Identification of Potential microRNA Panels for Male Non-Small Cell Lung Cancer Identification Using Microarray Datasets and Bioinformatics Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. NSCLC TCGA Data Analysis
2.2. Patient Cohort
2.3. Sample Processing and Microarray Evaluation
2.4. Functional Analysis and Target Gene Identification
2.5. miRNA Evaluation of Expression Levels in Tissue and Plasma Samples
2.6. EGFR, IGF-IR, and TGFβ1 Quantification in Serum Samples
3. Results
3.1. Clinical and Pathological Characteristics of the Cohorts
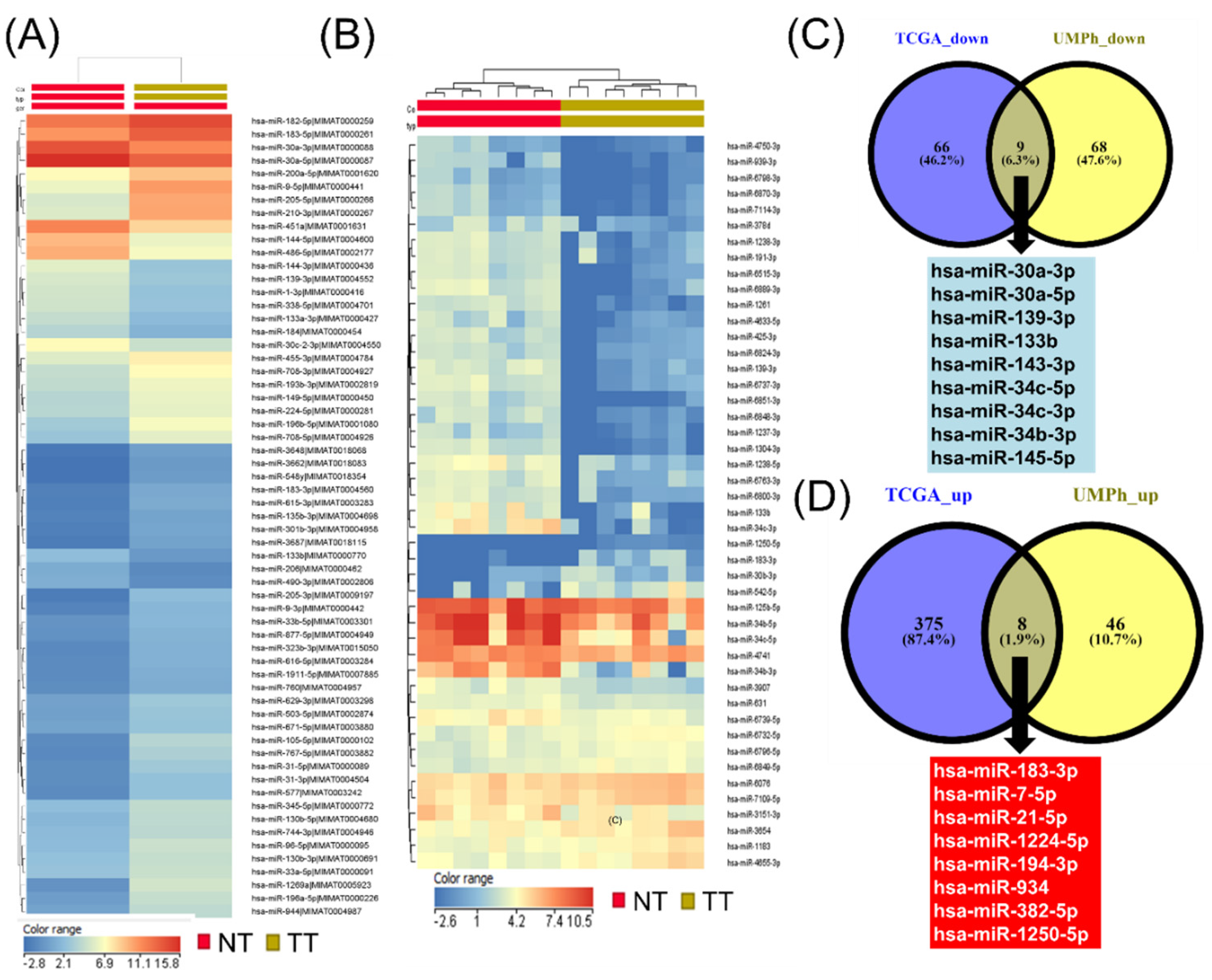
3.2. Evaluation of Tissue miRNAs’ Expression Levels in NSCLC Patients in TCGA and UMPh Cohort
3.3. Functional Analysis and Target Genes Identification
3.4. IPA miRNA–Gene Regulatory Network on Male NSCLC
3.5. RT-PCR Tissue Validation
3.6. RT-PCR Plasma Validation
3.7. EGFR, IGF-IR, and TGFβ1 Quantification in Serum Male NSCLC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://gco.iarc.fr/today/data/factsheets/populations/908-europe-fact-sheets.pdf (accessed on 23 October 2022).
