Microbiome Changes in Humans with Parkinson’s Disease after Photobiomodulation Therapy: A Retrospective Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Sample Collection
2.3. Microbiome Analysis
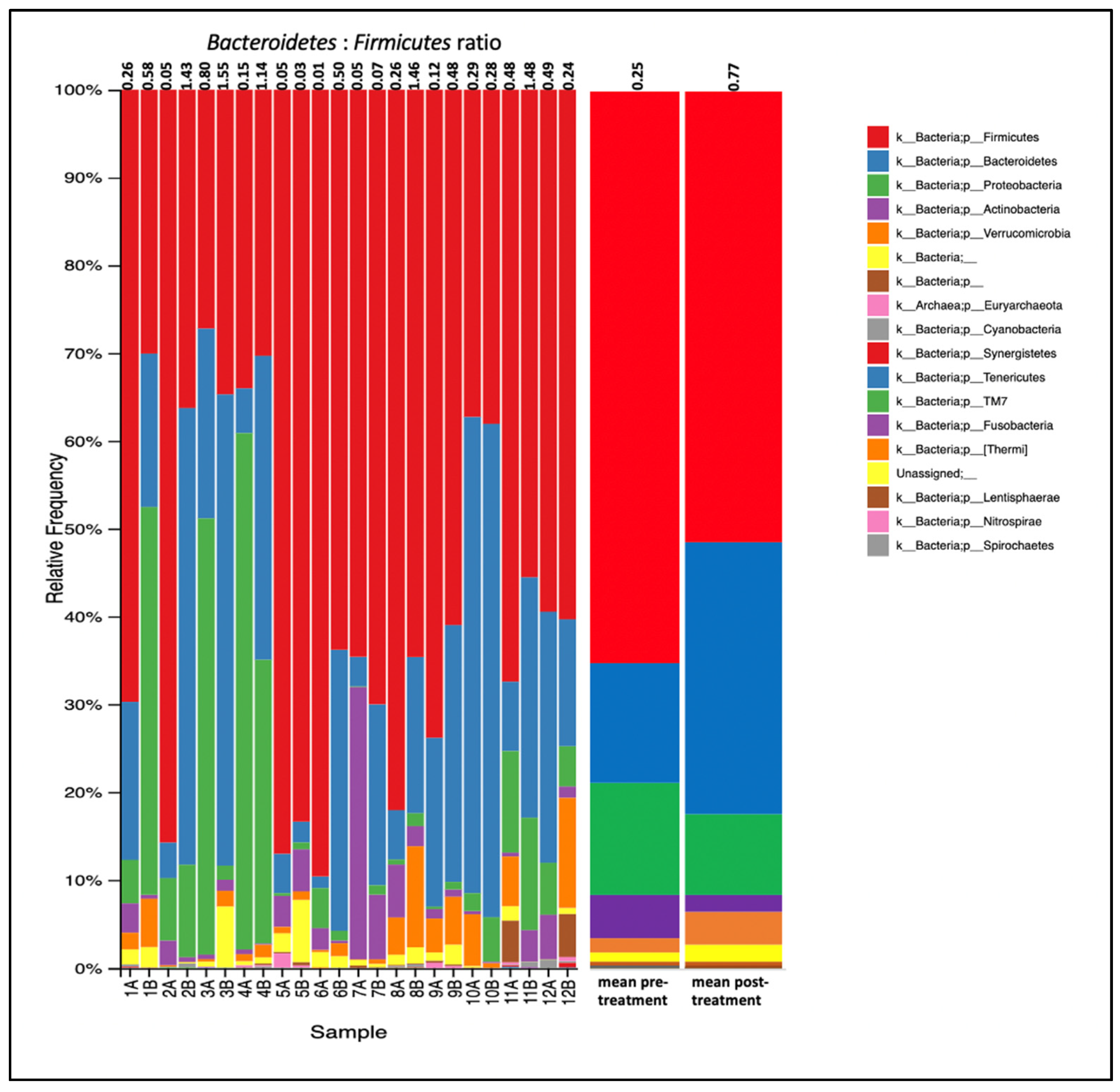
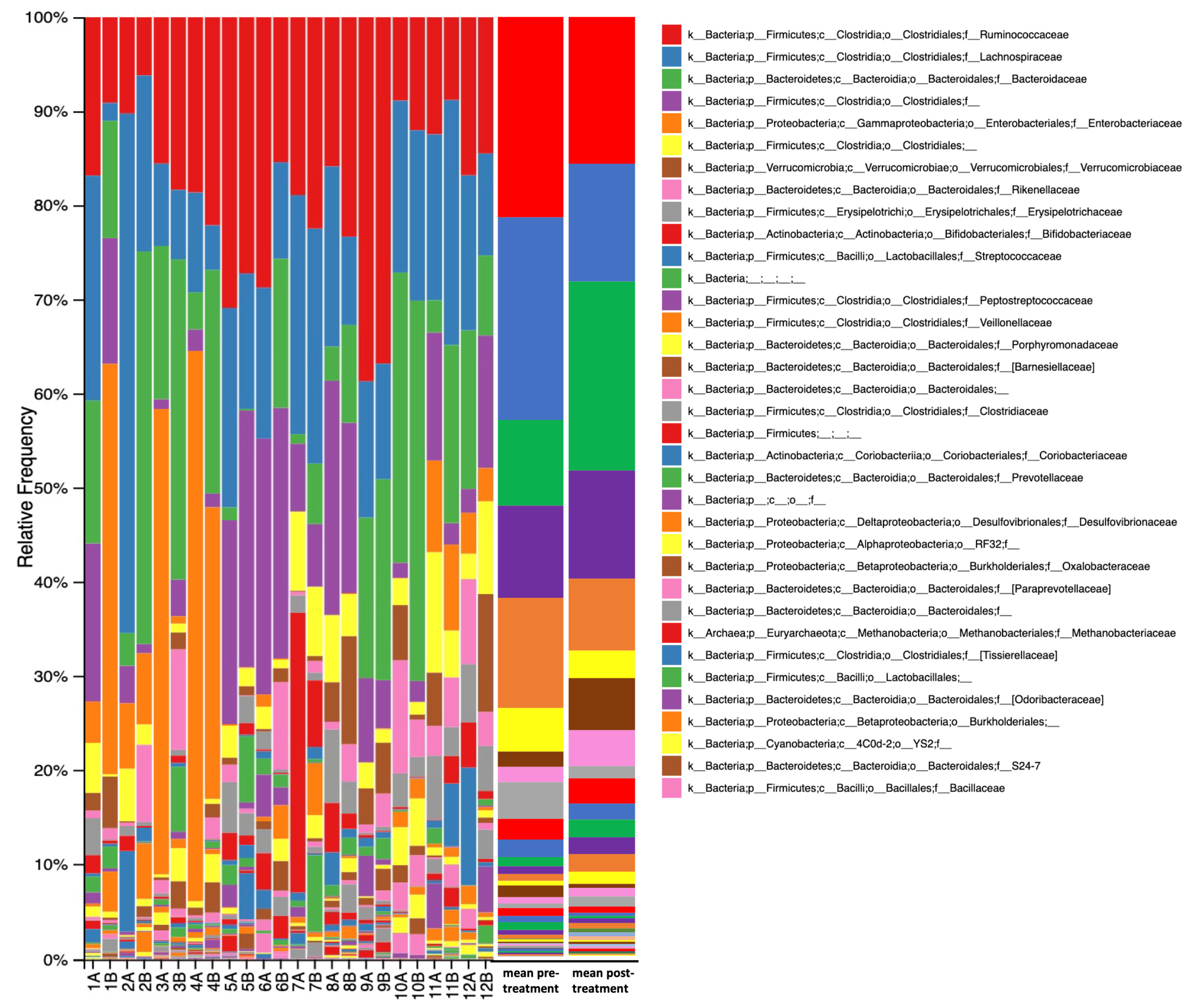
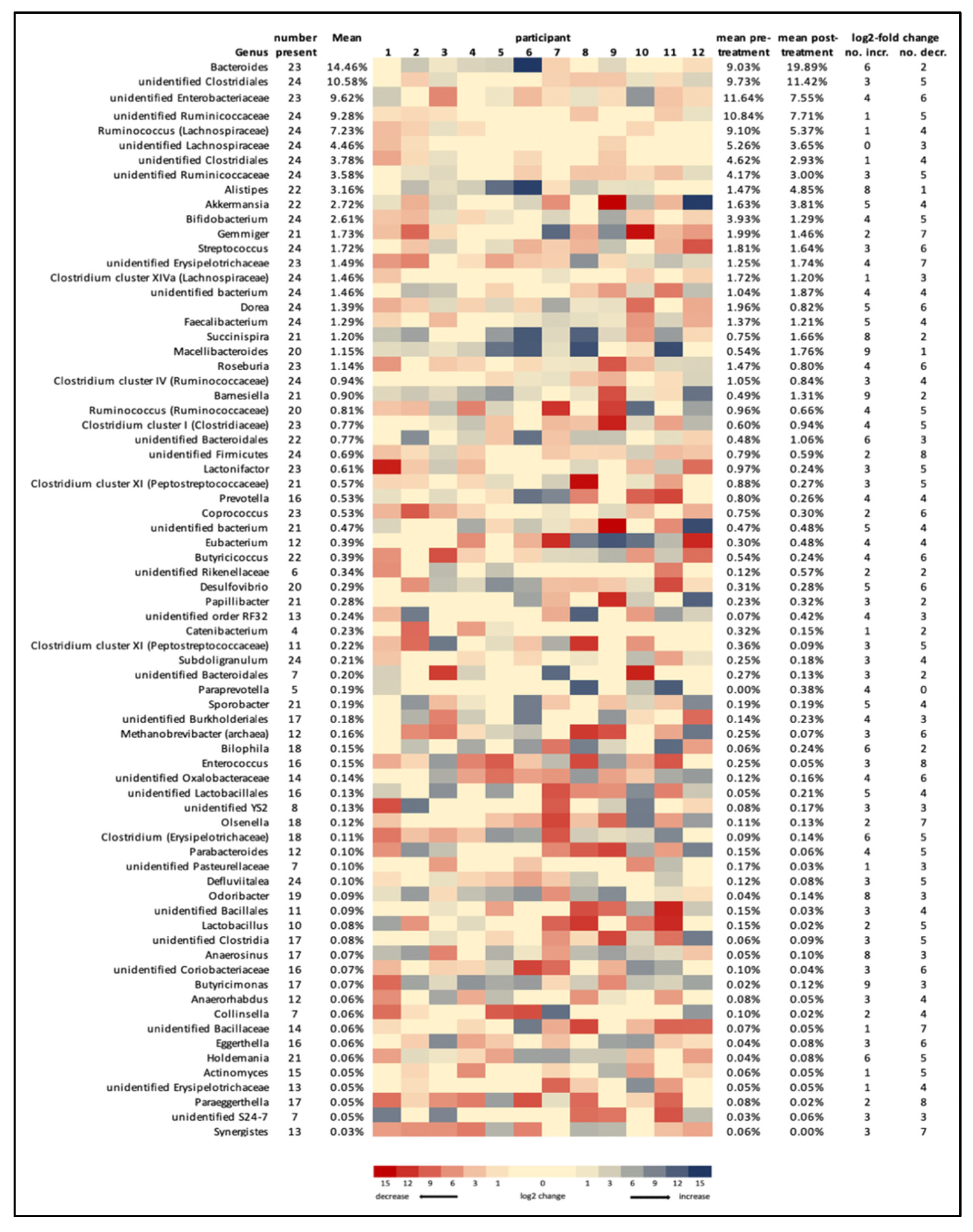
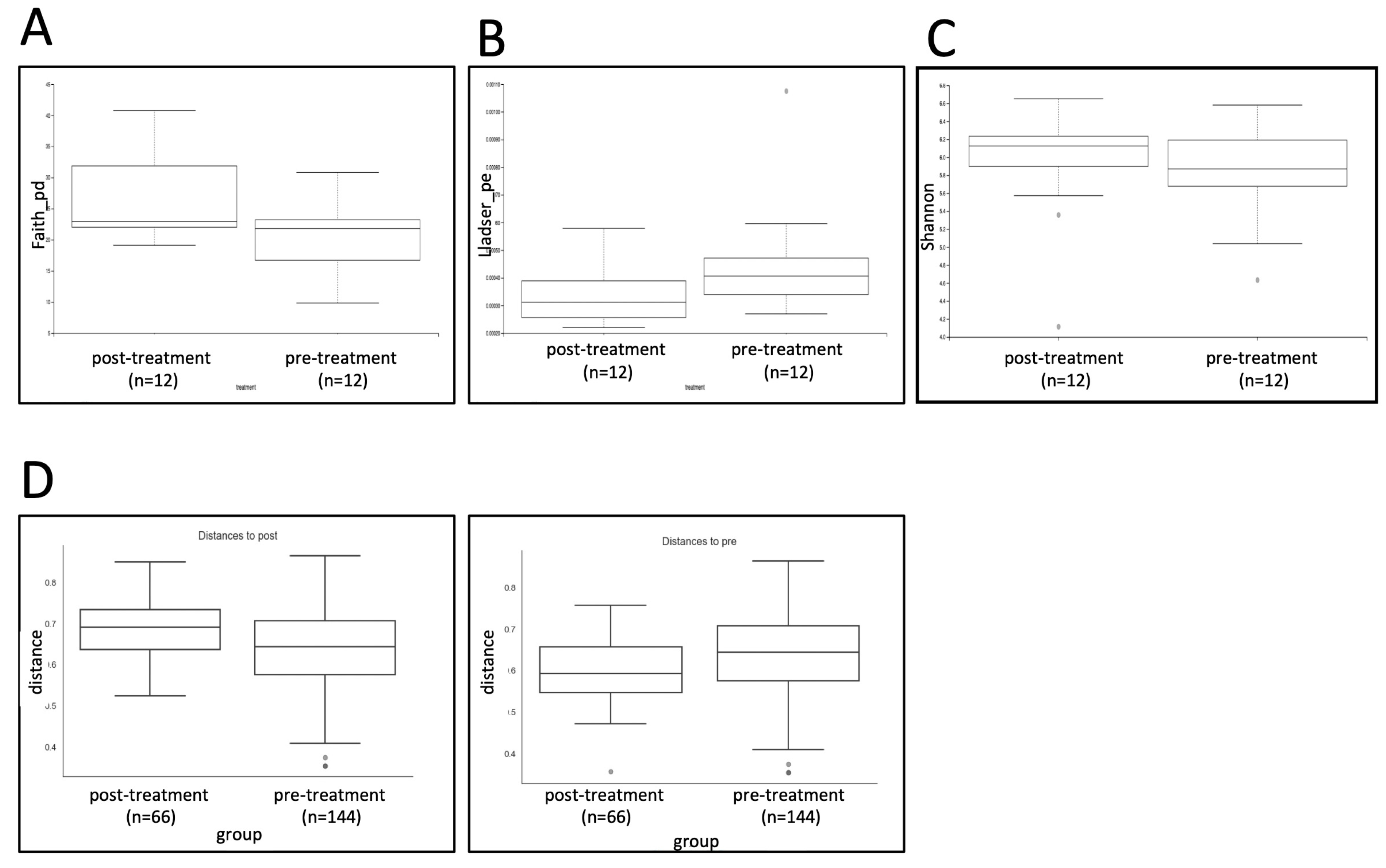
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giuffrè, M.; Moretti, R.; Campisciano, G.; Da Silveira, A.B.M.; Monda, V.M.; Comar, M.; Di Bella, S.; Antonello, R.M.; Luzzati, R.; Crocè, L.S. You Talking to Me? Says the Enteric Nervous System (ENS) to the Microbe. How Intestinal Microbes Interact with the ENS. J. Clin. Med. 2020, 9, 3705. [Google Scholar] [CrossRef]
- Wang, Y.F.; Zheng, L.J.; Liu, Y.; Ye, Y.B.; Luo, S.; Lu, G.M.; Gong, D.; Zhang, L.J. The gut microbiota-inflammation-brain axis in end-stage renal disease: Perspectives from default mode network. Theranostics 2019, 9, 8171–8181. [Google Scholar] [CrossRef] [PubMed]
- Giuffrè, M.; Campigotto, M.; Campisciano, G.; Comar, M.; Crocè, L.S. A story of liver and gut microbes: How does the intestinal flora affect liver disease? A review of the literature. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G889–G906. [Google Scholar] [CrossRef]
- van den Munckhof, I.C.; Kurilshikov, A.; Ter Horst, R.; Riksen, N.P.; Joosten, L.A.B.; Zhernakova, A.; Fu, J.; Keating, S.; Netea, M.G.; De Graaf, J.; et al. Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: A systematic review of human studies. Obes. Rev. 2018, 19, 1719–1734. [Google Scholar] [CrossRef] [Green Version]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.; Sandhu, K.V.; Bastiaanssen, T.F.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Bullich, C.; Keshavarzian, A.; Garssen, J.; Kraneveld, A.; Perez-Pardo, P. Gut Vibes in Parkinson’s Disease: The Microbiota-Gut-Brain Axis. Mov. Disord. Clin. Pract. 2019, 6, 639–651. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unger, M.M.; Spiegel, J.; Dillmann, K.U.; Grundmann, D.; Philippeit, H.; Bürmann, J.; Faßbender, K.; Schwiertz, A.; Schäfer, K.H. Short chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls. Park. Relat. Disord. 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Forsyth, C.B.; Shannon, K.M.; Kordower, J.H.; Voigt, R.M.; Shaikh, M.; Jaglin, J.A.; Estes, J.D.; Dodiya, H.B.; Keshavarzian, A. Increased intestinal permeability correlates with sigmoid mucosa alpha-synuclein staining and endotoxin exposure markers in early Parkinson’s disease. PLoS ONE 2011, 6, e28032. [Google Scholar] [CrossRef] [Green Version]
