Artificial Intelligence-Based Solution in Personalized Computer-Aided Arthroscopy of Shoulder Prostheses
Abstract
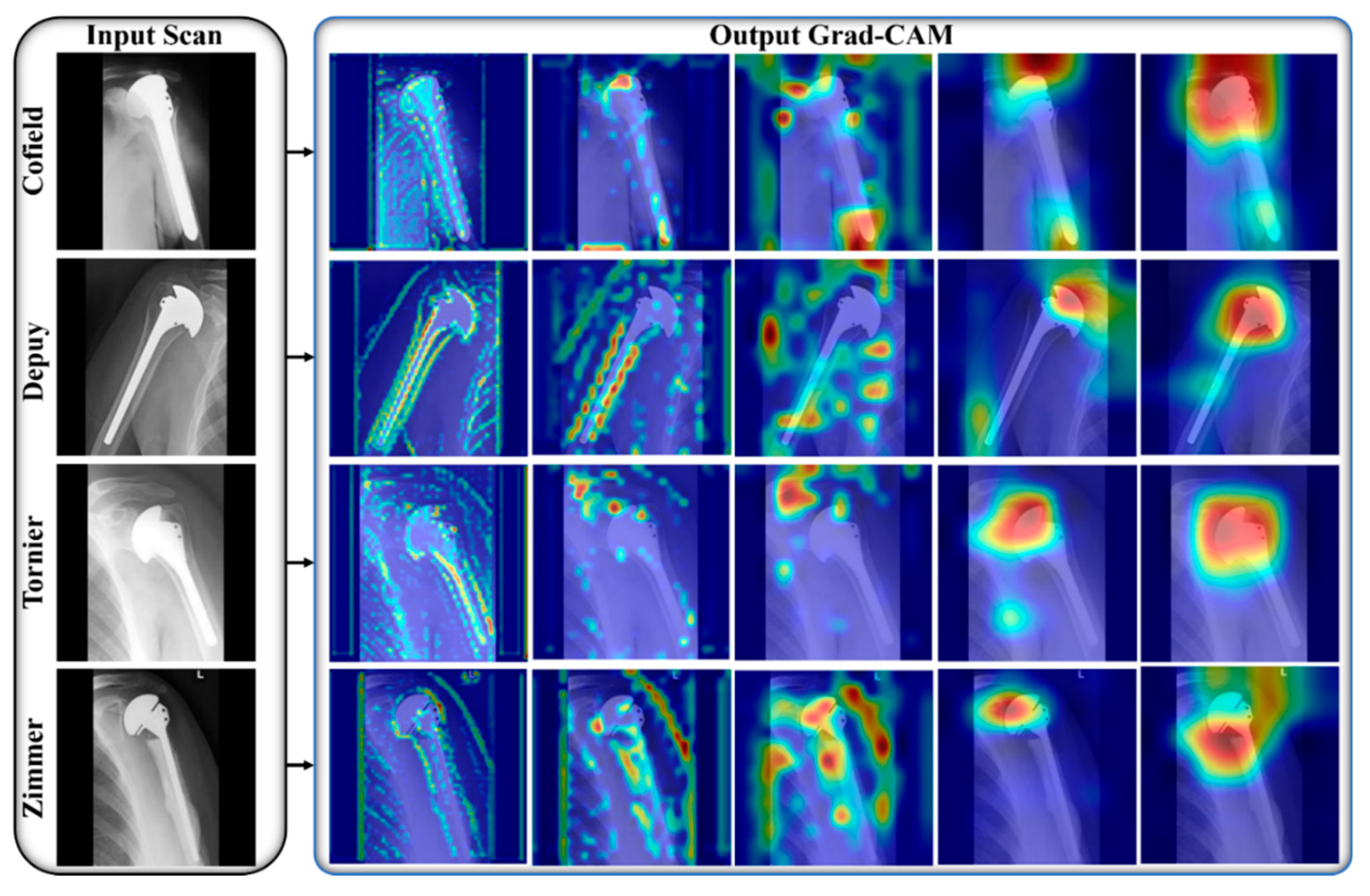
:1. Introduction
2. Materials and Methods
2.1. Dataset
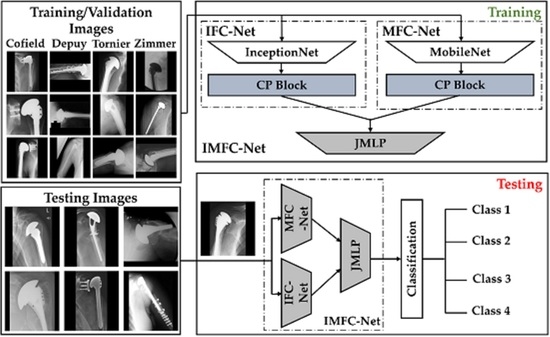
2.2. Overall Workflow
2.2.1. Model Design
- A.
