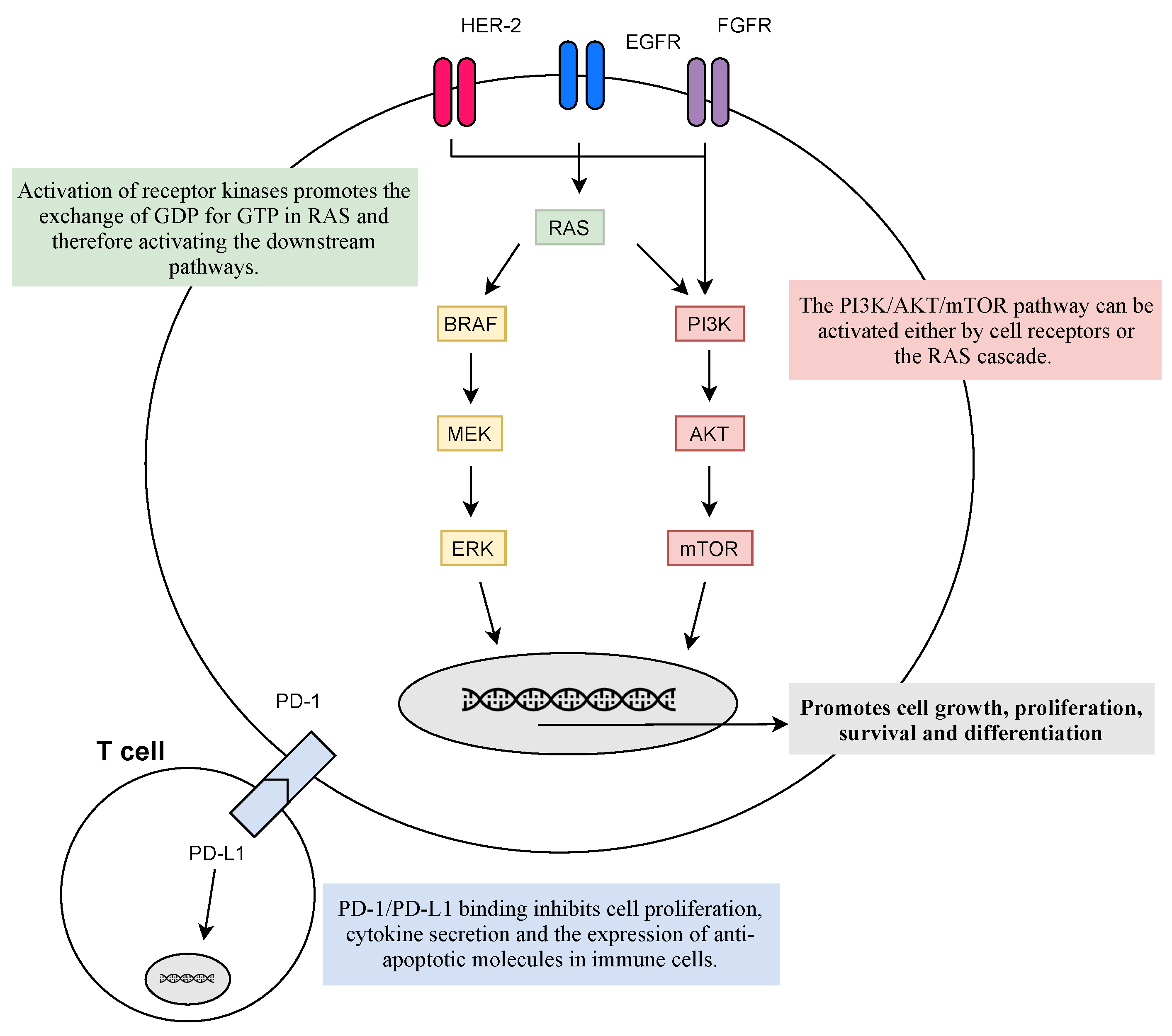
2.4. Main Biomarkers for Target Therapies
2.4.1. Oncogenes
2.4.2. Immunobiomarkers
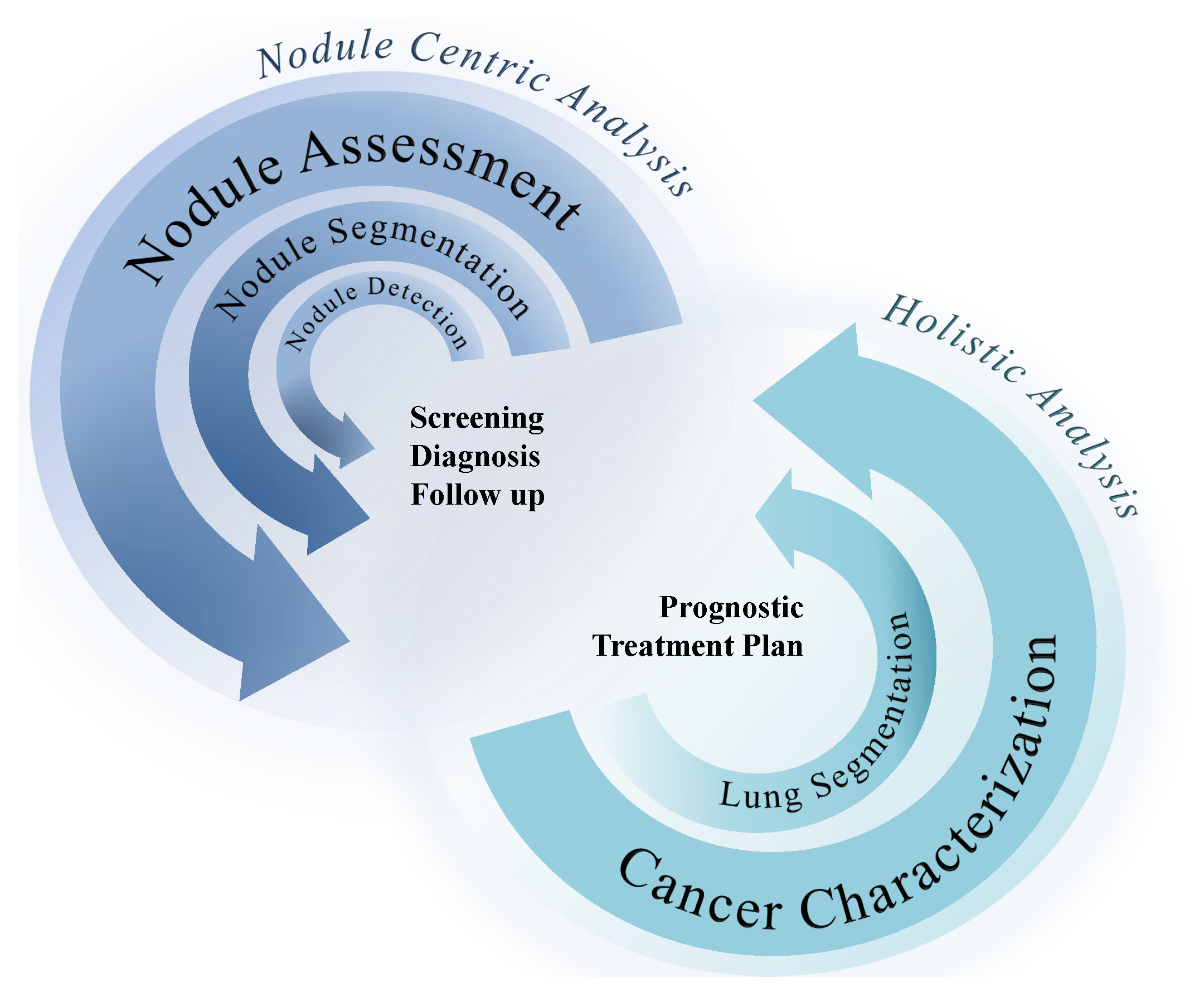
3. Computer-Aided Decision Systems
3.1. Nodule-Focused CADs
3.1.1. Nodule Detection and Segmentation
Nodule Detection
Nodule Segmentation
3.1.2. Nodule Classification
3.1.3. Interpretability Methods for Nodule-Focused CADs
3.1.4. Discussion and Future Work: Nodule Detection, Segmentation, and Classification
Improvements Needed
- Large and different public lung nodule databases for algorithm evaluation to provide replication of desired results and enhance the stringency of the algorithm so that lung nodule analysis tools can be validated mimicking real clinical scenarios.
- The ability to deal with pulmonary nodules based on location (isolated, juxtapleural, or juxta-vascular) and internal texture (solid, semi-solid, ground-glass opacity, and non-solid). In particular, the detection of ground glass optical and non-nodules is difficult and is explored by very few researchers.
- The ability to deal with pulmonary nodules with extremely small diameters. Most early-stage malignant tumors are smaller in size, and if these tumors are detected at an early stage, the survival chance of the individual can be increased.
- The ability to classify nodules not only as benign or malignant, but as benign, early-stage cancerous nodule, primary malignant, and metastasis malignant, decreasing the level of abstraction related to some clinical phenomena that must be considered.
- Develop a system capable of segmenting out large solid nodules attached to the pleural wall, which is quite challenging.
- Build a set of useful and efficient features based mainly on shape or geometry, intensity, and texture for better false-positive reduction.
- Develop a new CAD system based on powerful feature map visualization techniques to better analyze CNN’s decision and transfer it to radiologists.
- Fine-tune a pre-trained CNN model instead of training it from scratch to increase its robustness and surpass the limitation of annotated medical data.
- Develop in-depth research on GAN models, which can solve the problem of lack of medical databases.
- Design new CAD systems, including two or more of the CNN architectures to address the problem of overfitting that occurs during the training process due to imbalance in the datasets.
- Develop new deep learning techniques or optimize existing techniques to improve the performance of the CADe system, such as using a contracting path (to capture context) and a symmetric expanding path (to enable precise localization) to strengthen the use of available annotated samples, training multilayer networks efficiently by residual learning to gain accuracy from considerably increased depth.
- Promote cooperation and communication between academic institutions and medical organizations to combine real clinical requirements and the latest scientific achievements.