- Chiang, H.-L.; Lin, C.-H. Altered Gut Microbiome and Intestinal Pathology in Parkinson’s Disease. J. Mov. Disord. 2019, 12, 67–83. [Google Scholar] [CrossRef]
- Lin, C.H.; Chen, C.C.; Chiang, H.L.; Liou, J.M.; Chang, C.M.; Lu, T.P.; Chuang, E.Y.; Tai, Y.C.; Cheng, C.; Lin, H.Y.; et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflamm. 2019, 16, 129. [Google Scholar] [CrossRef] [PubMed]
- Caggiu, E.; Arru, G.; Hosseini, S.; Niegowska, M.; Sechi, G.; Zarbo, I.R.; Sechi, L.A. Inflammation, Infectious Triggers, and Parkinson’s Disease. Front. Neurol. 2019, 10, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmqvist, S.; Chutna, O.; Bousset, L.; Aldrin-Kirk, P.; Li, W.; Bjorklund, T.; Wang, Z.-Y.; Roybon, L.; Melki, R.; Li, J.-Y. Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol. 2014, 128, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Kwon, S.-H.; Kam, T.-I.; Panicker, N.; Karuppagounder, S.S.; Lee, S.; Lee, J.H.; Kim, W.R.; Kook, M.; Foss, C.A.; et al. Transneuronal Propagation of Pathologic α-Synuclein from the Gut to the Brain Models Parkinson’s Disease. Neuron 2019, 103, 627–641.e7. [Google Scholar] [CrossRef] [PubMed]
- Uemura, N.; Yagi, H.; Uemura, M.T.; Hatanaka, Y.; Yamakado, H.; Takahashi, R. Inoculation of α-synuclein preformed fibrils into the mouse gastrointestinal tract induces Lewy body-like aggregates in the brainstem via the vagus nerve. Mol. Neurodegener. 2018, 13, 21. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Fang, F.; Pedersen, N.L.; Tillander, A.; Ludvigsson, J.F.; Ekbom, A.; Svenningsson, P.; Chen, H.; Wirdefeldt, K. Vagotomy and Parkinson disease: A Swedish register–based matched-cohort study. Neurology 2017, 88, 1996–2002. [Google Scholar] [CrossRef] [Green Version]
- Fasano, A.; Visanji, N.P.; Liu, L.W.; Lang, A.E.; Pfeiffer, R.F. Gastrointestinal dysfunction in Parkinson’s disease. Lancet Neurol. 2015, 14, 625–639. [Google Scholar] [CrossRef]
- Scheperjans, F.; Derkinderen, P.; Borghammer, P. The Gut and Parkinson’s Disease: Hype or Hope? J. Parkinson’s Dis. 2018, 8, S31–S39. [Google Scholar] [CrossRef] [Green Version]
- Brudek, T. Inflammatory Bowel Diseases and Parkinson’s Disease. J. Park. Dis. 2019, 9, S331–S344. [Google Scholar] [CrossRef] [Green Version]
- Fu, P.; Gao, M.; Yung, K.K.L. Association of Intestinal Disorders with Parkinson’s Disease and Alzheimer’s Disease: A Systematic Review and Meta-Analysis. ACS Chem. Neurosci. 2019, 11, 395–405. [Google Scholar] [CrossRef]