- CP block of IFC-Net
- B.
- CP block of MFC-Net
- C.
- Feature Concatenation and Final Classification by JMLP
3. Results
3.1. Experimental Setup and Network Training
3.2. Our Results (Ablation Studies)
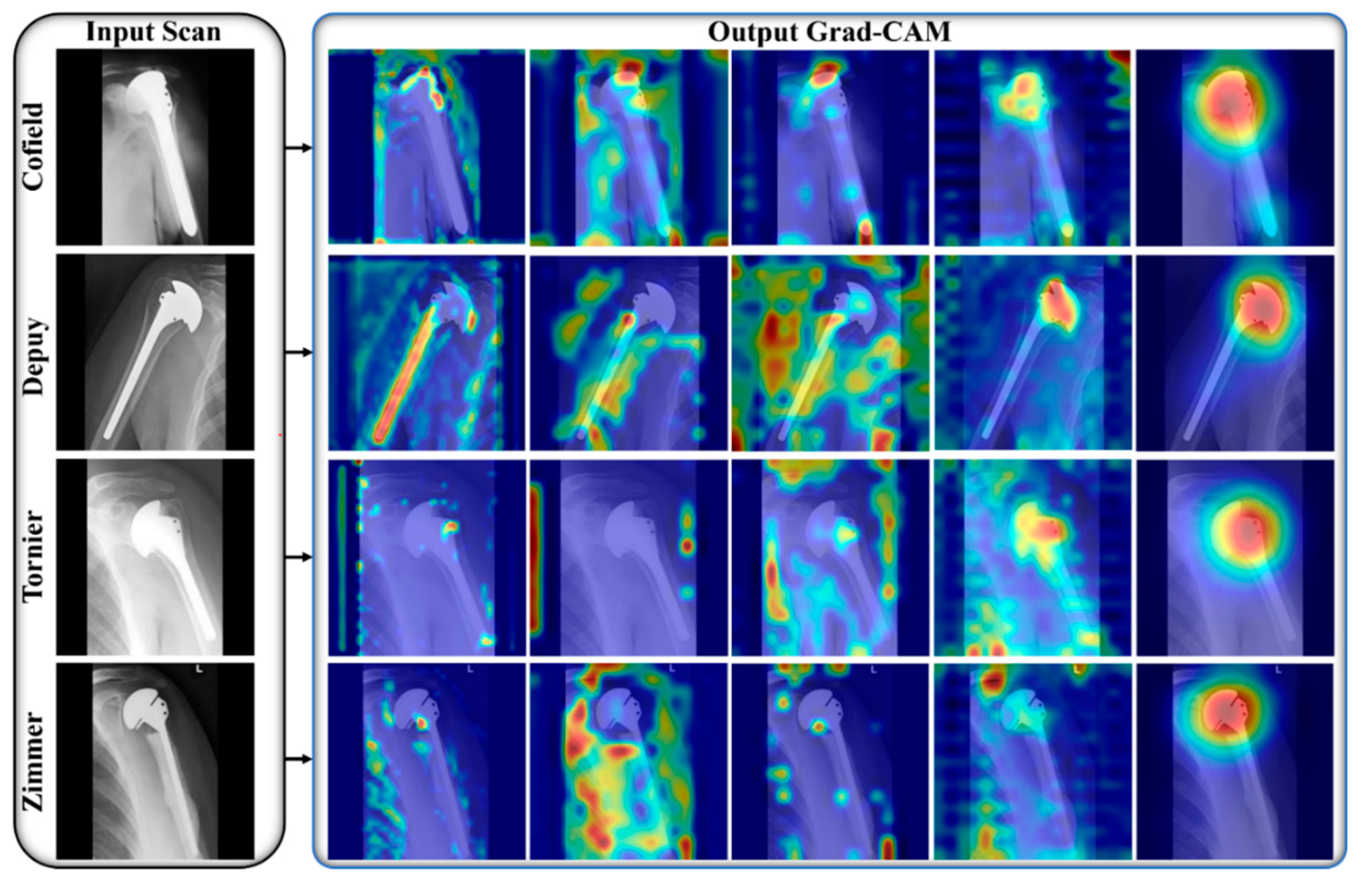
3.3. Comparisons
| Model | ACC ± Std | AP ± Std | AR ± Std | F1 ± Std |
|---|---|---|---|---|
| VGG-16 [19,20,22] | 72.97 ± 7.9 | 73.76 ± 8.39 | 69.16 ± 8.79 | 71.21 ± 7.7 |
| VGG-19 [19,20,22] | 67 ± 8.77 | 69.48 ± 10.15 | 61.42 ± 10.04 | 64.93 ± 9.8 |
| DarkNet-53 [32] | 51.66 ± 7.68 | 38.64 ± 8.55 | 39.44 ± 7.64 | 38.99 ± 7.98 |
| NASNet [19,20,33] | 70.8 ± 4.69 | 68.56 ± 6.59 | 62.78 ± 9.04 | 65.34 ± 6.95 |
| ResNet-18 [20,24,35] | 71.41 ± 5.81 | 69.9 ± 10.06 | 64.94 ± 7.41 | 67.03 ± 7.48 |
| ResNet-50 [19,20,24] | 80.26 ± 4.17 | 79.13 ± 5.45 | 74.93 ± 4.5 | 76.93 ± 4.52 |
| ResNet-101 [24] | 79.39 ± 6.44 | 79.27 ± 7.98 | 75.37 ± 8.37 | 77.14 ± 7.42 |
| DenseNet-201 [19,20,34] | 80.45 ± 4.77 | 78.68 ± 6.24 | 76.02 ± 5.9 | 77.26 ± 5.52 |
| Inception-V3 [23] | 76.23 ± 4.27 | 74.5 ± 6.01 | 68.48 ± 5.53 | 71.32 ± 5.46 |
| MobileNet-V2 [27] | 71.02 ± 6.56 | 67.51 ± 7.51 | 64.09 ± 8.8 | 65.7 ± 7.96 |
| DRE-Net [20] | 77.11 ± 5.35 | 78.14 ± 7.09 | 72.83 ± 5.33 | 75.21 ± 4.86 |
| Proposed (IMFC-Net) | 83.82 ± 3.12 | 82.36 ± 4.90 | 81.08 ± 5.27 | 81.56 ± 3.55 |
| Model | ACC ± Std | AP ± Std | AR ± Std | F1 ± Std |
|---|---|---|---|---|
| VGG-16 [19,20,22] | 64.48 ± 6.57 | 61.84 ± 8.7 | 59.54 ± 8.57 | 60.63 ± 8.5 |
| VGG-19 [19,20,22] | 66.77 ± 6.34 | 65.38 ± 8.79 | 61.57 ± 8.51 | 63.26 ± 7.87 |
| DarkNet-53 [32] | 48.12 ± 7.8 | 39.3 ± 7.69 | 36.87 ± 7.17 | 38.02 ± 7.37 |
| NASNet [19,20,33] | 58.18 ± 6.65 | 54.52 ± 8.66 | 49.06 ± 7.57 | 51.49 ± 7.61 |
| ResNet-18 [20,24,35] | 62.53 ± 8.02 | 59.72 ± 10.32 | 54.7 ± 10.34 | 56.97 ± 9.89 |
| ResNet-50 [19,20,24] | 70.17 ± 5.87 | 68.95 ± 6.78 | 63.37 ± 7.856 | 65.93 ± 6.76 |
| ResNet-101 [24] | 66.19 ± 6.31 | 65.89 ± 8.35 | 59.5 ± 8.04 | 62.42 ± 7.64 |
| DenseNet-201 [19,20,34] | 62.18 ± 6.55 | 53.95 ± 9.48 | 52.31 ± 8.08 | 53.04 ± 8.52 |