3.2. Lung Segmentation
3.2.1. Conventional Methods
3.2.2. Learning Methods
Discussion and Future Work: Lung Segmentation
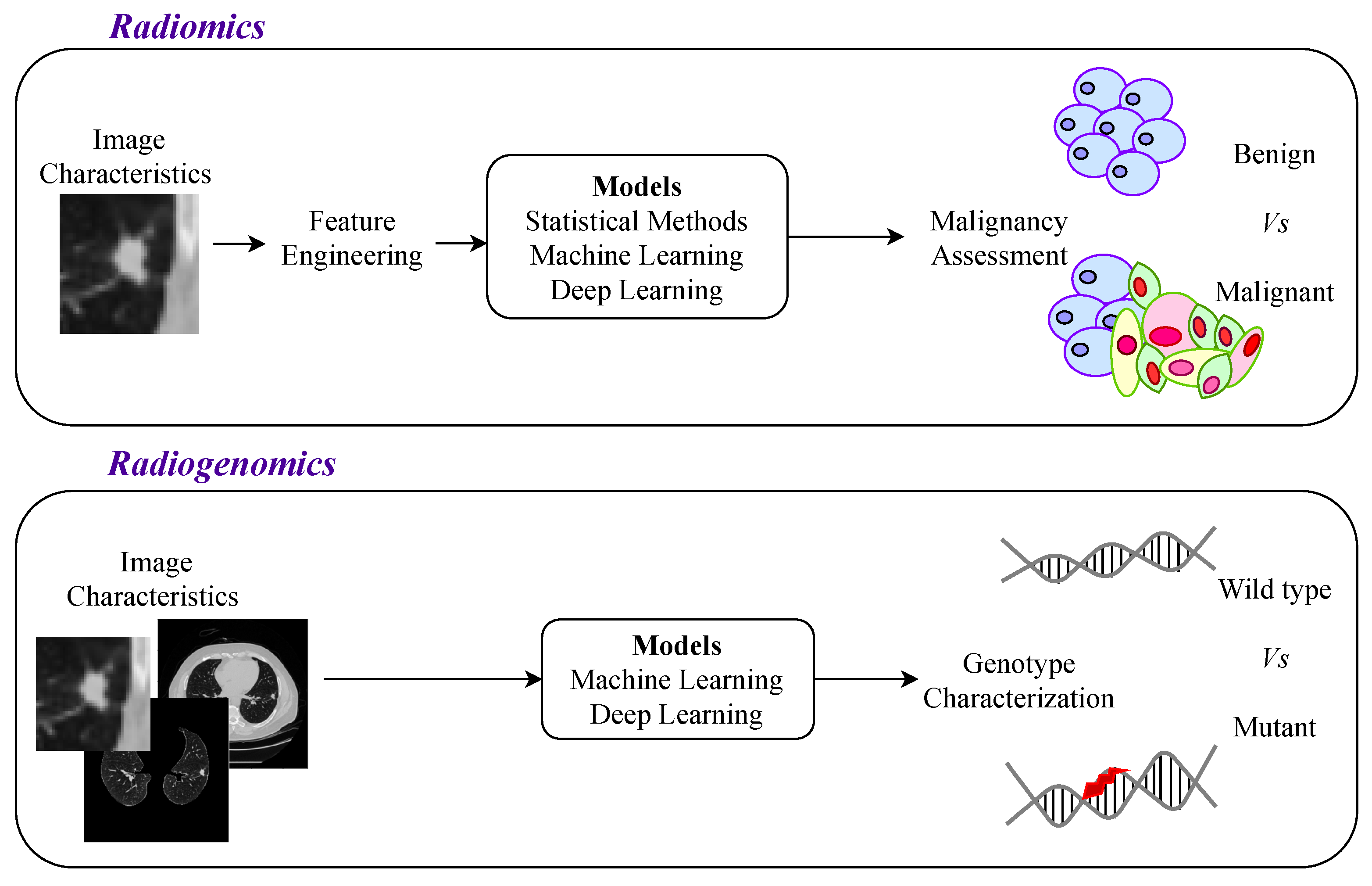
3.3. Genotype Prediction
3.3.1. Centered on Nodule
3.3.2. More Comprehensive Approaches
3.3.3. Discussion and Future Work: Genotype Prediction
3.4. PD-L1 Expression Prediction
Discussion and Future Work: PD-L1 Expression Assessment
4. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global epidemiology of lung cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef]
- World Health Organization. Latest global cancer data: Cancer burden rises to 18.1 million new cases and 9.6 million cancer deaths in 2018. In International Agency for Research on Cancer; World Health Organization: Geneva: Switzerland, 2018. [Google Scholar]
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung cancer statistics. Lung Cancer Pers. Med. 2016, 893, 1–19. [Google Scholar]
- Galvez-Nino, M.; Ruiz, R.; Pinto, J.A.; Roque, K.; Mantilla, R.; Raez, L.E.; Mas, L. Lung cancer in the young. Lung 2020, 198, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Zappa, C.; Mousa, S.A. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016, 5, 288. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, M.; Ding, X.J.; Cao, Y. Familial risk for lung cancer. Oncol. Lett. 2017, 13, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Bossé, Y.; Landi, M.T.; Gui, J.; Xiao, X.; Qian, D.; Joubert, P.; Lamontagne, M.; Li, Y.; Gorlov, I.; et al. Identification of susceptibility pathways for the role of chromosome 15q25. 1 in modifying lung cancer risk. Nat. Commun. 2018, 9, 3221. [Google Scholar] [CrossRef]
- Yokota, J.; Shiraishi, K.; Kohno, T. Genetic basis for susceptibility to lung cancer: Recent progress and future directions. Adv. Cancer Res. 2010, 109, 51–72. [Google Scholar]
- Okazaki, I.; Ishikawa, S.; Ando, W.; Sohara, Y. Lung adenocarcinoma in never smokers: Problems of primary prevention from aspects of susceptible genes and carcinogens. Anticancer. Res. 2016, 36, 6207–6224. [Google Scholar] [CrossRef]
- de Groot, P.M.; Wu, C.C.; Carter, B.W.; Munden, R.F. The epidemiology of lung cancer. Transl. Lung Cancer Res. 2018, 7, 220. [Google Scholar] [CrossRef]
- Rivera, G.A.; Wakelee, H. Lung cancer in never smokers. Lung Cancer Pers. Med. 2016, 893, 43–57. [Google Scholar]
- Malhotra, J.; Malvezzi, M.; Negri, E.; La Vecchia, C.; Boffetta, P. Risk factors for lung cancer worldwide. Eur. Respir. J. 2016, 48, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Cancer Facts & Figures. 2021. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021 (accessed on 15 April 2021).
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Lung Cancer. 2019. Available online: https://www.cancer.org/cancer/lung-cancer.html (accessed on 15 April 2021).
- Zhang, H.; Cai, W.; Wang, Y.; Liao, M.; Tian, S. CT and clinical characteristics that predict risk of EGFR mutation in non-small cell lung cancer: A systematic review and meta-analysis. Int. J. Clin. Oncol. 2019, 24, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, A.; Gul, M.; Chaudhry, A. Utility of computed tomography lung cancer screening and the management of computed tomography screen-detected findings. J. Thorac. Dis. 2018, 10, 1352. [Google Scholar] [CrossRef]
- Al Mohammad, B.; Brennan, P.C.; Mello-Thoms, C. A review of lung cancer screening and the role of computer-aided detection. Clin. Radiol. 2017, 72, 433–442. [Google Scholar] [CrossRef]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.W.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef]
- Junior, J.R.F.; Koenigkam-Santos, M.; Cipriano, F.E.G.; Fabro, A.T.; de Azevedo-Marques, P.M. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput. Methods Programs Biomed. 2018, 159, 23–30. [Google Scholar] [CrossRef]
- Ostridge, K.; Wilkinson, T.M. Present and future utility of computed tomography scanning in the assessment and management of COPD. Eur. Respir. J. 2016, 48, 216–228. [Google Scholar] [CrossRef]
- Bakr, S.; Gevaert, O.; Echegaray, S.; Ayers, K.; Zhou, M.; Shafiq, M.; Zheng, H.; Benson, J.A.; Zhang, W.; Leung, A.N.; et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data 2018, 5, 180202. [Google Scholar] [CrossRef]
- Ost, D.E.; Jim Yeung, S.C.; Tanoue, L.T.; Gould, M.K. Clinical and Organizational Factors in the Initial Evaluation of Patients With Lung Cancer. Chest 2013, 143, e121S–e141S. [Google Scholar] [CrossRef]
- Aggarwal, A.; Lewison, G.; Idir, S.; Peters, M.; Aldige, C.; Boerckel, W.; Boyle, P.; Trimble, E.L.; Roe, P.; Sethi, T.; et al. The state of lung cancer research: A global analysis. J. Thorac. Oncol. 2016, 11, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [CrossRef] [PubMed]
- Krist, A.H.; Davidson, K.W.; Mangione, C.M.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Kubik, M.; et al. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 325, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Latimer, K.M.; Mott, T.F. Lung cancer: Diagnosis, treatment principles, and screening. Am. Fam. Physician 2015, 91, 250–256. [Google Scholar]
- Loverdos, K.; Fotiadis, A.; Kontogianni, C.; Iliopoulou, M.; Gaga, M. Lung nodules: A comprehensive review on current approach and management. Ann. Thorac. Med. 2019, 14, 226–238. [Google Scholar] [CrossRef]
- Freitas, C.; Sousa, C.; Machado, F.; Serino, M.; Santos, V.; Cruz-Martins, N.; Teixeira, A.; Cunha, A.; Pereira, T.; Oliveira, H.P.; et al. The Role of Liquid Biopsy in Early Diagnosis of Lung Cancer. Front. Oncol. 2021, 11, 1130–1156. [Google Scholar] [CrossRef]
- Ganesha, D.V.; Naik, R.; Mufti, S.S.; Varayathu, H. Molecular Therapeutics of Non-Small Cell Lung Cancer (NSCLC) and Challenges in Repeat Tissue Biopsy. Adv. Lung Cancer 2021, 10, 21–39. [Google Scholar] [CrossRef]
- Tuzi, A.; Bolzacchini, E.; Suter, M.B.; Giaquinto, A.; Passaro, A.; Gobba, S.; Vallini, I.; Pinotti, G. Biopsy and re-biopsy in lung cancer: The oncologist requests and the role of endobronchial ultrasounds transbronchial needle aspiration. J. Thorac. Dis. 2017, 9, S405. [Google Scholar] [CrossRef][Green Version]
- Scrivener, M.; de Jong, E.E.; van Timmeren, J.E.; Pieters, T.; Ghaye, B.; Geets, X. Radiomics applied to lung cancer: A review. Transl. Cancer Res. 2016, 5, 398–409. [Google Scholar] [CrossRef]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 142, 321–346. [Google Scholar]
- Wu, Z.; Yang, Z.; Dai, Y.; Zhu, Q.; Chen, L.A. Update on liquid biopsy in clinical management of non-small cell lung cancer. Oncotargets Ther. 2019, 12, 5097. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, E.; Coco, S.; Genova, C.; Rossi, G.; Longo, L.; Grossi, F. Liquid biopsy in non-small cell lung cancer: Highlights and challenges. Cancers 2020, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhao, H. Liquid biopsy in tumors: Opportunities and challenges. Ann. Transl. Med. 2018, 6, S89. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Malapelle, U.; Troncone, G. Liquid biopsy and lung cancer. Acta Cytol. 2019, 63, 489–496. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. NCCN Guidelines Insights: Non–Small Cell Lung Cancer, Version 2.2021: Featured Updates to the NCCN Guidelines. J. Natl. Compr. Cancer Netw. 2021, 19, 254–266. [Google Scholar] [CrossRef]
- Carnio, S.; Novello, S.; Papotti, M.; Loiacono, M.; Scagliotti, G.V. Prognostic and predictive biomarkers in early stage non-small cell lung cancer: Tumor based approaches including gene signatures. Transl. Lung Cancer Res. 2013, 2, 372. [Google Scholar]
- Dine, J.; Gordon, R.; Shames, Y.; Kasler, M.K.; Barton-Burke, M. Immune checkpoint inhibitors: An innovation in immunotherapy for the treatment and management of patients with cancer. Asia-Pac. J. Oncol. Nurs. 2017, 4, 127. [Google Scholar] [CrossRef]
- N.I.H. National Cancer Institute, Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Lung and Bronchus Cancer; 2020. Available online: https://seer.cancer.gov/statfacts/html/lungb.html (accessed on 1 May 2021).