- Inamura, K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front. Oncol. 2017, 7, 193. [Google Scholar] [CrossRef] [Green Version]
- Capelletto, E.; Novello, S. Emerging new agents for the management of patients with non-small cell lung cancer. Drugs 2012, 72 (Suppl. 1), 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.M.; Weinberg, R.A.; Lees, J.A.; Guen, V.J. Emerging Mechanisms by which EMT Programs Control Stemness. Trends Cancer 2020, 6, 775–780. [Google Scholar] [CrossRef]
- Pinto, J.A.; Vallejos, C.S.; Raez, L.E.; Mas, L.A.; Ruiz, R.; Torres-Roman, J.S.; Rolfo, C. Gender and outcomes in non-small cell lung cancer: An old prognostic variable comes back for targeted therapy and immunotherapy? ESMO Open 2018, 3, e000344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boscaino, V.; Fiannaca, A.; La Paglia, L.; La Rosa, M.; Rizzo, R.; Urso, A. MiRNA therapeutics based on logic circuits of biological pathways. BMC Bioinform. 2019, 20 (Suppl. 9), 344. [Google Scholar] [CrossRef]
- Okamoto, A.; Sehouli, J.; Yanaihara, N.; Hirata, Y.; Braicu, I.; Kim, B.G.; Urashima, M. Somatic copy number alterations associated with Japanese or endometriosis in ovarian clear cell adenocarcinoma. PLoS ONE 2015, 10, e0116977. [Google Scholar] [CrossRef]
- Pop-Bica, C.; Gulei, D.; Cojocneanu-Petric, R.; Braicu, C.; Petrut, B.; Berindan-Neagoe, I. Understanding the Role of Non-Coding RNAs in Bladder Cancer: From Dark Matter to Valuable Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 1514. [Google Scholar] [CrossRef]
- Kato, M. Target RNA-directed microRNA degradation; which controls which? Noncoding RNA Investig. 2018, 2. [Google Scholar] [CrossRef] [PubMed]
- Petrek, H.; Yu, A.M. MicroRNAs in non-small cell lung cancer: Gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacol. Res. Perspect. 2019, 7, e00528. [Google Scholar] [CrossRef]
- Braicu, C.; Zimta, A.A.; Harangus, A.; Iurca, I.; Irimie, A.; Coza, O.; Berindan-Neagoe, I. The Function of Non-Coding RNAs in Lung Cancer Tumorigenesis. Cancers 2019, 11, 605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonea, L.; Buse, M.; Gulei, D.; Onaciu, A.; Simon, I.; Braicu, C.; Berindan-Neagoe, I. Decoding the Emerging Patterns Exhibited in Non-coding RNAs Characteristic of Lung Cancer with Regard to their Clinical Significance. Curr. Genom. 2018, 19, 258–278. [Google Scholar] [CrossRef] [PubMed]
- Guerau-de-Arellano, M.; Alder, H.; Ozer, H.G.; Lovett-Racke, A.; Racke, M.K. miRNA profiling for biomarker discovery in multiple sclerosis: From microarray to deep sequencing. J. Neuroimmunol. 2012, 248, 32–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braicu, C.; Buiga, R.; Cojocneanu, R.; Buse, M.; Raduly, L.; Pop, L.A.; Chira, S.; Budisan, L.; Jurj, A.; Ciocan, C.; et al. Connecting the dots between different networks: miRNAs associated with bladder cancer risk and progression. J. Exp. Clin. Cancer Res. 2019, 38, 433. [Google Scholar] [CrossRef]