| Inception-V3 [23] | 69.15 ± 5.29 | 67.37 ± 7.49 | 62.9 ± 6.98 | 65 ± 6.86 |
| MobileNet-V2 [27] | 64.06 ± 7.23 | 60.78 ± 10.95 | 57.35 ± 9.13 | 58.95 ± 9.78 |
| DRE-Net [20] | 56.27 ± 5.39 | 50.1 ± 7.52 | 47.95 ± 7.39 | 48.96 ± 7.29 |
| Proposed (IMFC-Net) | 81.93 ± 3.3 | 80.22 ± 4.7 | 77.98 ± 6.23 | 79.02 ± 4.98 |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goetti, P.; Denard, P.J.; Collin, P.; Ibrahim, M.; Hoffmeyer, P.; Lädermann, A. Shoulder Biomechanics in Normal and Selected Pathological Conditions. EFORT Open Rev. 2020, 5, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Kronberg, M.; Broström, L.A.; Söderlund, V. Retroversion of the Humeral Head in the Normal Shoulder and Its Relationship to the Normal Range of Motion. Clin. Orthop. Rel. Res. 1990, 253, 113–117. [Google Scholar] [CrossRef]
- Kadi, R.; Milants, A.; Shahabpour, M. Shoulder Anatomy and Normal Variants. J. Belg. Soc. Radiol. 2017, 101, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberson, T.A.; Bentley, J.C.; Griscom, J.T.; Kissenberth, M.J.; Tolan, S.J.; Hawkins, R.J.; Tokish, J.M. Outcomes of Total Shoulder Arthroplasty in Patients Younger than 65 Years: A Systematic Review. J. Shoulder Elb. Surg. 2017, 26, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Lo, I.K.Y.; Litchfield, R.B.; Griffin, S.; Faber, K.; Patterson, S.D.; Kirkley, A. Quality-of-Life Outcome Following Hemiarthroplasty or Total Shoulder Arthroplasty in Patients with Osteoarthritis: A Prospective, Randomized Trial. J. Bone Jt. Surg. Am. Vol. 2005, 87, 2178–2185. [Google Scholar] [CrossRef]
- Farley, K.X.; Wilson, J.M.; Daly, C.A.; Gottschalk, M.B.; Wagner, E.R. The Incidence of Shoulder Arthroplasty: Rise and Future Projections Compared to Hip and Knee Arthroplasty. J. Shoulder Elb. Surg. 2019, 3, 244. [Google Scholar] [CrossRef]
- Raiss, P.; Bruckner, T.; Rickert, M.; Walch, G. Longitudinal Observational Study of Total Shoulder Replacements with Cement: Fifteen to Twenty-Year Follow-Up. J. Bone Jt. Surg. Am. Vol. 2014, 96, 198–205. [Google Scholar] [CrossRef]