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef]
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.; Faivre-Finn, C.; Mok, T.; Reck, M.; Van Schil, P.; Hellmann, M.; et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv192–iv237. [Google Scholar] [CrossRef]
- Schrank, Z.; Chhabra, G.; Lin, L.; Iderzorig, T.; Osude, C.; Khan, N.; Kuckovic, A.; Singh, S.; Miller, R.J.; Puri, N. Current molecular-targeted therapies in NSCLC and their mechanism of resistance. Cancers 2018, 10, 224. [Google Scholar] [CrossRef]
- Jiang, W.; Cai, G.; Hu, P.C.; Wang, Y. Personalized medicine in non-small cell lung cancer: A review from a pharmacogenomics perspective. Acta Pharm. Sin. B 2018, 8, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Xie, C.; Yu, X.; Liu, J. EGFR TKI as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer. Oncotarget 2017, 8, 75712. [Google Scholar] [CrossRef] [PubMed]
- Holleman, M.S.; Al, M.J.; Zaim, R.; Groen, H.J.; Uyl-de Groot, C.A. Cost-effectiveness analysis of the first-line EGFR-TKIs in patients with non-small cell lung cancer harbouring EGFR mutations. Eur. J. Health Econ. 2020, 21, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Leighl, N.B.; Wu, Y.L.; Zhong, W.Z. Emerging therapies for non-small cell lung cancer. J. Hematol. Oncol. 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kim, J.; Qu, F.; Liu, S.; Wang, H.; Balagurunathan, Y.; Ye, Z.; Gillies, R.J. CT features associated with epidermal growth factor receptor mutation status in patients with lung adenocarcinoma. Radiology 2016, 280, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, G.; Pereira, T.; Dias, C.; Freitas, C.; Hespanhol, V.; Costa, J.L.; Cunha, A.; Oliveira, H.P. Identifying relationships between imaging phenotypes and lung cancer-related mutation status: EGFR and KRAS. Sci. Rep. 2020, 10, 3625. [Google Scholar] [CrossRef]
- Ma, W.; Gilligan, B.M.; Yuan, J.; Li, T. Current status and perspectives in translational biomarker research for PD-1/PD-L1 immune checkpoint blockade therapy. J. Hematol. Oncol. 2016, 9, 47. [Google Scholar] [CrossRef]
- Kerr, K.M.; Nicolson, M.C. Non–small cell lung cancer, PD-L1, and the pathologist. Arch. Pathol. Lab. Med. 2016, 140, 249–254. [Google Scholar] [CrossRef]
- Doroshow, D.B.; Sanmamed, M.F.; Hastings, K.; Politi, K.; Rimm, D.L.; Chen, L.; Melero, I.; Schalper, K.A.; Herbst, R.S. Immunotherapy in non–small cell lung cancer: Facts and hopes. Clin. Cancer Res. 2019, 25, 4592–4602. [Google Scholar] [CrossRef]
- Sharma, P. Immune checkpoint therapy and the search for predictive biomarkers. Cancer J. 2016, 22, 68. [Google Scholar] [CrossRef]
- Osmani, L.; Askin, F.; Gabrielson, E.; Li, Q.K. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): Moving from targeted therapy to immunotherapy. Semin. Cancer Biol. 2018, 52, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancte 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Lee, Y.T.; Tan, Y.J.; Oon, C.E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharmacol. 2018, 834, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Pakkala, S.; Ramalingam, S.S. Personalized therapy for lung cancer: Striking a moving target. JCI Insight 2018, 3, e120858. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.P.; Coelho, P.V.; Anazetti, M.; Simioni, P.U. Targeted therapies for the treatment of non-small-cell lung cancer: Monoclonal antibodies and biological inhibitors. Hum. Vaccines Immunother. 2017, 13, 843–853. [Google Scholar] [CrossRef]
- Liu, T.C.; Jin, X.; Wang, Y.; Wang, K. Role of epidermal growth factor receptor in lung cancer and targeted therapies. Am. J. Cancer Res. 2017, 7, 187–202. [Google Scholar]
- Dassonville, O.; Bozec, A.; Fischel, J.L.; Milano, G. EGFR targeting therapies: Monoclonal antibodies versus tyrosine kinase inhibitors. Similarities and differences. Crit. Rev. Oncol. 2007, 61, 53–61. [Google Scholar] [CrossRef]
- Uras, I.Z.; Moll, H.P.; Casanova, E. Targeting KRAS mutant non-small-cell lung cancer: Past, present and future. Int. J. Mol. Sci. 2020, 21, 4325. [Google Scholar] [CrossRef]
- Adderley, H.; Blackhall, F.H.; Lindsay, C.R. KRAS-mutant non-small cell lung cancer: Converging small molecules and immune checkpoint inhibition. EBioMedicine 2019, 41, 711–716. [Google Scholar] [CrossRef]
- Ghimessy, A.; Radeczky, P.; Laszlo, V.; Hegedus, B.; Renyi-Vamos, F.; Fillinger, J.; Klepetko, W.; Lang, C.; Dome, B.; Megyesfalvi, Z. Current therapy of KRAS-mutant lung cancer. Cancer Metastasis Rev. 2020, 39, 1159–1177. [Google Scholar] [CrossRef]
- Moore, A.R.; Rosenberg, S.C.; McCormick, F.; Malek, S. RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 2020, 19, 533–552. [Google Scholar] [CrossRef] [PubMed]
- Golding, B.; Luu, A.; Jones, R.; Viloria-Petit, A.M. The function and therapeutic targeting of anaplastic lymphoma kinase (ALK) in non-small cell lung cancer (NSCLC). Mol. Cancer 2018, 17, 52. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, E.E.; Kilvaer, T.K.; Rakaee, M.; Richardsen, E.; Hald, S.M.; Andersen, S.; Busund, L.T.; Bremnes, R.M.; Donnem, T. CTLA-4 expression in the non-small cell lung cancer patient tumor microenvironment: Diverging prognostic impact in primary tumors and lymph node metastases. Cancer Immunol. Immunother. 2017, 66, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Moya-Horno, I.; Viteri, S.; Karachaliou, N.; Rosell, R. Combination of immunotherapy with targeted therapies in advanced non-small cell lung cancer (NSCLC). Ther. Adv. Med. Oncol. 2018, 10, 1–12. [Google Scholar] [CrossRef]
- Pereira, T.; Freitas, C.; Costa, J.L.; Morgado, J.; Silva, F.; Negrão, E.; de Lima, B.F.; da Silva, M.C.; Madureira, A.J.; Ramos, I.; et al. Comprehensive Perspective for Lung Cancer Characterisation Based on AI Solutions Using CT Images. J. Clin. Med. 2021, 10, 118. [Google Scholar] [CrossRef]
- Liu, Y.; Kim, J.; Balagurunathan, Y.; Li, Q.; Garcia, A.L.; Stringfield, O.; Ye, Z.; Gillies, R.J. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin. Lung Cancer 2016, 17, 441–448. [Google Scholar] [CrossRef]
- Gevaert, O.; Echegaray, S.; Khuong, A.; Hoang, C.D.; Shrager, J.B.; Jensen, K.C.; Berry, G.J.; Guo, H.H.; Lau, C.; Plevritis, S.K.; et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep. 2017, 7, 41674. [Google Scholar] [CrossRef]
- Wilson, R.; Devaraj, A. Radiomics of pulmonary nodules and lung cancer. Transl. Lung Cancer Res. 2017, 6, 86. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Dias, C.; Pinheiro, G.; Cunha, A.; Oliveira, H.P. Radiogenomics: Lung Cancer-Related Genes Mutation Status Prediction. In Proceedings of the Iberian Conference on Pattern Recognition and Image Analysis, Madrid, Spain, 1–4 July 2019; pp. 335–345. [Google Scholar]
- Zhang, G.; Jiang, S.; Yang, Z.; Gong, L.; Ma, X.; Zhou, Z.; Bao, C.; Liu, Q. Automatic nodule detection for lung cancer in CT images: A review. Comput. Biol. Med. 2018, 103, 287–300. [Google Scholar] [CrossRef]
- Halder, A.; Dey, D.; Sadhu, A.K. Lung nodule detection from feature engineering to deep learning in thoracic CT images: A comprehensive review. J. Digit. Imaging 2020, 33, 655–677. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Liu, G.; Xue, Z. On the performance of lung nodule detection, segmentation and classification. Comput. Med. Imaging Graph. 2021, 89, 101886. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Wu, F.; Yang, B.; Ma, J.; Kong, D.; Chen, Z.; Long, D. Pulmonary nodule detection using hybrid two-stage 3D CNNs. Med. Phys. 2020, 47, 3376–3388. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Ma, H.; Zhang, J. Automatic detection of pulmonary nodules in CT images based on 3D Res-I network. Vis. Comput. 2020, 10, 1917–1929. [Google Scholar] [CrossRef]
- Zheng, S.; Cornelissen, L.J.; Cui, X.; Jing, X.; Veldhuis, R.N.; Oudkerk, M.; van Ooijen, P.M. Deep convolutional neural networks for multi-planar lung nodule detection: Improvement in small nodule identification. Med. Phys. 2020, 48, 733–744. [Google Scholar] [CrossRef]
- Xu, Y.M.; Zhang, T.; Xu, H.; Qi, L.; Zhang, W.; Zhang, Y.D.; Gao, D.S.; Yuan, M.; Yu, T.F. Deep learning in CT images: Automated pulmonary nodule detection for subsequent management using convolutional neural network. Cancer Manag. Res. 2020, 12, 2979. [Google Scholar] [CrossRef]
- Ye, Y.; Tian, M.; Liu, Q.; Tai, H.M. Pulmonary Nodule Detection Using V-Net and High-Level Descriptor Based SVM Classifier. IEEE Access 2020, 8, 176033–176041. [Google Scholar] [CrossRef]
- Jain, P.; Shivwanshi, R.R.; Nirala, N.; Gupta, S. SumNet Convolution Neural network based Automated pulmonary nodule detection system. In Proceedings of the 2020 IEEE International Conference on Advent Trends in Multidisciplinary Research and Innovation (ICATMRI), Buldhana, India, 30 December 2020; pp. 1–6. [Google Scholar]