- Stokholm, M.G.; Danielsen, E.H.; Hamilton-Dutoit, S.; Borghammer, P. Pathological α-synuclein in gastrointestinal tissues from prodromal Parkinson disease patients. Ann. Neurol. 2016, 79, 940–949. [Google Scholar] [CrossRef]
- Hamblin, M.R. Mechanisms and Mitochondrial Redox Signaling in Photobiomodulation. Photochem. Photobiol. 2018, 94, 199–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, P.; Kim, J.Y.; Riveros, C.; Camp, A.; Johnstone, D.M. Elucidating the time course of the transcriptomic response to photobiomodulation through gene co-expression analysis. J. Photochem. Photobiol. B Biol. 2020, 208, 111916. [Google Scholar] [CrossRef]
- Hamblin, M.R. Mechanisms and applications of the anti-inflammatory effects of photobiomodulation. AIMS Biophys. 2017, 4, 337–361. [Google Scholar] [CrossRef]
- Bicknell, B.; Liebert, A.; Johnstone, D.; Kiat, H. Photobiomodulation of the microbiome: Implications for metabolic and inflammatory diseases. Lasers Med. Sci. 2018, 34, 317–327. [Google Scholar] [CrossRef]
- Liebert, A.; Bicknell, B.; Laakso, E.-L.; Heller, G.; Jalilitabaei, P.; Tilley, S.; Mitrofanis, J.; Kiat, H. Improvements in clinical signs of Parkinson’s disease using photobiomodulation: A prospective proof-of-concept study. BMC Neurol. 2021, 21, 256. [Google Scholar] [CrossRef]
- Liebert, A.; Bicknell, B.; Laakso, E.-L.; Jalilitabaei, P.; Tilley, S.; Kiat, H.; Mitrofanis, J. Remote Photobiomodulation Treatment for the Clinical Signs of Parkinson’s Disease: A Case Series Conducted During COVID-19. Photobiomodul. Photomed. Laser Surg. 2021. [Google Scholar] [CrossRef]
- Bhidayasiri, R.; Tarsy, D. Parkinson’s disease: Hoehn and Yahr scale. In Movement Disorders: A Video Atlas; Humana: Totowa, NJ, USA, 2012; pp. 4–5. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Dillon, M.R.; Zhang, Y.; Rideout, J.R.; Bolyen, E.; Li, H.; Albert, P.S.; Caporaso, J.G. q2-longitudinal: Longitudinal and Paired-Sample Analyses of Microbiome Data. mSystems 2018, 3, e00219-18. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Bicknell, B.; Laakso, E.-L.; Liebert, A.; Kiat, H. Modifying the microbiome as a potential mechanism of photobiomodulation: A case report. Photobiomodul. Photomed. Laser Surg. 2022; in press. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef]
- Vaiserman, A.; Romanenko, M.; Piven, L.; Moseiko, V.; Lushchak, O.; Kryzhanovska, N.; Guryanov, V.; Koliada, A. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 2020, 20, 221. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, X.; Yang, S.; Meng, F.; Wang, X.; Wei, H.; Chen, T. Evaluation of the Microbial Diversity in Amyotrophic Lateral Sclerosis Using High-Throughput Sequencing. Front. Microbiol. 2016, 7, 1479. [Google Scholar] [CrossRef] [Green Version]