- Teusink, M.J.; Pappou, I.P.; Schwartz, D.G.; Cottrell, B.J.; Frankle, M.A. Results of Closed Management of Acute Dislocation after Reverse Shoulder Arthroplasty. J. Shoulder Elb. Surg. 2015, 24, 621–627. [Google Scholar] [CrossRef]
- Farley, K.X.; Wilson, J.M.; Kumar, A.; Gottschalk, M.B.; Daly, C.; Sanchez-Sotelo, J.; Wagner, E.R. Prevalence of Shoulder Arthroplasty in the United States and the Increasing Burden of Revision Shoulder Arthroplasty. J. Bone Jt. Surg. Am. Vol. 2021, 6, e20.00156. [Google Scholar] [CrossRef]
- Goyal, N.; Patel, A.R.; Yaffe, M.A.; Luo, M.Y.; Stulberg, S.D. Does Implant Design Influence the Accuracy of Patient Specific Instrumentation in Total Knee Arthroplasty? J. Arthroplast. 2015, 30, 1526–1530. [Google Scholar] [CrossRef]
- Burns, L.R.; Housman, M.G.; Booth, R.E.J.; Koenig, A. Implant Vendors and Hospitals: Competing Influences over Product Choice by Orthopedic Surgeons. Health Care Manag. Rev. 2009, 34, 2–18. [Google Scholar] [CrossRef]
- Wilson, N.A.; Jehn, M.; York, S.; Davis, C.M. Revision Total Hip and Knee Arthroplasty Implant Identification: Implications for Use of Unique Device Identification 2012 AAHKS Member Survey Results. J. Arthroplast. 2014, 29, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Hendel, M.D.; Bryan, J.A.; Barsoum, W.K.; Rodriguez, E.J.; Brems, J.J.; Evans, P.J.; Iannotti, J.P. Comparison of Patient-Specific Instruments with Standard Surgical Instruments in Determining Glenoid Component Position: A Randomized Prospective Clinical Trial. J. Bone Jt. Surg. Am. Vol. 2012, 94, 2167–2175. [Google Scholar] [CrossRef] [PubMed]
- Dy, C.J.; Bozic, K.J.; Padgett, D.E.; Pan, T.J.; Marx, R.G.; Lyman, S. Is Changing Hospitals for Revision Total Joint Arthroplasty Associated With More Complications? Clin. Orthop. Rel. Res. 2014, 472, 2006–2015. [Google Scholar] [CrossRef] [Green Version]
- Branovacki, G. Ortho Atlas—Hip Arthroplasty—U.S. Femoral Implants 1938–2008; Ortho Atlas Publishing: Chicago, IL, USA, 2008. [Google Scholar]