- Luo, X.; Song, T.; Wang, G.; Chen, J.; Chen, Y.; Li, K.; Metaxas, D.N.; Zhang, S. SCPM-Net: An Anchor-free 3D Lung Nodule Detection Network using Sphere Representation and Center Points Matching. arXiv 2021, arXiv:2104.05215. [Google Scholar] [CrossRef]
- Bhaskar, N.; Ganashree, T. Lung Nodule Detection from CT scans using Gaussian Mixture Convolutional AutoEncoder and Convolutional Neural Network. Ann. Rom. Soc. Cell Biol. 2021, 25, 6524–6531. [Google Scholar]
- Khehrah, N.; Farid, M.S.; Bilal, S.; Khan, M.H. Lung Nodule Detection in CT Images Using Statistical and Shape-Based Features. J. Imaging 2020, 6, 6. [Google Scholar] [CrossRef]
- Kuo, C.F.J.; Huang, C.C.; Siao, J.J.; Hsieh, C.W.; Huy, V.Q.; Ko, K.H.; Hsu, H.H. Automatic lung nodule detection system using image processing techniques in computed tomography. Biomed. Signal Process. Control 2020, 56, 101659. [Google Scholar] [CrossRef]
- Mukherjee, J.; Kar, M.; Chakrabarti, A.; Das, S. A soft-computing based approach towards automatic detection of pulmonary nodule. Biocybern. Biomed. Eng. 2020, 40, 1036–1051. [Google Scholar] [CrossRef]
- Paing, M.P.; Hamamoto, K.; Tungjitkusolmun, S.; Visitsattapongse, S.; Pintavirooj, C. Automatic detection of pulmonary nodules using three-dimensional chain coding and optimized random forest. Appl. Sci. 2020, 10, 2346. [Google Scholar] [CrossRef]
- El-Regaily, S.A.; Salem, M.A.M.; Aziz, M.H.A.; Roushdy, M.I. Multi-view Convolutional Neural Network for lung nodule false positive reduction. Expert Syst. Appl. 2020, 162, 113017. [Google Scholar] [CrossRef]
- Mittapalli, P.S.; Thanikaiselvan, V. Multiscale CNN with compound fusions for false positive reduction in lung nodule detection. Artif. Intell. Med. 2021, 113, 102017. [Google Scholar] [CrossRef]
- Vipparla, V.K.; Chilukuri, P.K.; Kande, G.B. Attention Based Multi-Patched 3D-CNNs with Hybrid Fusion Architecture for Reducing False Positives during Lung Nodule Detection. J. Comput. Commun. 2021, 9, 1. [Google Scholar] [CrossRef]
- Baker, A.A.A.; Ghadi, Y. A novel CAD system to automatically detect cancerous lung nodules using wavelet transform and SVM. Int. J. Electr. Comput. Eng. 2020, 10, 4745. [Google Scholar]
- Halder, A.; Chatterjee, S.; Dey, D. Morphological Filter Aided GMM Technique for Lung Nodule Detection. In Proceedings of the 2020 IEEE Applied Signal Processing Conference (ASPCON), Kolkata, India, 7–9 October 2020; pp. 198–202. [Google Scholar]
- Mahersia, H.; Boulehmi, H.; Hamrouni, K. CAD system for lung nodules detection using wavelet-based approach and intelligent classifiers. In Proceedings of the 2020 17th International Multi-Conference on Systems, Signals & Devices (SSD), Monastir, Tunisia, 20–23 July 2020; pp. 173–178. [Google Scholar]
- Drokin, I.; Ericheva, E. Deep Learning on Point Clouds for False Positive Reduction at Nodule Detection in Chest CT Scans. arXiv 2020, arXiv:2005.03654. [Google Scholar]
- Harsono, I.W.; Liawatimena, S.; Cenggoro, T.W. Lung nodule detection and classification from thorax CT-scan using RetinaNet with transfer learning. J. King Saud-Univ.-Comput. Inf. Sci. 2020, 34, 567–577. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Liu, C.; Hu, S.C.; Wang, C.; Lafata, K.; Yin, F.F. Automatic detection of pulmonary nodules on CT images with YOLOv3: Development and evaluation using simulated and patient data. Quant. Imaging Med. Surg. 2020, 10, 1917. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Fulzele, P.; Sreedevi, I. Hybrid Model for Lung Nodule Segmentation based on Support Vector Machine and k-Nearest Neighbor. In Proceedings of the 2020 Fourth International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 11–13 March 2020; pp. 170–175. [Google Scholar]
- Hancock, M.C.; Magnan, J.F. Level set image segmentation with velocity term learned from data with applications to lung nodule segmentation. arXiv 2021, arXiv:1910.03191. [Google Scholar]
- Savic, M.; Ma, Y.; Ramponi, G.; Du, W.; Peng, Y. Lung Nodule Segmentation with a Region-Based Fast Marching Method. Sensors 2021, 21, 1908. [Google Scholar] [CrossRef]
- Xiao, Z.; Liu, B.; Geng, L.; Zhang, F.; Liu, Y. Segmentation of Lung Nodules Using Improved 3D-UNet Neural Network. Symmetry 2020, 12, 1787. [Google Scholar] [CrossRef]
- Kumar, S.; Raman, S. Lung nodule segmentation using 3-dimensional convolutional neural networks. In Soft Computing for Problem Solving; Springer: Berlin/Heidelberg, Germany, 2020; pp. 585–596. [Google Scholar]
- Rocha, J.; Cunha, A.; Mendonça, A.M. Conventional filtering versus u-net based models for pulmonary nodule segmentation in ct images. J. Med. Syst. 2020, 44, 81. [Google Scholar] [CrossRef]
- Singadkar, G.; Mahajan, A.; Thakur, M.; Talbar, S. Deep deconvolutional residual network based automatic lung nodule segmentation. J. Digit. Imaging 2020, 33, 678–684. [Google Scholar] [CrossRef]
- Naik, A.; Edla, D.R. Lung nodule classification on computed tomography images using deep learning. Wirel. Pers. Commun. 2021, 116, 655–690. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Lu, H.; Zhang, L.; Pan, J.; Bao, Y.; Su, J.; Qian, D. Feature-shared adaptive-boost deep learning for invasiveness classification of pulmonary subsolid nodules in CT images. Med. Phys. 2020, 47, 1738–1749. [Google Scholar] [CrossRef]
- Lin, Z.; Zheng, J.; Hu, W. Using 3D Convolutional Networks with Shortcut Connections for Improved Lung Nodules Classification. In Proceedings of the 2020 2nd International Conference on Big Data Engineering, Shanghai, China, 29–31 May 2020; pp. 42–49. [Google Scholar]
- Shah, G.; Thammasudjarit, R.; Thakkinstian, A.; Suwatanapongched, T. NoduleNet: A Lung Nodule Classification Using Deep Learning. Ramathibodi Med. J. 2020, 43, 11–19. [Google Scholar] [CrossRef]
- Tong, C.; Liang, B.; Su, Q.; Yu, M.; Hu, J.; Bashir, A.K.; Zheng, Z. Pulmonary nodule classification based on heterogeneous features learning. IEEE J. Sel. Areas Commun. 2020, 39, 574–581. [Google Scholar] [CrossRef]
- Huang, X.; Lei, Q.; Xie, T.; Zhang, Y.; Hu, Z.; Zhou, Q. Deep transfer convolutional neural network and extreme learning machine for lung nodule diagnosis on CT images. Knowl.-Based Syst. 2020, 204, 106230. [Google Scholar] [CrossRef]
- Lyu, J.; Bi, X.; Ling, S.H. Multi-level cross residual network for lung nodule classification. Sensors 2020, 20, 2837. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Sun, X.; Zhao, Z.; Wang, H.; Pan, S.; Schuller, B. Classification of lung nodules based on deep residual networks and migration learning. Comput. Intell. Neurosci. 2020, 2020, 8975078. [Google Scholar] [CrossRef]
- Lin, C.J.; Li, Y.C. Lung Nodule Classification Using Taguchi-Based Convolutional Neural Networks for Computer Tomography Images. Electronics 2020, 9, 1066. [Google Scholar] [CrossRef]
- Bansal, G.; Chamola, V.; Narang, P.; Kumar, S.; Raman, S. Deep3DSCan: Deep residual network and morphological descriptor based framework for lung cancer classification and 3D segmentation. IET Image Process. 2020, 14, 1240–1247. [Google Scholar] [CrossRef]
- Zhang, G.; Yang, Z.; Gong, L.; Jiang, S.; Wang, L.; Zhang, H. Classification of lung nodules based on CT images using squeeze-and-excitation network and aggregated residual transformations. Radiol. Med. 2020, 125, 374–383. [Google Scholar] [CrossRef]
- Ali, I.; Muzammil, M.; Haq, I.U.; Khaliq, A.A.; Abdullah, S. Efficient Lung Nodule Classification Using Transferable Texture Convolutional Neural Network. IEEE Access 2020, 8, 175859–175870. [Google Scholar] [CrossRef]
- Xiao, N.; Qiang, Y.; Bilal Zia, M.; Wang, S.; Lian, J. Ensemble classification for predicting the malignancy level of pulmonary nodules on chest computed tomography images. Oncol. Lett. 2020, 20, 401–408. [Google Scholar] [CrossRef]
- Jiang, H.; Gao, F.; Xu, X.; Huang, F.; Zhu, S. Attentive and ensemble 3D dual path networks for pulmonary nodules classification. Neurocomputing 2020, 398, 422–430. [Google Scholar] [CrossRef]
- Naik, A.; Edla, D.R.; Kuppili, V. A combination of FractalNet and CNN for Lung Nodule Classification. In Proceedings of the 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kharagpur, India, 1–3 July 2020; pp. 1–7. [Google Scholar]
- Liu, H.; Cao, H.; Song, E.; Ma, G.; Xu, X.; Jin, R.; Liu, C.; Hung, C.C. Multi-model Ensemble Learning Architecture Based on 3D CNN for Lung Nodule Malignancy Suspiciousness Classification. J. Digit. Imaging 2020, 33, 1242–1256. [Google Scholar] [CrossRef] [PubMed]
- Zhai, P.; Tao, Y.; Chen, H.; Cai, T.; Li, J. Multi-Task Learning for Lung Nodule Classification on Chest CT. IEEE Access 2020, 8, 180317–180327. [Google Scholar] [CrossRef]