- Zhai, C.D.; Zheng, J.J.; An, B.C.; Huang, H.F.; Tan, Z.C. Intestinal microbiota composition in patients with amyotrophic lateral sclerosis: Establishment of bacterial and archaeal communities analyses. Chin. Med. J. 2019, 132, 1815–1822. [Google Scholar] [CrossRef]
- Saji, N.; Niida, S.; Murotani, K.; Hisada, T.; Tsuduki, T.; Sugimoto, T.; Kimura, A.; Toba, K.; Sakurai, T. Analysis of the relationship between the gut microbiome and dementia: A cross-sectional study conducted in Japan. Sci. Rep. 2019, 9, 1008. [Google Scholar] [CrossRef]
- Gerhardt, S.; Mohajeri, M.H. Changes of Colonic Bacterial Composition in Parkinson’s Disease and Other Neurodegenerative Diseases. Nutrients 2018, 10, 708. [Google Scholar] [CrossRef] [Green Version]
- Brown, E.G.; Goldman, S.M. Modulation of the Microbiome in Parkinson’s Disease: Diet, Drug, Stool Transplant, and Beyond. Neurotherapeutics 2020, 17, 1406–1417. [Google Scholar] [CrossRef]
- Borody, T.; Bienenstock, J. The Role of Fecal Microbiota Transplantation in Neurological Diseases. In The Microbiome and the Brain; CRC Press: Boca Raton, FL, USA, 2019; pp. 161–177. [Google Scholar] [CrossRef]
- Cassano, P.; Caldieraro, M.A.; Norton, R.; Mischoulon, D.; Trinh, N.-H.; Nyer, M.; Dording, C.; Hamblin, M.R.; Campbell, B.; Iosifescu, D.V. Reported Side Effects, Weight and Blood Pressure, After Repeated Sessions of Transcranial Photobiomodulation. Photobiomodul. Photomed. Laser Surg. 2019, 37, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, J.; Tan, E.K. Parkinson’s disease: Etiopathogenesis and treatment. J. Neurol. Neurosurg. Psychiatry 2020, 91, 795–808. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, V.; Skladnev, N.V.; Kim, J.Y.; Mitrofanis, J.; Stone, J.; Johnstone, D.M. Pre-conditioning with Remote Photobiomodulation Modulates the Brain Transcriptome and Protects Against MPTP Insult in Mice. Neuroscience 2019, 400, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Blatt, A.; Elbaz-Greener, G.A.; Tuby, H.; Maltz, L.; Siman-Tov, Y.; Ben-Aharon, G.; Copel, L.; Eisenberg, I.; Efrati, S.; Jonas, M.; et al. Low-Level Laser Therapy to the Bone Marrow Reduces Scarring and Improves Heart Function Post-Acute Myocardial Infarction in the Pig. Photomed. Laser Surg. 2016, 34, 516–524. [Google Scholar] [CrossRef]