- Cojocneanu, R.; Braicu, C.; Raduly, L.; Jurj, A.; Zanoaga, O.; Magdo, L.; Irimie, A.; Muresan, M.-S.; Ionescu, C.; Grigorescu, M.; et al. Plasma and Tissue Specific miRNA Expression Pattern and Functional Analysis Associated to Colorectal Cancer Patients. Cancers 2020, 12, 843. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, C.; Das, S. Profiling cell-free and circulating miRNA: A clinical diagnostic tool for different cancers. Tumour Biol. 2016, 37, 5705–5714. [Google Scholar] [CrossRef]
- Pirlog, R.; Chiroi, P.; Rusu, I.; Jurj, A.M.; Budisan, L.; Pop-Bica, C.; Braicu, C.; Crisan, D.; Sabourin, J.-C.; Berindan-Neagoe, I. Cellular and Molecular Profiling of Tumor Microenvironment and Early-Stage Lung Cancer. Int. J. Mol. Sci. 2022, 23, 5346. [Google Scholar] [CrossRef]
- Peng, X.X.; Yu, R.; Wu, X.; Wu, S.Y.; Pi, C.; Chen, Z.H.; Zhou, Q. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J. Immunother. Cancer 2020, 8, e000376. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Xu, P.; Mi, Y.; Wang, W.; Pan, X.; Wu, X.; He, Q.; Liu, H.; Tang, W.; An, H. Plasma MiRNA alterations between NSCLC patients harboring Del19 and L858R EGFR mutations. Oncotarget 2016, 7, 54965–54972. [Google Scholar] [CrossRef] [Green Version]
- Gasparini, P.; Cascione, L.; Landi, L.; Carasi, S.; Lovat, F.; Tibaldi, C.; Croce, C.M. microRNA classifiers are powerful diagnostic/prognostic tools in ALK-, EGFR-, and KRAS-driven lung cancers. Proc. Natl. Acad. Sci. USA 2015, 112, 14924–14929. [Google Scholar] [CrossRef]
- Liang, G.; Meng, W.; Huang, X.; Zhu, W.; Yin, C.; Wang, C.; Fassan, M.; Yu, Y.; Kudo, M.; Xiao, S.; et al. miR-196b-5p-mediated downregulation of TSPAN12 and GATA6 promotes tumor progression in non-small cell lung cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 4347–4357. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell. Mol. Biol. Lett. 2019, 24, 60. [Google Scholar] [CrossRef] [PubMed]
- Pirlog, R.; Cismaru, A.; Nutu, A.; Berindan-Neagoe, I. Field Cancerization in NSCLC: A New Perspective on MicroRNAs in Macrophage Polarization. Int. J. Mol. Sci. 2021, 22, 746. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhang, Q.; Ma, X.; Wang, J.; Liang, T. miRNA and mRNA expression analysis reveals potential sex-biased miRNA expression. Sci. Rep. 2017, 7, 39812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, C.; Yang, W.; Shi, J.; Zhou, Y.; Yang, J.; Cui, Q.; Zhou, Y. Identification and Analysis of Human Sex-biased MicroRNAs. Genom. Proteom. Bioinform. 2018, 16, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhou, Y.; Zeng, P.; Xu, G.; Wang, G.; Cui, Q. Identification and analysis of the human sex-biased genes. Brief. Bioinform. 2018, 19, 188–198. [Google Scholar] [CrossRef]