- Paul, R.; Schabath, M.; Gillies, R.; Hall, L.; Goldgof, D. Convolutional Neural Network ensembles for accurate lung nodule malignancy prediction 2 years in the future. Comput. Biol. Med. 2020, 122, 103882. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, C.; Li, D.; Niu, J. Combining multi-scale feature fusion with multi-attribute grading, a CNN model for benign and malignant classification of pulmonary nodules. J. Digit. Imaging 2020, 33, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Chae, K.J.; Choi, Y.; Jin, G.Y.; Ko, S.B. Novel convolutional neural network architecture for improved pulmonary nodule classification on computed tomography. Multidimens. Syst. Signal Process. 2020, 31, 1163–1183. [Google Scholar] [CrossRef]
- Xu, X.; Wang, C.; Guo, J.; Gan, Y.; Wang, J.; Bai, H.; Zhang, L.; Li, W.; Yi, Z. MSCS-DeepLN: Evaluating lung nodule malignancy using multi-scale cost-sensitive neural networks. Med. Image Anal. 2020, 65, 101772. [Google Scholar] [CrossRef]
- Afshar, P.; Oikonomou, A.; Naderkhani, F.; Tyrrell, P.N.; Plataniotis, K.N.; Farahani, K.; Mohammadi, A. 3D-MCN: A 3D multi-scale capsule network for lung nodule malignancy prediction. Sci. Rep. 2020, 10, 7948. [Google Scholar] [CrossRef]
- Xia, K.; Chi, J.; Gao, Y.; Jiang, Y.; Wu, C. Adaptive Aggregated Attention Network for Pulmonary Nodule Classification. Appl. Sci. 2021, 11, 610. [Google Scholar] [CrossRef]
- Bao, L.; Bao, T.; Zheng, Y.; Xia, J. A Simple Residual Network for Lung Nodule Classification. In Proceedings of the Fourth International Conference on Biological Information and Biomedical Engineering, Chengdu, China, 21–23 July 2020; pp. 1–5. [Google Scholar]
- Zia, M.B.; Zhao, J.J.; Zhou, X.; Xiao, N.; Wang, J.; Khan, A. Classification of malignant and benign lung nodule and prediction of image label class using multi-deep model. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 35–41. [Google Scholar] [CrossRef][Green Version]
- Veasey, B.; Farhangi, M.M.; Frigui, H.; Broadhead, J.; Dahle, M.; Pezeshk, A.; Seow, A.; Amini, A.A. Lung nodule malignancy classification based on NLSTx Data. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1870–1874. [Google Scholar]
- Onishi, Y.; Teramoto, A.; Tsujimoto, M.; Tsukamoto, T.; Saito, K.; Toyama, H.; Imaizumi, K.; Fujita, H. Investigation of pulmonary nodule classification using multi-scale residual network enhanced with 3DGAN-synthesized volumes. Radiol. Phys. Technol. 2020, 13, 160–169. [Google Scholar] [CrossRef]
- Kuang, Y.; Lan, T.; Peng, X.; Selasi, G.E.; Liu, Q.; Zhang, J. Unsupervised multi-discriminator generative adversarial network for lung nodule malignancy classification. IEEE Access 2020, 8, 77725–77734. [Google Scholar] [CrossRef]
- Silva, F.; Pereira, T.; Frade, J.; Mendes, J.; Freitas, C.; Hespanhol, V.; Costa, J.L.; Cunha, A.; Oliveira, H.P. Pre-Training Autoencoder for Lung Nodule Malignancy Assessment Using CT Images. Appl. Sci. 2020, 10, 7837. [Google Scholar] [CrossRef]
- Lima, L.; Vieira, T.; Costa, E.; Azevedo-Marques, P.; Oliveira, M. Using Support Vector Machine and Features Selection on Classification of Early Lung Nodules. In Anais do XX Simpósio Brasileiro de Computação Aplicada à Saúde; SBC: Porto Alegre, Brazil, 2020; pp. 60–71. [Google Scholar]
- Reyes, M.; Meier, R.; Pereira, S.; Silva, C.A.; Dahlweid, F.M.; von Tengg-Kobligk, H.; Summers, R.M.; Wiest, R. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiol. Artif. Intell. 2020, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Knapič, S.; Malhi, A.; Salujaa, R.; Främling, K. Explainable Artificial Intelligence for Human Decision-Support System in Medical Domain. arXiv 2021, arXiv:2105.02357. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A survey on explainable artificial intelligence (xai): Toward medical xai. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4793–4813. [Google Scholar] [CrossRef]
- van der Velden, B.H.; Kuijf, H.J.; Gilhuijs, K.G.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. arXiv 2021, arXiv:2107.10912. [Google Scholar]
- Kokhlikyan, N.; Miglani, V.; Martin, M.; Wang, E.; Alsallakh, B.; Reynolds, J.; Melnikov, A.; Kliushkina, N.; Araya, C.; Yan, S.; et al. Captum: A unified and generic model interpretability library for PyTorch. arXiv 2020, arXiv:2009.07896. [Google Scholar]
- Li, Y.; Gu, D.; Wen, Z.; Jiang, F.; Liu, S. Classify and explain: An interpretable convolutional neural network for lung cancer diagnosis. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1065–1069. [Google Scholar]
- Jiang, H.; Shen, F.; Gao, F.; Han, W. Learning efficient, explainable and discriminative representations for pulmonary nodules classification. Pattern Recognit. 2021, 113, 107825. [Google Scholar] [CrossRef]
- Shaziya, H.; Shyamala, K.; Zaheer, R. Comprehensive Review of Automatic Lung Segmentation Techniques on Pulmonary CT Images. In Proceedings of the 2019 Third International Conference on Inventive Systems and Control (ICISC), Coimbatore, India, 10–11 January 2019; pp. 540–545. [Google Scholar] [CrossRef]
- El-Baz, A.; Beache, G.M.; Gimel’Farb, G.; Suzuki, K.; Okada, K.; Elnakib, A.; Soliman, A.; Abdollahi, B. Computer-aided diagnosis systems for lung cancer: Challenges and methodologies. Int. J. Biomed. Imaging 2013, 2013, 46. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Dai, Y.; Zhang, P. Supervised recursive segmentation of volumetric CT images for 3D reconstruction of lung and vessel tree. Comput. Methods Programs Biomed. 2015, 122, 316–329. [Google Scholar] [CrossRef]
- Lai, J.; Wei, Q. Automatic lung fields segmentation in CT scans using morphological operation and anatomical information. Biomed. Mater. Eng. 2014, 24, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Ma, J.; Zhao, M.; Liu, Y.; Feng, Y.; Zhang, M.; He, L.; Suzuki, K. Many Is Better Than One: An Integration of Multiple Simple Strategies for Accurate Lung Segmentation in CT Images. Biomed Res. Int. 2016, 2016, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, X.; Zhang, P.; Chen, J. Global optimal hybrid geometric active contour for automated lung segmentation on CT images. Comput. Biol. Med. 2017, 91, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Rebouças Filho, P.P.; Cortez, P.C.; da Silva Barros, A.C.; Victor, V.H.; Tavares, R.S.M. Novel and powerful 3D adaptive crisp active contour method applied in the segmentation of CT lung images. Med. Image Anal. 2017, 35, 503–516. [Google Scholar] [CrossRef]
- Chen, C.; Xiao, R.; Zhang, T.; Lu, Y.; Guo, X.; Wang, J.; Chen, H.; Wang, Z. Pathological lung segmentation in chest CT images based on improved random walker. Comput. Methods Programs Biomed. 2021, 200, 105864. [Google Scholar] [CrossRef]
- Oliveira, B.; Queirós, S.; Morais, P.; Torres, H.R.; Gomes-Fonseca, J.; Fonseca, J.C.; Vilaça, J.L. A novel multi-atlas strategy with dense deformation field reconstruction for abdominal and thoracic multi-organ segmentation from computed tomography. Med. Image Anal. 2018, 45, 108–120. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar] [CrossRef]
- Dong, X.; Lei, Y.; Wang, T.; Thomas, M.; Tang, L.; Curran, W.J.; Liu, T.; Yang, X. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med. Phys. 2019, 46, 2157–2168. [Google Scholar] [CrossRef]
- Feng, X.; Qing, K.; Tustison, N.J.; Meyer, C.H.; Chen, Q. Deep convolutional neural network for segmentation of thoracic organs-at-risk using cropped 3D images. Med. Phys. 2019, 46, 2169–2180. [Google Scholar] [CrossRef]
- Park, B.; Park, H.; Lee, S.M.; Seo, J.B.; Kim, N. Lung Segmentation on HRCT and Volumetric CT for Diffuse Interstitial Lung Disease Using Deep Convolutional Neural Networks. J. Digit. Imaging 2019, 32, 1019–1026. [Google Scholar] [CrossRef]
- Hofmanninger, J.; Prayer, F.; Pan, J.; Rohrich, S.; Prosch, H.; Langs, G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur. Radiol. Exp. 2020, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.J.; Yoon, S.H.; Lee, J.H.; Kim, K.H.; Choi, H.I.; Park, S.J.; Goo, J.M. Automated Lung Segmentation on Chest Computed Tomography Images with Extensive Lung Parenchymal Abnormalities Using a Deep Neural Network. Korean J. Radiol. 2020, 21, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, K.; Hao, X.; Xue, X.; An, H.; Zhang, H. A Novel U-Like Network for the Segmentation of Thoracic Organs. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging Workshops (ISBI Workshops), Iowa City, IA, USA, 4 April 2020. [Google Scholar] [CrossRef]
- Nemoto, T.; Futakami, N.; Yagi, M.; Kumabe, A.; Takeda, A.; Kunieda, E.; Shigematsu, N. Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi. J. Radiat. Res. 2020, 61, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, C.; Coleman, S.; Kerr, D. DENSE-INception U-net for medical image segmentation. Comput. Methods Programs Biomed. 2020, 192, 105395. [Google Scholar] [CrossRef]
- Vu, C.C.; Siddiqui, Z.A.; Zamdborg, L.; Thompson, A.B.; Quinn, T.J.; Castillo, E.; Guerrero, T.M. Deep convolutional neural networks for automatic segmentation of thoracic organs-at-risk in radiation oncology – use of non-domain transfer learning. J. Appl. Clin. Med. Phys. 2020, 21, 108–113. [Google Scholar] [CrossRef]