- Al Amir Dache, Z.; Otandault, A.; Tanos, R.; Pastor, B.; Meddeb, R.; Sanchez, C.; Arena, G.; Lasorsa, L.; Bennett, A.; Grange, T.; et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB J. 2020, 34, 3616–3630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.; Brandli, A.; Mitrofanis, J.; Stone, J.; Purushothuman, S.; Johnstone, D.M. Remote tissue conditioning—An emerging approach for inducing body-wide protection against diseases of ageing. Ageing Res. Rev. 2017, 37, 69–78. [Google Scholar] [CrossRef]
- Lubomski, M.; Tan, A.H.; Lim, S.-Y.; Holmes, A.J.; Davis, R.L.; Sue, C.M. Parkinson’s disease and the gastrointestinal microbiome. J. Neurol. 2019, 267, 2507–2523. [Google Scholar] [CrossRef]
- Baldini, F.; Hertel, J.; Sandt, E.; Thinnes, C.C.; Neuberger-Castillo, L.; Pavelka, L.; Betsou, F.; Krüger, R.; Thiele, I.; Aguayo, G.; et al. Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biol. 2020, 18, 62. [Google Scholar] [CrossRef]
- Li, F.; Wang, P.; Chen, Z.; Sui, X.; Xie, X.; Zhang, J. Alteration of the fecal microbiota in North-Eastern Han Chinese population with sporadic Parkinson’s disease. Neurosci. Lett. 2019, 707, 134297. [Google Scholar] [CrossRef]
- Romano, S.; Savva, G.M.; Bedarf, J.R.; Charles, I.G.; Hildebrand, F.; Narbad, A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. npj Park. Dis. 2021, 7, 27. [Google Scholar] [CrossRef]
- Wallen, Z.D.; Appah, M.; Dean, M.N.; Sesler, C.L.; Factor, S.A.; Molho, E.; Zabetian, C.P.; Standaert, D.G.; Payami, H. Characterizing dysbiosis of gut microbiome in PD: Evidence for overabundance of opportunistic pathogens. npj Park. Dis. 2020, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Uhr, G.T.; Dohnalová, L.; Thaiss, C.A. The Dimension of Time in Host-Microbiome Interactions. mSystems 2019, 4, e00216-18. [Google Scholar] [CrossRef] [Green Version]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The Long-Term Stability of the Human Gut Microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef] [Green Version]
- Fu, B.C.; Randolph, T.W.; Lim, U.; Monroe, K.R.; Cheng, I.; Wilkens, L.R.; Le Marchand, L.; Lampe, J.W.; Hullar, M.A. Temporal Variability and Stability of the Fecal Microbiome: The Multiethnic Cohort Study. Cancer Epidemiol. Biomark. Prev. 2018, 28, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Sunagawa, S.; Schloissnig, S.; Arumugam, M.; Forslund, K.; Mitreva, M.; Tap, J.; Zhu, A.; Waller, A.; Mende, D.R.; Kultima, J.R.; et al. Individuality and temporal stability of the human gut microbiome. Cent. Asian J. Glob. Health 2014, 2, 120. [Google Scholar] [CrossRef] [Green Version]