- Mahomed, N.N.; Barrett, J.A.; Katz, J.N.; Phillips, C.B.; Losina, E.; Lew, R.A.; Guadagnoli, E.; Harris, W.H.; Poss, R.; Baron, J.A. Rates and Outcomes of Primary and Revision Total Hip Replacement in the United States Medicare Population. J. Bone Jt. Surg. Am. Vol. 2003, 85, 27–32. [Google Scholar] [CrossRef]
- IMFC-Net for Shoulder Prostheses Recognition. Available online: http://dm.dgu.edu/link.html (accessed on 28 October 2021).
- Stark, M.B.C.G. Automatic Detection and Segmentation of Shoulder Implants in X-ray Images. Master’s Thesis, San Francisco State University, San Francisco, CA, USA, 2018. [Google Scholar]
- Urban, G.; Porhemmat, S.; Stark, M.; Feeley, B.; Okada, K.; Baldi, P. Classifying Shoulder Implants in X-Ray Images Using Deep Learning. Comp. Struct. Biotechnol. J. 2020, 18, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Sultan, H.; Owais, M.; Park, C.; Mahmood, T.; Haider, A.; Park, K.R. Artificial Intelligence-Based Recognition of Different Types of Shoulder Implants in X-Ray Scans Based on Dense Residual Ensemble-Network for Personalized Medicine. J. Pers. Med. 2021, 11, 482. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, Y.; Zhang, X.; Wang, S. Two-Stage Selective Ensemble of CNN via Deep Tree Training for Medical Image Classification. IEEE Trans. Cybern. 2021, 1–14. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; Volume 1, pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Owais, M.; SikYoon, H.; Mahmood, T.; Haider, A.; Sultan, H.; Park, K.R. Light-Weighted Ensemble Network with Multilevel Activation Visualization for Robust Diagnosis of COVID19 Pneumonia from Large-Scale Chest Radiographic Database. Appl. Soft. Comput. 2021, 108, 107490. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.-F. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Heaton, J. Artificial Intelligence for Humans, Vol 3: Neural Networks and Deep Learning; Heaton Research Inc.: St. Louis, MO, USA, 2015. [Google Scholar]
- Deep Learning Toolbox. Available online: https://www.mathworks.com/products/deep-learning.html (accessed on 7 October 2021).