- Pérez-Díez, I.; Hidalgo, M.; Malmierca-Merlo, P.; Andreu, Z.; Romera-Giner, S.; Farràs, R.; de la Iglesia-Vayá, M.; Provencio, M.; Romero, A.; García-García, F. Functional Signatures in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis of Sex-Based Differences in Transcriptomic Studies. Cancers 2021, 13, 143. [Google Scholar] [CrossRef]
- Fuentes, N.; Roy, A.; Mishra, V.; Cabello, N.; Silveyra, P. Sex-specific microRNA expression networks in an acute mouse model of ozone-induced lung inflammation. Biol. Sex Differ. 2018, 9, 18. [Google Scholar] [CrossRef] [Green Version]
- Li, E.W.; Bai, Y. Computational Identification of Sex-Biased Biomarker MicroRNAs and Genes Associated with Immune Infiltration in Breast Cancer. Genes 2021, 12, 570. [Google Scholar] [CrossRef]
- Vlachos, I.; Kostoulas, N.; Vergoulis, T.; Georgakilas, G.; Reczko, M.; Maragkakis, M.; Paraskevopoulou, M.D.; Prionidis, K.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA miRPath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012, 40, W498–W504. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/next-generation-sequencing/informatics-and-data/interpretation-content-databases/ingenuity-pathway-analysis/ (accessed on 23 October 2022).
- Mazzara, S.; Rossi, R.L.; Grifantini, R.; Donizetti, S.; Abrignani, S.; Bombaci, M. CombiROC: An interactive web tool for selecting accurate marker combinations of omics data. Sci. Rep. 2017, 7, 45477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peter, M.E. Targeting of mRNAs by multiple miRNAs: The next step. Oncogene 2010, 29, 2161–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garofalo, M.; Jeon, Y.J.; Nuovo, G.J.; Middleton, J.; Secchiero, P.; Joshi, P.; Alder, H.; Nazaryan, N.; Di Leva, G.; Romano, G.; et al. MiR-34a/c-Dependent PDGFR-α/β Downregulation Inhibits Tumorigenesis and Enhances TRAIL-Induced Apoptosis in Lung Cancer. PLoS One 2013, 8, e67581. [Google Scholar] [CrossRef] [PubMed]
- Stahlhut, C.; Slack, F.J. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle 2015, 14, 2171–2180. [Google Scholar] [CrossRef] [Green Version]
- Tafsiri, E.; Darbouy, M.; Shadmehr, M.B.; Zagryazhskaya, A.; Alizadeh, J.; Karimipoor, M. Expression of miRNAs in non-small-cell lung carcinomas and their association with clinicopathological features. Tumour. Biol. 2015, 36, 1603–1612. [Google Scholar] [CrossRef] [PubMed]
- Veena, M.S.; Raychaudhuri, S.; Basak, S.K.; Venkatesan, N.; Kumar, P.; Biswas, R.; Chakrabarti, R.; Lu, J.; Su, T.; Gallagher-Jones, M.; et al. Dysregulation of hsa-miR-34a and hsa-miR-449a leads to overexpression of PACS-1 and loss of DNA damage response (DDR) in cervical cancer. J. Biol. Chem. 2020, 295, 17169–17186. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, E.J.; Lee, S.; Tan, X.; Liu, X.; Park, S.; Kang, K.; Yoon, J.-S.; Ko, Y.H.; Kurie, J.M.; et al. MiR-34a and miR-34b/c have distinct effects on the suppression of lung adenocarcinomas. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Cheng, J.; Chen, B.; Liu, Q.; Xu, D.; Zhang, Y. Circulating microRNA-34 family low expression correlates with poor prognosis in patients with non-small cell lung cancer. J. Thorac. Dis. 2017, 9, 3735–3746. [Google Scholar] [CrossRef] [Green Version]