- Wang, Z.; Zou, Y.; Liu, P.X. Hybrid dilation and attention residual U-Net for medical image segmentation. Comput. Biol. Med. 2021, 134, 104449. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Yu, B.; Lei, B.; Ying, H.; Zhang, X.; Chen, D.Z.; Wu, J. Cascaded SE-ResUnet for segmentation of thoracic organs at risk. Neurocomputing 2021, 453, 357–368. [Google Scholar] [CrossRef]
- Jalali, Y.; Fateh, M.; Rezvani, M.; Abolghasemi, V.; Anisi, M.H. ResBCDU-net: A deep learning framework for lung CT image segmentation. Sensors 2021, 21, 268. [Google Scholar] [CrossRef]
- Khanna, A.; Londhe, N.D.; Gupta, S.; Semwal, A. A deep Residual U-Net convolutional neural network for automated lung segmentation in computed tomography images. Biocybern. Biomed. Eng. 2020, 40, 1314–1327. [Google Scholar] [CrossRef]
- Tan, J.; Jing, L.; Huo, Y.; Li, L.; Akin, O.; Tian, Y. LGAN: Lung segmentation in CT scans using generative adversarial network. Comput. Med. Imaging Graph. 2021, 87, 101817. [Google Scholar] [CrossRef]
- Pawar, S.P.; Talbar, S.N. LungSeg-Net: Lung field segmentation using generative adversarial network. Biomed. Signal Process. Control 2021, 64, 102296. [Google Scholar] [CrossRef]
- Xu, R.; Wang, Y.; Liu, T.; Ye, X.; Lin, L.; Chen, Y.W.; Kido, S.; Tomiyama, N. BG-Net: Boundary-Guided Network for Lung Segmentation on Clinical CT Images. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 8782–8788. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, R.; Xie, W.; Pang, M. Pathological lung segmentation based on random forest combined with deep model and multi-scale superpixels. Neural Process. Lett. 2020, 52, 1631–1649. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Luís, L.F.; Holanda, G.B.; Alves, S.S.; Francisco, F.H.; Han, T.; Rebouças Filho, P.P. An effective approach for CT lung segmentation using mask region-based convolutional neural networks. Artif. Intell. Med. 2020, 103, 101792. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Nunes, V.X.; De Freitas Souza, L.F.; Marques, A.G.; Silva, I.C.L.; Junior, M.A.A.F.; Sun, J.; Filho, P.P. Internet of medical things - Based on deep learning techniques for segmentation of lung and stroke regions in CT Scans. IEEE Access 2020, 8, 71117–71135. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar] [CrossRef]
- Carper, M.B.; Claudio, P.P. Clinical potential of gene mutations in lung cancer. Clin. Transl. Med. 2015, 4, 33. [Google Scholar] [CrossRef]
- Cheng, Z.; Shan, F.; Yang, Y.; Shi, Y.; Zhang, Z. CT characteristics of non-small cell lung cancer with epidermal growth factor receptor mutation: A systematic review and meta-analysis. BMC Med. Imaging 2017, 17, 5. [Google Scholar] [CrossRef]
- Zou, J.; Lv, T.; Zhu, S.; Lu, Z.; Shen, Q.; Xia, L.; Wu, J.; Song, Y.; Liu, H. Computed tomography and clinical features associated with epidermal growth factor receptor mutation status in stage I/II lung adenocarcinoma. Thorac. Cancer 2017, 8, 260–270. [Google Scholar] [CrossRef]
- Koyasu, S.; Nishio, M.; Isoda, H.; Nakamoto, Y.; Togashi, K. Usefulness of gradient tree boosting for predicting histological subtype and EGFR mutation status of non-small cell lung cancer on 18F FDG-PET/CT. Ann. Nucl. Med. 2019, 34, 49–57. [Google Scholar] [CrossRef]
- Li, X.Y.; Xiong, J.F.; Jia, T.Y.; Shen, T.L.; Hou, R.P.; Zhao, J.; Fu, X.L. Detection of epithelial growth factor receptor (EGFR) mutations on CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks. J. Thorac. Dis. 2018, 10, 6624–6635. [Google Scholar] [CrossRef]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, J.; Ni, B.; Bi, D.; Sun, Y.; Xu, M.; Zhu, X.; Li, C.; Jin, L.; Gao, P.; et al. Toward automatic prediction of EGFR mutation status in pulmonary adenocarcinoma with 3D deep learning. Cancer Med. 2019, 8, 3532–3543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Qi, S.; Pan, X.; Li, C.; Yao, Y.; Qian, W.; Guan, Y. Deep CNN Model Using CT Radiomics Feature Mapping Recognizes EGFR Gene Mutation Status of Lung Adenocarcinoma. Front. Oncol. 2021, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Moreno, S.; Bonfante, M.; Zurek, E.; Cherezov, D.; Goldgof, D.; Hall, L.; Schabath, M. A Radiogenomics Ensemble to Predict EGFR and KRAS Mutations in NSCLC. Tomography 2021, 7, 14. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Kha, Q.H.; Nguyen, V.H.; Chen, Y.C.; Cheng, S.J.; Chen, C.Y. Machine Learning-Based Radiomics Signatures for EGFR and KRAS Mutations Prediction in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2021, 22, 9254. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Fan, J.; Li, Y.; Cao, Y.; Gu, J.; Jia, X.; Wang, Y.; Shi, H. Value of CT features for predicting EGFR mutations and ALK positivity in patients with lung adenocarcinoma. Sci. Rep. 2021, 11, 5679. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xu, Z.; Liu, G.; Jiang, B.; de Bock, G.H.; Groen, H.J.; Vliegenthart, R.; Xie, X. Simultaneous identification of egfr, kras, erbb2, and tp53 mutations in patients with non-small cell lung cancer by machine learning-derived three-dimensional radiomics. Cancers 2021, 13, 1814. [Google Scholar] [CrossRef]
- Cheng, J.; Liu, J.; Jiang, M.; Yue, H.; Wu, L.; Wang, J. Prediction of Egfr Mutation Status in Lung Adenocarcinoma Using Multi-Source Feature Representations. ICASSP 2021, 1350–1354. [Google Scholar] [CrossRef]
- Cao, Y.; Xu, H.; Liao, M.; Qu, Y.; Xu, L.; Zhu, D.; Wang, B.; Tian, S. Associations between clinical data and computed tomography features in patients with epidermal growth factor receptor mutations in lung adenocarcinoma. Int. J. Clin. Oncol. 2018, 23, 249–257. [Google Scholar] [CrossRef]
- Rizzo, S.; Raimondi, S.; de Jong, E.E.; van Elmpt, W.; De Piano, F.; Petrella, F.; Bagnardi, V.; Jochems, A.; Bellomi, M.; Dingemans, A.M.; et al. Genomics of non-small cell lung cancer (NSCLC): Association between CT-based imaging features and EGFR and K-RAS mutations in 122 patients—An external validation. Eur. J. Radiol. 2019, 110, 148–155. [Google Scholar] [CrossRef]
- Xiong, J.; Li, X.; Lu, L.; Schwartz, L.H.; Fu, X.; Zhao, J.; Zhao, B. Implementation Strategy of a CNN Model Affects the Performance of CT Assessment of EGFR Mutation Status in Lung Cancer Patients. IEEE Access 2019, 7, 64583–64591. [Google Scholar] [CrossRef]
- Silva, F.; Pereira, T.; Morgado, J.; Frade, J.; Mendes, J.; Freitas, C.; Negrão, E.; De Lima, B.F.; Da Silva, M.C.; Madureira, A.J.; et al. EGFR Assessment in Lung Cancer CT Images: Analysis of Local and Holistic Regions of Interest Using Deep Unsupervised Transfer Learning. IEEE Access 2021, 9, 58667–58676. [Google Scholar] [CrossRef]
- Morgado, J.; Pereira, T.; Silva, F.; Freitas, C.; Negrão, E.; de Lima, B.F.; da Silva, M.C.; Madureira, A.J.; Ramos, I.; Hespanhol, V.; et al. Machine Learning and Feature Selection Methods for EGFR Mutation Status Prediction in Lung Cancer. Appl. Sci. 2021, 11, 3273. [Google Scholar] [CrossRef]
- Toyokawa, G.; Takada, K.; Okamoto, T.; Shimokawa, M.; Kozuma, Y.; Matsubara, T.; Haratake, N.; Takamori, S.; Akamine, T.; Katsura, M.; et al. Computed Tomography Features of Lung Adenocarcinomas With Programmed Death Ligand 1 Expression. Clin. Lung Cancer 2017, 18, e375–e383. [Google Scholar] [CrossRef]
- Wu, T.; Zhou, F.; Soodeen-Lalloo, A.K.; Yang, X.; Shen, Y.; Ding, X.; Shi, J.; Dai, J.; Shi, J. The Association Between Imaging Features of TSCT and the Expression of PD-L1 in Patients With Surgical Resection of Lung Adenocarcinoma. Clin. Lung Cancer 2019, 20, e195–e207. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, Y.L.; Feng, Y.; Yang, X.Y.; Zhang, J.; Chang, D.D.; Wu, X.; Tian, X.; Tang, K.J.; Xie, C.M.; et al. A CT-derived deep neural network predicts for programmed death ligand-1 expression status in advanced lung adenocarcinomas. Ann. Transl. Med. 2020, 8, 930. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; He, B.; Mu, W.; Liu, K.; Liu, L.; Zeng, H.; Liu, Y.; Jiang, L.; Zhou, P.; Huang, Z.; et al. Assessing PD-L1 expression in non-small cell lung cancer and predicting responses to immune checkpoint inhibitors using deep learning on computed tomography images. Theranostics 2021, 11, 2098. [Google Scholar] [CrossRef]
- Jiang, M.; Sun, D.; Guo, Y.; Guo, Y.; Xiao, J.; Wang, L.; Yao, X. Assessing PD-L1 Expression Level by Radiomic Features From PET/CT in Nonsmall Cell Lung Cancer Patients: An Initial Result. Acad. Radiol. 2020, 27, 171–179. [Google Scholar] [CrossRef]
- Jiang, Z.; Dong, Y.; Yang, L.; Lv, Y.; Dong, S.; Yuan, S.; Li, D.; Liu, L. CT-Based Hand-crafted Radiomic Signatures Can Predict PD-L1 Expression Levels in Non-small Cell Lung Cancer: A Two-Center Study. J. Digit. Imaging 2021, 34, 1073–1085. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, J.; Shen, L.; Chen, J.; Xia, L.; Ni, B.; Ge, L.; Wang, Y.; Lu, S. A multi-omics-based serial deep learning approach to predict clinical outcomes of single-agent anti-PD-1/PD-L1 immunotherapy in advanced stage non-small-cell lung cancer. Am. J. Transl. Res. 2021, 13, 743. [Google Scholar]