- Ruder, S. An Overview of Gradient Descent Optimization Algorithms. arXiv 2017, arXiv:1609.04747. [Google Scholar]
- Livingston, E.H. Who Was Student and Why Do We Care so Much about His T-Test?1. J. Surg. Res. 2004, 118, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Yi, P.H.; Kim, T.K.; Wei, J.; Li, X.; Hager, G.D.; Sair, H.I.; Fritz, J. Automated Detection and Classification of Shoulder Arthroplasty Models Using Deep Learning. Skelet. Radiol. 2020, 49, 1623–1632. [Google Scholar] [CrossRef]
- Owais, M.; Lee, Y.W.; Mahmood, T.; Haider, A.; Sultan, H.; Park, K.R. Multilevel Deep-Aggregated Boosted Network to Recognize COVID-19 Infection from Large-Scale Heterogeneous Radiographic Data. IEEE J. Biomed. Health Inform. 2021, 25, 1881–1891. [Google Scholar] [CrossRef]
- Owais, M.; Arsalan, M.; Mahmood, T.; Kim, Y.H.; Park, K.R. Comprehensive Computer-Aided Decision Support Framework to Diagnose Tuberculosis From Chest X-Ray Images: Data Mining Study. JMIR Med. Inf. 2020, 8, 89–111. [Google Scholar] [CrossRef]
- Mahmood, T.; Owais, M.; Noh, K.J.; Yoon, H.S.; Koo, J.H.; Haider, A.; Sultan, H.; Park, K.R. Accurate Segmentation of Nuclear Regions with Multi-Organ Histopathology Images Using Artificial Intelligence for Cancer Diagnosis in Personalized Medicine. J. Pers. Med. 2021, 11, 515. [Google Scholar] [CrossRef]
- Arsalan, M.; Kim, D.S.; Owais, M.; Park, K.R. OR-Skip-Net: Outer Residual Skip Network for Skin Segmentation in Non-Ideal Situations. Expert Syst. Appl. 2020, 141, 112922. [Google Scholar] [CrossRef]
- Owais, M.; Baek, N.R.; Park, K.R. Domain-Adaptive Artificial Intelligence-Based Model for Personalized Diagnosis of Trivial Lesions Related to COVID-19 in Chest Computed Tomography Scans. J. Pers. Med. 2021, 11, 1008. [Google Scholar] [CrossRef] [PubMed]
- Morais, P.; Queirós, S.; Moreira, A.H.J.; Ferreira, A.; Ferreira, E.; Duque, D.; Rodrigues, N.F.; Vilaça, J.L. Computer-Aided Recognition of Dental Implants in X-Ray Images. In Proceedings of the SPIE 9414, Medical Imaging: Computer-Aided Diagnosis, Orlando, FL, USA, 21–26 February 2015; Volume 9414. [Google Scholar]