- Tu, L.; Long, X.; Song, W.; Lv, Z.; Zeng, H.; Wang, T.; Liu, X.; Dong, J.; Xu, P. MiR-34c acts as a tumor suppressor in non-small cell lung cancer by inducing endoplasmic reticulum stress through targeting HMGB1. OncoTargets Ther. 2019, 12, 5729–5739. [Google Scholar] [CrossRef]
- Navarro, F.; Lieberman, J. miR-34 and p53: New Insights into a Complex Functional Relationship. PloS ONE 2015, 10, e0132767. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Fu, J.; Chen, H.; Cheng, J.; Fu, J. Roles of MicroRNA-34a in Epithelial to Mesenchymal Transition, Competing Endogenous RNA Sponging and Its Therapeutic Potential. Int. J. Mol. Sci. 2019, 20, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Kudo, M.; Huang, X.; Sui, H.; Tian, H.; Croce, C.M.; Cui, R. Frontiers of MicroRNA Signature in Non-small Cell Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 643942. [Google Scholar] [CrossRef]
- Wang, H.; Ma, Z.; Liu, X.; Zhang, C.; Hu, Y.; Ding, L.; Qi, P.; Wang, J.; Lu, S.; Li, Y. MiR-183-5p is required for non-small cell lung cancer progression by repressing, P.T.E.N. Biomed. Pharmacother. 2019, 111, 1103–1111. [Google Scholar] [CrossRef]
- Meng, F.; Zhang, L. miR-183-5p functions as a tumor suppressor in lung cancer through PIK3CA inhibition. Exp. Cell Res. 2019, 374, 315–322. [Google Scholar] [CrossRef]
- Ciardiello, F.; De Vita, F.; Orditura, M.; Tortora, G. The role of EGFR inhibitors in nonsmall cell lung cancer. Curr. Opin. Oncol. 2004, 16, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Bal, A.; Aggarwal, A.N.; Das, A.; Behera, D. Clinical outcomes in non-small-cell lung cancer in relation to expression of predictive and prognostic biomarkers. Future Oncol. 2010, 6, 741–767. [Google Scholar] [CrossRef]
- Barker, A.J.; Gibson, K.H.; Grundy, W.; Godfrey, A.A.; Barlow, J.J.; Healy, M.P.; Woodburn, J.R.; Ashton, S.E.; Curry, B.J.; Scarlett, L.; et al. Studies leading to the identification of ZD1839 (IRESSA): An orally active, selective epidermal growth factor receptor tyrosine kinase inhibitor targeted to the treatment of cancer. Bioorg. Med. Chem. Lett. 2001, 11, 1911–1914. [Google Scholar] [CrossRef]
- Iams, W.T.; Lovly, C.M. Molecular Pathways: Clinical Applications and Future Direction of Insulin-like Growth Factor-1 Receptor Pathway Blockade. Clin. Cancer Res. 2015, 21, 4270–4277. [Google Scholar] [CrossRef] [Green Version]
- Morgillo, F.; Kim, W.Y.; Kim, E.S.; Ciardiello, F.; Hong, W.K.; Lee, H.Y. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin. Cancer Res. 2007, 13, 2795–2803. [Google Scholar] [CrossRef]