- Qayyum, A.; Qadir, J.; Bilal, M.; Al-Fuqaha, A. Secure and Robust Machine Learning for Healthcare: A Survey. IEEE Rev. Biomed. Eng. 2021, 14, 156–180. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Xia, B.R.; Jin, W.L.; Lou, G. Circulating tumor cells in precision oncology: Clinical applications in liquid biopsy and 3D organoid model. Cancer Cell Int. 2019, 19, 341. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.L.; Moreira, C.; Bruza, P.; Ouyang, C.; Jorge, J. Counterfactuals and Causability in Explainable Artificial Intelligence: Theory, Algorithms, and Applications. arXiv 2021, arXiv:2103.04244. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef]
- Holzinger, A. Interactive machine learning for health informatics: When do we need the human-in-the-loop? Brain Inform. 2016, 3, 119–131. [Google Scholar] [CrossRef]
- Pereira, T.; Morgado, J.; Silva, F.; Pelter, M.M.; Dias, V.R.; Barros, R.; Freitas, C.; Negrão, E.; de Lima, B.F.; da Silva, M.C.; et al. Sharing biomedical data: Strengthening ai development in healthcare. Healthcare 2021, 9, 827. [Google Scholar] [CrossRef]
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Tan et al. [78] | 2020 | LIDC-IDRI | 3D CNNs, based on FCN, DenseNet, and U-Net | TPR = 97.5 |
| Mukherjee et al. [88] | 2020 | LIDC-IDRI | Ensemble stacking | ACC = 99.5 TPR = 99.2 TNR = 98.8 FPR = 1.09 FNR = 0.85 |
| Shi et al. [79] | 2020 | LUNA16 | 3D Res-I and U-Net network | TPR = 96.4 FROC = 83.7 |
| Khehrah et al. [86] | 2020 | LIDC-IDRI | SVM | ACC = 92 TPR = 93.7 TNR = 91.2 PPV = 83.3 MCC = 83.8 |
| Kuo et al. [87] | 2020 | LIDC-IDRI Private (320 patients) | SVM | TPR = 92.1 |
| Zheng et al. [80] | 2020 | LIDC-IDRI | 3D multiscale dense CNNs | TPR = 94.2 (1.0 FP/scan), 96.0 (2.0 FPs/image) |
| Paing et al. [89] | 2020 | LIDC-IDRI | Optimized random forest | ACC = 93.1 TPR = 94.9 TNR = 91.4 |
| Liu et al. [100] | 2020 | LIDC-IDRI | CNN algorithm: You Only Look Once v3 | TPR = 87.3 |
| Harsono et al. [97] | 2020 | LIDC-IDRI Private (546 patients) | I3DR-Net | mAP = 49.6 (LIDC), 22.9 (private) AUC = 81.8 (LIDC), 70.4 (private) |
| Xu et al. [81] | 2020 | LUNA16 | 3D CNN networks: V-Net and multi-level contextual 3D CNNs | TPR = 93.1 (1.64 FP/scan) CPM = 75.7 |
| Drokin and Ericheva [96] | 2020 | LIDC-IDRI | Algorithm for sampling points from a point cloud | FROC = 85.9 |
| El-Regaily et al. [90] | 2020 | LIDC-IDRI | Multi-view CNN | ACC = 91.0 TPR = 96.0 TNR = 87.3 F-score = 78.7 |
| Ye et al. [82] | 2020 | LUNA16 | Three modified V-Nets with multilevel receptive fields | ACC = 66.7 TPR = 81.1 PPV = 78.1 F-score = 78.7 |
| Baker and Ghadi [93] | 2020 | LIDC-IDRI | SVM | NRR = 94.5 FPR = 7 cluster/image |
| Halder et al. [94] | 2020 | LIDC-IDRI | SVM | ACC = 88.2 TPR = 86.9 TNR = 86.9 |
| Jain et al. [83] | 2020 | LUNA16 | SumNet | ACC = 94.1 TNR = 94.0 DSC = 93.0 |
| Mahersia et al. [95] | 2020 | LIDC-IDRI | SVM, Bayesian back-propagation neuronal classifier and neuro-fuzzy classifier | NRR = 97.9 (neuronal classifier), 97.3 (SVM), 94.2 (neuro-fuzzy classifier) |
| Mittapalli and Thanikaiselvan [91] | 2021 | LUNA16 | Multiscale CNN with Compound Fusions | CPM = 94.8 |
| Vipparla et al. [92] | 2021 | LUNA16 | 3D Attention-based CNN architectures: MP-ACNN1, MP-ACNN2 and MP-ACNN3 | CPM = 93.1 |
| Luo et al. [84] | 2021 | LUNA16 | SCPM-Net | TPR = 92.2 (1 FPs/image), 93.9 (2 FPs/image), 96.4 (8FPs/image) |
| Bhaskar and Ganashree [85] | 2021 | DSB-2017 | Gaussian mixture convolutional auto encoder + 3D deep CNN | ACC = 74.0 |
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Sharma et al. [101] | 2020 | SPIE-AAPM Lung CT Challenge | SVM + k-NN | ACC = 93.9 TPR = 94.5 GM = 94.2 |
| Xiao et al. [104] | 2020 | LUNA16 | 3D-UNet + Res2Net Neural Network | TPR = 99.1 DSC = 95.3 |
| Singadkar et al. [107] | 2020 | LIDC-IDRI | Deep deconvolutional residual network | DSC = 95.0 JI = 88.7 |
| Kumar and Raman [105] | 2020 | LUNA16 | V-Net (3D CNN) | DSC = 96.1 |
| Rocha et al. [106] | 2020 | LIDC-IDRI | Sliding Band Filter + U-Net + SegU-Net | DSC = 66.3 (SBF), 83.0 (U-Net), 82.3 (SegU-Net) |
| Hancock and Magnan [102] | 2021 | LIDC-IDRI | Level set machine learning method | DSC = 83.6 JI = 71.8 |
| Savic et al. [103] | 2021 | LIDC-IDRI Private—phantom (108 patients) | Algorithm based on the fast marching method | DSC = 93.3 (solid round nodules), 90.1 (solid irregular nodules), 79.9 (non-solid nodules), 61.4 (cavity nodules) |
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Wang et al. [109] | 2020 | Private (1478 patients) | Adaptive-boost deep learning strategy with multiple 3D CNN-based weak classifiers | ACC = 73.4 TPR = 70.5 TNR = 76.2 PPV = 83.8 AUC = 82.0 F-score = 71.6 |
| Xiao et al. [120] | 2020 | LIDC-IDRI | ResNet-18 + Denoising autoencoder classifier + handcrafted features | ACC = 93.1 TPR = 81.7 PPV = 83.8 AUC = 82.0 |
| Wang et al. [127] | 2020 | LUNGx | ConvNet | ACC = 90.4 TPR = 88.7 TNR = 92.4 AUC = 94.8 |
| Lin et al. [110] | 2020 | LUNA16 | GVGG + ResCon network | TPR = 92.5 TNR = 96.8 PPV = 93.6 F-score = 93.0 |
| Onishi et al. [134] | 2020 | Private (60 patients) | M-Scale 3D CNN | TPR = 90.9 TNR = 74.1 |
| Zhao et al. [126] | 2020 | LIDC-IDRI | Multi-stream multi-task network | ACC = 93.9 TPR = 92.6 TNR = 96.2 AUC = 97.9 |
| Zia et al. [132] | 2020 | LIDC-IDRI | Multi-deep model | ACC = 90.7 TPR = 90.7 TNR = 90.8 |
| Jiang et al. [121] | 2020 | LUNA16 | Ensemble of 3D Dual Path Networks | ACC = 90.2 TPR = 92.0 FPR = 11.1 F-score = 90.4 |
| Bao et al. [131] | 2020 | LIDC-IDRI | Global-local residual network | ACC = 90.4 TPR = 90.1 PPV = 89.9 AUC = 96.1 |
| Shah et al. [111] | 2020 | LUNA16 | NoduleNet (transfer learning from VGG16 and VGG19 models) | ACC = 95.0 TPR = 84.0 TNR = 97.0 |
| Tong et al. [112] | 2020 | LIDC-IDRI | 3D-ResNet + SVM with RBF and polynomial kernels | ACC = 90.6 TPR = 87.5 TNR = 94.1 |
| Xu et al. [128] | 2020 | LIDC-IDRI | Multi-scale cost-sensitive methods | ACC = 92.6 TPR = 85.6 TNR = 95.9 PPV = 90.4 AUC = 94.0 F-score = 87.9 |
| Huang et al. [113] | 2020 | LIDC-IDRI | Deep transfer convolutional neural network + Extreme learning machine | ACC = 94.6 TPR = 93.7 TNR = 95.1 AUC = 94.9 |
| Naik et al. [122] | 2020 | LUNA16 | FractalNet + CNN | ACC = 94.1 TPR = 97.5 TNR = 86.8 AUC = 98.0 |
| Zhang et al. [118] | 2020 | LUNA16 | 3D squeeze-and-excitation network and aggregated residual transformations | ACC = 91.7 AUC = 95.6 |
| Liu et al. [123] | 2020 | LIDC-IDRI | Multi-model ensemble learning architecture based on 3D CNNs: VggNet, ResNet, and InceptionNet | ACC = 90.6 TPR = 83.7 TNR = 93.9 AUC = 93.0 |
| Afshar et al. [129] | 2020 | LIDC-IDRI | 3D Multi-scale Capsule Network | ACC = 93.1 TPR = 94.9 TNR = 90.0 AUC = 96.4 |
| Lyu et al. [114] | 2020 | LIDC-IDRI | Multi-level cross ResNet | ACC = 92.2 TPR = 92.1 TNR = 91.5 AUC = 97.1 |
| Wu et al. [115] | 2020 | LIDC-IDRI | Deep residual network (ResNet + residual learning + migration learning) | ACC = 98.2 TPR = 97.7 TNR = 98.3 PPV = 98.5 F-score = 98.1 FPR = 1.60 |
| Lin and Li [116] | 2020 | LIDC-IDRI | Taguchi-based AlexNet CNN | ACC = 99.6 |
| Kuang et al. [135] | 2020 | LIDC-IDRI | Combination of a multi-discriminator generative adversarial network and an encoder | ACC = 95.3 TPR = 94.1 TNR = 90.8 AUC = 94.3 |
| Lima et al. [137] | 2020 | LIDC-IDRI | SVM with Gaussian kernel + Relief + Evolutionary Genetic Algorithm | AUC = 85.6 |
| Veasey et al. [133] | 2020 | NLST | Recurrent neural network with 2D CNN | PPV = 55.9 (t0), 66.9 (t1) AUC = 80.6 (t0), 83.5 (t1) |
| Bansal et al. [117] | 2020 | LUNA16 | Deep3DSCan | TPR = 87.1 TNR = 89.7 AUC = 88.3 F-score = 88.5 |
| Zhai et al. [124] | 2020 | LUNA16 LIDC-IDRI | Multi-task learning CNN | TPR = 84.0 (LUNA16), 95.6 (LIDC-IDRI) TNR = 96.8 (LUNA16), 88.9 (LIDC-IDRI) AUC = 97.3 (LUNA16), 95.6 (LIDC-IDRI) |
| Paul et al. [125] | 2020 | NLST | Ensemble of CNNs | ACC = 90.3 AUC = 96.0 TPR = 73.0 FNR = 27.0 |
| Ali et al. [119] | 2020 | LIDC-IDRI LUNGx | Transferable texture CNN | ACC = 96.6 (LIDC-IDRI), 90.9 (LUNGx) TPR = 96.1 (LIDC-IDRI), 91.4 (LUNGx) TNR = 97.4 (LIDC-IDRI), 90.5 (LUNGx) AUC = 99.1 (LIDC-IDRI), 94.1 (LUNGx) |
| Silva et al. [136] | 2020 | LIDC-IDRI | Transfer learning (convolutional autoencoder) | AUC = 93.6 PPV = 79.4 TPR = 84.8 F-score = 81.7 |
| Xia et al. [130] | 2021 | LIDC-IDRI | Gradient boosting machine algorithm | ACC = 91.9 TPR = 91.3 F-score = 91.0 FPR = 8.00 |
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Lai and Wei [148] | 2014 | Private (10 patients) | Filtering process + morphological operations (threshold, region filling, closing) | TPR = 97.0 TNR = 99.0 AAE = 1.58 |