- Aho, V.T.; Houser, M.C.; Pereira, P.A.; Chang, J.; Rudi, K.; Paulin, L.; Hertzberg, V.; Auvinen, P.; Tansey, M.G. Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson’s disease. Mol. Neurodegener. 2021, 16, 6. [Google Scholar] [CrossRef]
- Minato, T.; Maeda, T.; Fujisawa, Y.; Tsuji, H.; Nomoto, K.; Ohno, K.; Hirayama, M. Progression of Parkinson’s disease is associated with gut dysbiosis: Two-year follow-up study. PLoS ONE 2017, 12, e0187307. [Google Scholar] [CrossRef] [Green Version]
- Cilia, R.; Piatti, M.; Cereda, E.; Bolliri, C.; Caronni, S.; Ferri, V.; Cassani, E.; Bonvegna, S.; Ferrarese, C.; Zecchinelli, A.L.; et al. Does Gut Microbiota Influence the Course of Parkinson’s Disease? A 3-Year Prospective Exploratory Study in de novo Patients. J. Park. Dis. 2021, 11, 159–170. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Fernández, J.; Redondo-Blanco, S.; Gutiérrez-Del-Río, I.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Colon microbiota fermentation of dietary prebiotics towards short-chain fatty acids and their roles as anti-inflammatory and antitumour agents: A review. J. Funct. Foods 2016, 25, 511–522. [Google Scholar] [CrossRef]
- Senghor, B.; Sokhna, C.; Ruimy, R.; Lagier, J.-C. Gut microbiota diversity according to dietary habits and geographical provenance. Hum. Microbiome J. 2018, 7–8, 1–9. [Google Scholar] [CrossRef]
- Hegelmaier, T.; Lebbing, M.; Duscha, A.; Tomaske, L.; Tönges, L.; Holm, J.B.; Nielsen, H.B.; Gatermann, S.G.; Przuntek, H.; Haghikia, A. Interventional Influence of the Intestinal Microbiome Through Dietary Intervention and Bowel Cleansing Might Improve Motor Symptoms in Parkinson’s Disease. Cells 2020, 9, 376. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, S.; Goto, S.; Tsuji, H.; Okuno, T.; Asahara, T.; Nomoto, K.; Shibata, A.; Fujisawa, Y.; Minato, T.; Okamoto, A.; et al. Intestinal Dysbiosis and Lowered Serum Lipopolysaccharide-Binding Protein in Parkinson’s Disease. PLoS ONE 2015, 10, e0142164. [Google Scholar] [CrossRef] [Green Version]
- Petrov, V.A.; Saltykova, I.V.; Zhukova, I.A.; Alifirova, V.M.; Zhukova, N.G.; Dorofeeva, Y.B.; Tyakht, A.V.; Kovarsky, B.A.; Alekseev, D.G.; Kostryukova, E.S.; et al. Analysis of Gut Microbiota in Patients with Parkinson’s Disease. Bull. Exp. Biol. Med. 2017, 162, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Kulagina, E.; Efimov, B.A.; Maximov, P.Y.; Kafarskaia, L.I.; Chaplin, A.; Shkoporov, A. Species Composition ofBacteroidalesOrder Bacteria in the Feces of Healthy People of Various Ages. Biosci. Biotechnol. Biochem. 