- Fidler, M.J.; Shersher, D.D.; Borgia, J.A.; Bonomi, P. Targeting the insulin-like growth factor receptor pathway in lung cancer: Problems and pitfalls. Ther. Adv. Med. Oncol. 2012, 4, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef] [Green Version]
- Zou, M.; Wang, F.; Gao, R.; Wu, J.; Ou, Y.; Chen, X.; Chen, Y. Autophagy inhibition of hsa-miR-19a-3p/19b-3p by targeting TGF-beta R II during TGF-beta1-induced fibrogenesis in human cardiac fibroblasts. Sci. Rep. 2016, 6, 24747. [Google Scholar] [CrossRef]
- Bhowmick, N.A.; Ghiassi, M.; Bakin, A.; Aakre, M.; Lundquist, C.A.; Engel, M.E.; Moses, H.L. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 2001, 12, 27–36. [Google Scholar] [CrossRef]
- Seike, M.; Goto, A.; Okano, T.; Bowman, E.D.; Schetter, A.J.; Horikawa, I.; Mathe, E.A.; Jen, J.; Yang, P.; Sugimura, H.; et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. USA 2009, 106, 12085–12090. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, Y.; Guo, Q.; Song, P.; Zhang, Q.; Guo, C.; Zeng, H.; Guan, Y.; Liu, X.; Zhao, C. Correlation study and significance of the EGFR expression in serum, lymph nodes and tumor tissue of NSCLC. Thorac Cancer 2014, 5, 31–37. [Google Scholar] [CrossRef] [PubMed]







| Samples Parameters | LUAD (n = 210) | LUSC (n = 257) | |
|---|---|---|---|
| Age | Median, Range ♂ | 67, 41–88 | 68, 41–90 |
| Unknown | 9 | 4 | |
| T stage | T1 | 56 | 52 |
| T2 | 121 | 153 | |
| T3 | 23 | 43 | |
| T4 | 9 | 9 | |
| Tx | 1 | - | |
| N stage | N0 | 131 | 168 |
| N1 | 48 | 63 | |
| N2 | 28 | 21 | |
| N3 | - | - | |
| Nx | 2 | 5 | |
| N unknown | 1 | - | |
| M stage | M0 | 142 | 199 |
| M1 | 12 | 3 | |
| Mx | 55 | 55 | |
| M unknown | 1 | - | |
| Tumor stage | I | 99 | 117 |
| II | 63 | 95 | |
| III | 32 | 39 | |
| IV | 12 | 3 | |
| Unknown | 4 | 3 | |
| Smoking status | Never smoker | 19 | 6 |
| Current smoker | 61 | 86 | |
| Quit > 15 years | 63 | 39 | |
| Quit ≤ 15 years | 57 | 113 | |
| Quit (unknown) | 3 | 4 | |
| Unknown | 7 | 9 | |
| Demographics | LUAD n = 4 | LUSC n = 4 | |
|---|---|---|---|
| No. of Patients (%) | No. of Patients (%) | ||
| Age | 50–59 | 1 (25) | 0 (0) |
| 60–69 | 2 (50) | 3 (75) | |
| 70–79 | 1 (25) | 1 (25) | |
| Sex | M | 4 (100) | 4 (100) |
| Stage | IB | 0 (0) | 1 (25) |
| IIA | 2 (50) | 1 (25) | |
| IIB | 1 (25) | 2 (50) | |
| IIIA | 1 (25) | 0 (0) | |
| Smoking status | Never smoker | 2 (50) | 0 (0) |
| Former smoker | 0 (0) | 2 (50) | |
| Current smoker | 2 (50) | 2 (50) | |
| Characteristics | LUAD n = 28 | LUSC n = 34 | |
|---|---|---|---|
| No. of Patients (%) | No. of Patients (%) | ||
| Age | 50–59 | 8 (28.6) | 9 (26.5) |
| 60–69 | 13 (46.4) | 14 (41.1) | |
| 70–79 | 6 (21.4) | 9 (26.5) | |
| 80–89 | 1 (3.6) | 2 (5.9) | |
| Sex | M | 28 (100) | 34 (100) |