- Sukegawa, S.; Yoshii, K.; Hara, T.; Yamashita, K.; Nakano, K.; Yamamoto, N.; Nagatsuka, H.; Furuki, Y. Deep Neural Networks for Dental Implant System Classification. Biomolecules 2020, 10, 984. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-T.; Lee, J.-B.; Jeong, S.-N. A Performance Comparison between Automated Deep Learning and Dental Professionals in Classification of Dental Implant Systems from Dental Imaging: A Multi-Center Study. Diagnostics 2020, 10, 910. [Google Scholar] [CrossRef] [PubMed]
- Sukegawa, S.; Yoshii, K.; Hara, T.; Matsuyama, T.; Yamashita, K.; Nakano, K.; Takabatake, K.; Kawai, H.; Nagatsuka, H.; Furuki, Y. Multi-Task Deep Learning Model for Classification of Dental Implant Brand and Treatment Stage Using Dental Panoramic Radiograph Images. Biomolecules 2021, 11, 815. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Jeong, S.-N. Efficacy of Deep Convolutional Neural Network Algorithm for the Identification and Classification of Dental Implant Systems, Using Panoramic and Periapical Radiographs. Medicine (Baltimore) 2020, 99, e20787. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-E.; Nam, N.-E.; Shim, J.-S.; Jung, Y.-H.; Cho, B.-H.; Hwang, J.J. Transfer Learning via Deep Neural Networks for Implant Fixture System Classification Using Periapical Radiographs. J. Clin. Med. 2020, 9, 1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, Y.-J.; Yoo, J.-I.; Cha, Y.-H.; Park, C.H.; Kim, J.-T. Machine Learning–Based Identification of Hip Arthroplasty Designs. J. Orthop. Transl. 2020, 21, 13–17. [Google Scholar] [CrossRef]
- Karnuta, J.M.; Haeberle, H.S.; Luu, B.C.; Roth, A.L.; Molloy, R.M.; Nystrom, L.M.; Piuzzi, N.S.; Schaffer, J.L.; Chen, A.F.; Iorio, R.; et al. Artificial Intelligence to Identify Arthroplasty Implants From Radiographs of the Hip. J. Arthroplast. 2021, 36, S290–S294.e1. [Google Scholar] [CrossRef]
- Borjali, A.; Chen, A.F.; Muratoglu, O.K.; Morid, M.A.; Varadarajan, K.M. Detecting Total Hip Replacement Prosthesis Design on Plain Radiographs Using Deep Convolutional Neural Network. J. Orthop. Res. 2020, 38, 1465–1471. [Google Scholar] [CrossRef] [Green Version]
- Borjali, A.; Chen, A.F.; Bedair, H.S.; Melnic, C.M.; Muratoglu, O.K.; Morid, M.A.; Varadarajan, K.M. Comparing the Performance of a Deep Convolutional Neural Network with Orthopedic Surgeons on the Identification of Total Hip Prosthesis Design from Plain Radiographs. Med. Phys. 2021, 48, 2327–2336. [Google Scholar] [CrossRef]