| Li et al. [147] | 2015 | Private (15 patients) | Edge-based recursive geometric active contour (GAC) model | OV = 98.0 |
| Shi et al. [149] | 2016 | Private (23 patients) | Histogram thresholding + region growing and random walk | OR = 1.87 UR = 2.36 ABD = 0.620 mm |
| Zhang et al. [150] | 2017 | LIDC-IDRI | Region- and edge-based GAC (REGAC) method | DSC = 97.7 HD-95 = 2.50 mm |
| Rebouças Filho et al. [151] | 2017 | Private (40 patients) | 3D ACACM | F-score = 99.2 (ACACM), 97.6 (RG), 97.4 (OsiriX), 97.2 (LSCPM) |
| Oliveira et al. [153] | 2018 | VISCERAL Anatomy3 | Multi-atlas alignment + label fusion (voting and statistical selection) | DSC = 97.4 (LL), 97.9 (RL) HD-95 = 4.65 mm (LL), 2.81 mm (RL) |
| Chen et al. [152] | 2021 | LOLA11 Private (65 patients) | Random walker | (Private) DSC = 98.6 (LL), 98.5 (RL) (LOLA11) DSC = 97.4 |
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Dong et al. [155] | 2019 | LCTSC | U-net generator with a FCN discriminator | DSC = 97.0 |
| Feng et al. [156] | 2019 | LCTSC | Two-stage segmentation process with 3D U-net | DSC = 97.2 (RL), 97.9 (LL) |
| Park et al. [157] | 2019 | LCTSC Private (30 patients) | U-net | DSC = 98.8 JSC = 97.7 MSD = 0.270 mm HSD = 25.5 mm |
| Hofmanninger et al. [158] | 2020 | LCTSC, LTRC, VISCERAL, VESSEL12 Private (5300 patients) | U-net, ResUNet, Dilated residual network-D-22, DeepLab v3+ | (merged dataset) DSC = 98.0 HD95 = 3.14 mm MSD = 0.620 mm |
| Yoo et al. [159] | 2020 | HUG-ILD Private (203 patients) | 2D and 3D U-net | (Private - 2D; 3D) DSC = 99.6; 99.4 TPR = 99.5; 99.1 PPV = 99.6; 99.7 HD = 17.7 px; 18.7 px (HUG-ILD - 2D; 3D) DSC = 98.4; 95.3 TPR = 98.7; 98.0 PPV = 98.1; 92.8 HD = 7.66 px; 15.6 px |
| Khanna et al. [167] | 2020 | LUNA16 VESSEL12 2HUG-ILD | ResUNet + false positive removal algorithm | (LUNA16) DSC = 96.6 JI = 93.4 TPR = 97.5 (VESSEL12) DSC = 98.3 JI = 97.9 TPR = 98.8 (HUG-ILD) DSC = 98.1 JI = 96.3 TPR = 98.3 |
| Shi et al. [160] | 2020 | StructSeg 2019 | TA-Net | DSC = 96.8 (LL), 97.1 (RL) HD = 0.188 mm (LL), 0.171 mm (RL) |
| Nemoto et al. [161] | 2020 | NSCLC-Radiomics | 2D and 3D U-net | DSC = 99.0 (2D/3D U-net) |
| Zhang et al. [162] | 2020 | Lung dataset (Kaggle “Finding and Measuring Lungs in CT Data” competition) | Dense-Inception U-net (DIU-net) | DSC = 98.6 JI = 98.7 ACC = 99.4 TPR = 98.5 TNR = 99.8 F-score = 98.5 AUC = 99.0 |
| Vu et al. [163] | 2020 | Private (168 patients) | U-net with pre-trained VGG16 | DSC = 97.0 (RL and LL) HD-95 = 5.10 mm (RL), 4.09 mm (LL) |
| Liu et al. [171] | 2020 | HUG-ILD | Random forest fusion classification of deep, texture and intensity features | DSC = 96.4 JI = 91.1 OR = 5.04 UR = 4.76 |
| Hu et al. [172] | 2020 | Private (39 patients) | Mask R-CNN + supervised and unsupervised classifiers | DSC = 97.3 ACC = 97.7 TPR = 96.6 TNR = 97.1 |
| Han et al. [173] | 2020 | Private | Xception + VGG with SVM-RBF Detectron2 + contour fine-tuning | DSC = 97.0 ACC = 99.0 TPR = 96.5 TNR = 99.4 |
| Xu et al. [170] | 2021 | Private (217 patients) COVID-19-CT-Seg HUG-ILD VESSEL12 | Boundary-Guided Network (BG-Net) | DSC = 98.6 (Private), 96.5 (StructSeg), 98.9 (HUG-ILD), 99.5 (VESSEL12) HD = 2.77 mm (Private), 1.39 mm (StructSeg), 0.665 mm (HUD-ILD), 1.40 mm (VESSEL12) |
| Jalali et al. [166] | 2021 | LIDC-IDRI | ResBCDU-Net | DSC = 97.1 |
| Wang et al. [164] | 2021 | Lung dataset (Kaggle “Finding and Measuring Lungs in CT Data” competition) | HDA-ResUNet | DSC = 97.9 JI = 96.0 ACC = 99.3 |
| Tan et al. [168] | 2021 | LIDC-IDRI QIN lung CT dataset | LGAN | (LIDC-IDRI) IOU = 92.3 HD = 3.38 mm (QIN) IOU = 93.8 HD = 2.68 mm |
| Pawar and Talbar [169] | 2021 | HUG-ILD | LungSeg-Net | DSC = 96.3 (Fibrosis), 96.5 (Ground glass), 91.4 (Reticulation), 97.6 (Consolidation), 97.8 (Emphysema), 99.0 (Nodules) JI = 93.7 (Fibrosis), 93.9 (Ground glass), 86.9 (Reticulation), 95.3 (Consolidation), 96.2 (Emphysema), 98.0 (Nodules) |
| Cao et al. [165] | 2021 | StructSeg 2019 | C-SE-ResUNet | DCS = 97.0 (LL) 96.6 (RL) |
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Zou et al. [177] | 2017 | Private (209 patients) | Multivariable analyses | EGFR: AUC = 73.7 |
| Cheng et al. [176] | 2017 | Private (2146 patients) | Weighted mean difference, inverse variance | EGFR: OR = 49.0 |
| Li et al. [179] | 2018 | Private (1010 patients) | Random forest/CNNs | EGFR: AUC = 83.4 |
| Koyasu et al. [178] | 2019 | NSCLC-radiogenomics | XGBoost/random forest | EGFR: AUC = 65.9 |
| Wang et al. [180] | 2019 | Private (844 patients) | CNNs | EGFR: AUC = 85.0 |
| Zhao et al. [181] | 2019 | TCIA and private (879 patients) | 3D DenseNet | EGFR: AUC = 75.8 |
| Moreno et al. [183] | 2021 | NSCLC-radiogenomics | SCAV with ML/CNN | EGFR: AUC = 82.0 (CNN) KRAS: AUC = 73.9 (CNN) |
| Zhang et al. [182] | 2021 | Private (914 patients) | Machine learning (SVM/RF/MLP) Deep learning (SE-CNN/CNN/1D-CNN/AlexNet/Fine-tuned VG16/Fine-tuned VGG19) | EGFR: AUC = 91.0 (SE-CNN) AUC = 83.6 (SVM) |
| Le et al. [184] | 2021 | NSCLC-radiogenomics | LR / KNN / RF / XGBoost | EGFR: ACC = 77.8 KRAS: ACC = 83.3 |
| Cheng et al. [187] | 2021 | Private (670 patients) | Pre-trained 3D DenseNet | EGFR: AUC = 76.0 ACC = 72.5 F-score = 71.3 |
| Zhang et al. [186] | 2021 | Private (134 patients) | Logistic regression | EGFR: AUC = 78.0 KRAS: AUC = 81.0 ERBB2: AUC = 87.0 TP53: AUC = 84.0 |
| Han et al. [185] | 2021 | Private (827 patients) | Logistic Regression | EGFR: AUC = 75.8 ALK: AUC = 73.9 |
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Gevaert et al. [71] | 2017 | Private (186 patients) | Decision Tree | EGFR: AUC = 89.0 |
| Cao et al. [188] | 2018 | Private (156 patients) | Principal component analysis | EGFR: TPR = 72.3 TNR = 78.5 |
| Rizzo et al. [189] | 2019 | Private (122 patients) | Univariate analysis | EGFR: AUC = 82.0 KRAS: AUC = 67.0 |
| Pinheiro et al. [50] | 2019 | NSCLC-radiogenomics | Gradient tree boosting | EGFR: AUC = 74.6 |
| Xiong et al. [190] | 2019 | Private (1010 patients) | ResNet 101 | EGFR: AUC = 83.8 |
| Silva et al. [191] | 2021 | LIDC-IDRI NSCLC-radiogenomics | Convolutional autoencoder | EGFR: AUC = 68.0 |
| Morgado et al. [192] | 2021 | NSCLC-radiogenomics | LR, Elastic Net, Linear SVM, RBG SVM, RF, and XGBoost | EGFR: AUC = 73.7 (Linear SVM) AUC = 73.3 (Elastic Net) AUC = 72.5 (LR) |
| Authors | Year | Dataset | Methods | Performance Results (%) |
|---|---|---|---|---|
| Toyokawa et al. [193] | 2017 | Private (394 patients) | Fisher’s exact test Univariate/multivariate LR (CT features) | PD-L1+ statistical association: (p < 0.01)—convergence, notching, spiculation, cavitation |
| Wu et al. [194] | 2019 | Private (350 patients) | Univariate/multivariate LR Fisher’s exact test Mann–Whitney U test | AUC = 78.3 TPR = 81.1 TNR = 64.1 |
| Zhu et al. [195] | 2020 | Private (127 patients) | Univariate/multivariate LR 3D DenseNet | AUC = 78.0 ACC = 77.8 TPR = 77.8 TNR = 77.4 |
| Jiang et al. [197] | 2020 | Private (399 patients) | Random forest Logistic regression | AUC = 97.0 (≥1%) AUC = 80.0 (≥50%) |
| Tian et al. [196] | 2021 | Private (939 patients) | Fully connected classifier | AUC = 76.0 |
| Yang et al. [199] | 2021 | Private (200 patients) | Simple temporal attention (SimTA) module | AUC = 77.0 (SimTA60) AUC = 80.0 (SimTA90) AUC = 69.0 (RNN) AUC = 64.0 (Radiomics) |
| Jiang et al. [198] | 2021 | Private (125 patients) | Random forest Decision tree Logistic regression AdaBoost Support vector machine | (Internal validation) AUC = 96.0 TNR = 80.0 TPR = 98.5 (External validation) AUC = 85.0 TNR = 63.6 TPR = 91.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, F.; Pereira, T.; Neves, I.; Morgado, J.; Freitas, C.; Malafaia, M.; Sousa, J.; Fonseca, J.; Negrão, E.; Flor de Lima, B.; et al. Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges. J. Pers. Med. 2022, 12, 480. https://doi.org/10.3390/jpm12030480
Silva F, Pereira T, Neves I, Morgado J, Freitas C, Malafaia M, Sousa J, Fonseca J, Negrão E, Flor de Lima B, et al. Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges. Journal of Personalized Medicine. 2022; 12(3):480. https://doi.org/10.3390/jpm12030480
Chicago/Turabian StyleSilva, Francisco, Tania Pereira, Inês Neves, Joana Morgado, Cláudia Freitas, Mafalda Malafaia, Joana Sousa, João Fonseca, Eduardo Negrão, Beatriz Flor de Lima, and et al. 2022. "Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges" Journal of Personalized Medicine 12, no. 3: 480. https://doi.org/10.3390/jpm12030480
APA StyleSilva, F., Pereira, T., Neves, I., Morgado, J., Freitas, C., Malafaia, M., Sousa, J., Fonseca, J., Negrão, E., Flor de Lima, B., Correia da Silva, M., Madureira, A. J., Ramos, I., Costa, J. L., Hespanhol, V., Cunha, A., & Oliveira, H. P. (2022). Towards Machine Learning-Aided Lung Cancer Clinical Routines: Approaches and Open Challenges. Journal of Personalized Medicine, 12(3), 480. https://doi.org/10.3390/jpm12030480