2012, 76, 169–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiippala, K.; Barreto, G.; Burrello, C.; Diaz-Basabe, A.; Suutarinen, M.; Kainulainen, V.; Bowers, J.R.; Lemmer, D.; Engelthaler, D.M.; Eklund, K.K.; et al. Novel Odoribacter splanchnicus Strain and Its Outer Membrane Vesicles Exert Immunoregulatory Effects in vitro. Front. Microbiol. 2020, 11, 575455. [Google Scholar] [CrossRef]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- Lang, S.; Martin, A.; Farowski, F.; Wisplinghoff, H.; Vehreschild, M.J.; Liu, J.; Krawczyk, M.; Nowag, A.; Kretzschmar, A.; Herweg, J.; et al. High Protein Intake Is Associated With Histological Disease Activity in Patients With NAFLD. Hepatol. Commun. 2020, 4, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lee, I.-S.; Braun, C.; Enck, P. Effect of Probiotics on Central Nervous System Functions in Animals and Humans: A Systematic Review. J. Neurogastroenterol. Motil. 2016, 22, 589–605. [Google Scholar] [CrossRef]
- Karunaratne, T.B.; Okereke, C.; Seamon, M.; Purohit, S.; Wakade, C.; Sharma, A. Niacin and Butyrate: Nutraceuticals Targeting Dysbiosis and Intestinal Permeability in Parkinson’s Disease. Nutrients 2020, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Rani, L.; Mondal, A.C. Unravelling the role of gut microbiota in Parkinson’s disease progression: Pathogenic and therapeutic implications. Neurosci. Res. 2021, 168, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, Y.; Fujisawa, T. Analysis of Clostridium cluster XI bacteria in human feces. Biosci. Microbiota Food Health 2019, 38, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Cao, J.; Gong, C.; Amakye, K.; Yao, M.; Ren, J. Exploring the microbiota-Alzheimer’s disease linkage using short-term antibiotic treatment followed by fecal microbiota transplantation. Brain Behav. Immun. 2021, 96, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wu, J.; Dong, X.; Yin, H.; Shi, X.; Su, S.; Che, B.; Li, Y.; Yang, J. Gut flora-targeted photobiomodulation therapy improves senile dementia in an Aß-induced Alzheimer’s disease animal model. J. Photochem. Photobiol. B Biol. 2021, 216, 112152. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yang, J.; Dong, C.; Fu, Y.; Liu, H. Gut microbiome-mediated changes in bone metabolism upon infrared light exposure in rats. J. Photochem. Photobiol. B Biol. 2021, 217, 112156. [Google Scholar] [CrossRef]