| T | T2 | 3 (10.7) | 3 (8.8) |
| T3 | 10 (35.7) | 9 (26.5) | |
| T4 | 15 (53.6) | 22 (64.7) | |
| N | N0 | 4 (14.3) | 3 (8.8) |
| N1 | 7 (25) | 6 (17.6) | |
| N2 | 14 (50) | 21 (61.7) | |
| N3 | 3 (10.7) | 4 (11.8) | |
| M | M0 | 12 (42.8) | 26 (76.5) |
| M1 | 16 (57.2) | 8 (23.5) | |
| Stage | II | 2 (7.2) | 2 (5.9) |
| III | 10 (35.7) | 25 (73.5) | |
| IV | 16 (57.1) | 7 (20.6) | |
| Smoking status | Never smoker | 6 (21.4) | 0 (0) |
| Current smoker | 13 (46.4) | 17 (50) | |
| Former smoker | 9 (32.2) | 17 (50) | |
| Characteristics | LUAD n = 4 | LUSC n = 15 | |
|---|---|---|---|
| No. of Patients (%) | No. of Patients (%) | ||
| Age | 50–59 | 2 (50) | 5 (33.3) |
| 60–69 | 1 (25) | 5 (33.3) | |
| 70–79 | 1 (25) | 4 (26.6) | |
| 80–89 | 0 (0) | 1 (6.7) | |
| Sex | M | 4 (100) | 15 (100) |
| Stage | II | 1 (25) | 0 (0) |
| III | 2 (50) | 12 (80) | |
| IV | 1 (25) | 3 (20) | |
| T | T2 | 3 (75) | 2 (13.3) |
| T3 | 1 (25) | 3 (20) | |
| T4 | 0 (0) | 10 (66.7) | |
| N | N0 | 2 (50) | 0 (0) |
| N1 | 0 (0) | 2 (13.3) | |
| N2 | 1 (25) | 11 (73.4) | |
| N3 | 1 (25) | 2 (13.3) | |
| M | M0 | 3 (75) | 11 (73.4) |
| M1 | 1 (25) | 4 (26.7) | |
| Smoking status | Never smoker | 1 (25) | 0 (0) |
| Current smoker | 1 (25) | 7 (46.7) | |
| Former smoker | 2 (50) | 8 (53.3) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haranguș, A.; Lajos, R.; Budisan, L.; Zanoaga, O.; Ciocan, C.; Bica, C.; Pirlog, R.; Simon, I.; Simon, M.; Braicu, C.; et al. Identification of Potential microRNA Panels for Male Non-Small Cell Lung Cancer Identification Using Microarray Datasets and Bioinformatics Methods. J. Pers. Med. 2022, 12, 2056. https://doi.org/10.3390/jpm12122056
Haranguș A, Lajos R, Budisan L, Zanoaga O, Ciocan C, Bica C, Pirlog R, Simon I, Simon M, Braicu C, et al. Identification of Potential microRNA Panels for Male Non-Small Cell Lung Cancer Identification Using Microarray Datasets and Bioinformatics Methods. Journal of Personalized Medicine. 2022; 12(12):2056. https://doi.org/10.3390/jpm12122056
Chicago/Turabian StyleHaranguș, Antonia, Raduly Lajos, Livia Budisan, Oana Zanoaga, Cristina Ciocan, Cecilia Bica, Radu Pirlog, Ioan Simon, Marioara Simon, Cornelia Braicu, and et al. 2022. "Identification of Potential microRNA Panels for Male Non-Small Cell Lung Cancer Identification Using Microarray Datasets and Bioinformatics Methods" Journal of Personalized Medicine 12, no. 12: 2056. https://doi.org/10.3390/jpm12122056
APA StyleHaranguș, A., Lajos, R., Budisan, L., Zanoaga, O., Ciocan, C., Bica, C., Pirlog, R., Simon, I., Simon, M., Braicu, C., & Berindan-Neagoe, I. (2022). Identification of Potential microRNA Panels for Male Non-Small Cell Lung Cancer Identification Using Microarray Datasets and Bioinformatics Methods. Journal of Personalized Medicine, 12(12), 2056. https://doi.org/10.3390/jpm12122056