- Yoshimura, T.M.; Sabino, C.P.; Ribeiro, M. Photobiomodulation reduces abdominal adipose tissue inflammatory infiltrate of diet-induced obese and hyperglycemic mice. J. Biophotonics 2016, 9, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Lubomski, M.; Xu, X.; Holmes, A.J.; Yang, J.Y.H.; Sue, C.M.; Davis, R.L. The impact of device-assisted therapies on the gut microbiome in Parkinson’s disease. J. Neurol. 2021. [Google Scholar] [CrossRef] [PubMed]

| Participant | Sex | Age | Hoehn and Yahr Stage | Height | Weight | MDS UPDRS Score | Dominant Hand | Affected Side | Sample Collection | |
|---|---|---|---|---|---|---|---|---|---|---|
| Pre- Treatment | Post- Treatment | |||||||||
| 1 | M | 71 | 2 | 178 | 78.7 | 89 | R | L | 6 January 2019 | 31 March 2019 |
| 2 | F | 74 | 2 | 165 | 68.3 | 31 | R | L | 6 January 2019 | 31 March 2019 |
| 3 | F | 78 | 3 | 156 | 76.0 | 57 | R | L | 6 January 2019 | 31 March 2019 |
| 4 | M | 75 | 2 | 177 | 75.5 | 52 | R | L | 6 January 2019 | 31 March 2019 |
| 5 | M | 67 | 2 | 173 | 78.6 | 53 | R | L | 6 January 2019 | 31 March 2019 |
| 6 | M | 63 | 1 | 175 | 76.1 | 36 | L | L | 6 January 2019 | 31 March 2019 |
| 7 | F | 53 | 2 | 150 | 48.3 | 53 | R | L | 31 March 2019 | 7 July 2019 |
| 8 | F | 72 | 2 | 160 | 61.7 | 70 | R | L | 31 March 2019 | 7 July 2019 |
| 9 | F | 57 | 2 | 169 | 53.0 | 42 | R | R | 31 March 2019 | 7 July 2019 |
| 10 | M | 69 | 1 | 180 | 77.6 | 29 | R | L | 31 March 2019 | 7 July 2019 |
| 11 | F | 61 | 2 | 167 | 67.7 | 36 | L | R | 31 March 2019 | 7 July 2019 |
| 12 | F | 71 | 2 | 163 | 61.4 | 67 | R | R | 31 March 2019 | 7 July 2019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bicknell, B.; Liebert, A.; McLachlan, C.S.; Kiat, H. Microbiome Changes in Humans with Parkinson’s Disease after Photobiomodulation Therapy: A Retrospective Study. J. Pers. Med. 2022, 12, 49. https://doi.org/10.3390/jpm12010049
Bicknell B, Liebert A, McLachlan CS, Kiat H. Microbiome Changes in Humans with Parkinson’s Disease after Photobiomodulation Therapy: A Retrospective Study. Journal of Personalized Medicine. 2022; 12(1):49. https://doi.org/10.3390/jpm12010049
Chicago/Turabian StyleBicknell, Brian, Ann Liebert, Craig S. McLachlan, and Hosen Kiat. 2022. "Microbiome Changes in Humans with Parkinson’s Disease after Photobiomodulation Therapy: A Retrospective Study" Journal of Personalized Medicine 12, no. 1: 49. https://doi.org/10.3390/jpm12010049
APA StyleBicknell, B., Liebert, A., McLachlan, C. S., & Kiat, H. (2022). Microbiome Changes in Humans with Parkinson’s Disease after Photobiomodulation Therapy: A Retrospective Study. Journal of Personalized Medicine, 12(1), 49. https://doi.org/10.3390/jpm12010049