- Bredow, J.; Wenk, B.; Westphal, R.; Wahl, F.; Budde, S.; Eysel, P.; Oppermann, J. Software-Based Matching of x-Ray Images and 3D Models of Knee Prostheses. Technol. Health Care 2014, 22, 895–900. [Google Scholar] [CrossRef] [Green Version]
- Yi, P.H.; Wei, J.; Kim, T.K.; Sair, H.I.; Hui, F.K.; Hager, G.D.; Fritz, J.; Oni, J.K. Automated Detection & Classification of Knee Arthroplasty Using Deep Learning. Knee 2020, 27, 535–542. [Google Scholar] [CrossRef]
- Karnuta, J.M.; Luu, B.C.; Roth, A.L.; Haeberle, H.S.; Chen, A.F.; Iorio, R.; Schaffer, J.L.; Mont, M.A.; Patterson, B.M.; Krebs, V.E.; et al. Artificial Intelligence to Identify Arthroplasty Implants from Radiographs of the Knee. J. Arthroplast. 2021, 36, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Belete, S.C.; Batta, V.; Kunz, H. Automated Classification of Total Knee Replacement Prosthesis on Plain Film Radiograph Using a Deep Convolutional Neural Network. Inform. Med. Unlocked 2021, 25, 100669. [Google Scholar] [CrossRef]
- Yan, S.; Ramazanian, T.; Sagheb, E.; Fu, S.; Sohn, S.; Lewallen, D.G.; Liu, H.; Kremers, W.K.; Chaudhary, V.; Taunton, M.; et al. DeepTKAClassifier: Brand Classification of Total Knee Arthroplasty Implants Using Explainable Deep Convolutional Neural Networks. In Proceedings of the Advances in Visual Computing Part 2, San Diego, CA, USA, 5–7 October 2020; Bebis, G., Yin, Z., Kim, E., Bender, J., Subr, K., Kwon, B.C., Zhao, J., Kalkofen, D., Baciu, G., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 154–165. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]

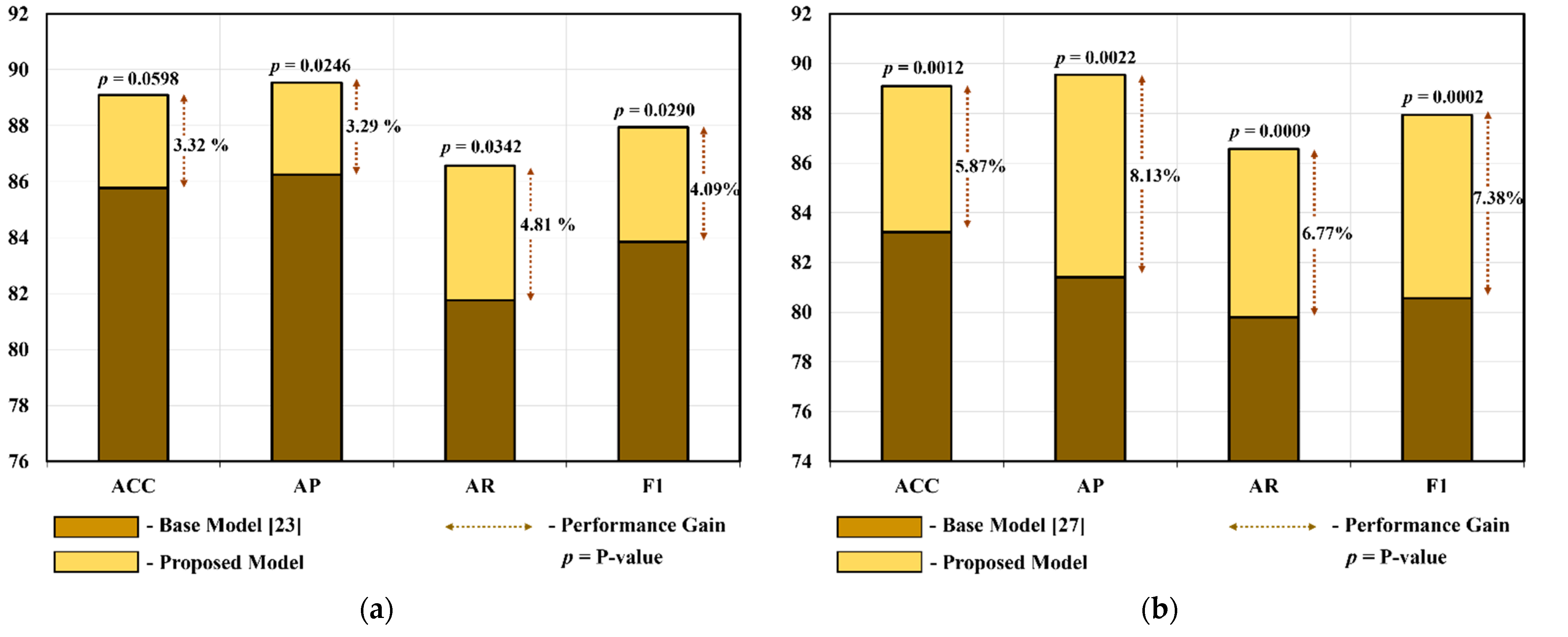

| Model | Performance without CP Block | Performance with CP Block | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC ± Std | AP ± Std | AR ± Std | F1 ± Std | ACC ± Std | AP ± Std | AR ± Std | F1 ± Std | |
| Inception-V3 [23] for IFC-Net | 85.77 ± 4.55 | 86.25 ± 5.14 | 81.76 ± 4.76 | 83.85 ± 3.93 | 87.22 ± 4.47 | 87.66 ± 5.39 | 84.32 ± 4.15 | 85.88 ± 3.98 |
| MobileNet-V2 [27] for MFC-Net | 83.22 ± 3.96 | 81.41 ± 4.38 | 79.8 ± 7.3 | 80.56 ± 5.7 | 83.86 ± 4.88 | 83.29 ± 4.98 | 80.54 ± 8.8 | 81.79 ± 6.67 |
| Model | ACC ± Std | AP ± Std | AR ± Std | F1 ± Std |
|---|---|---|---|---|
| MFC-Net | 83.86 ± 4.88 | 83.29 ± 4.98 | 80.54 ± 8.8 | 81.79 ± 6.67 |
| IFC-Net | 87.22 ± 4.47 | 87.66 ± 5.39 | 84.32 ± 4.15 | 85.88 ± 3.98 |
| Proposed (IMFC-Net) | 89.09 ± 4.55 | 89.54 ± 3.82 | 86.57 ± 7.63 | 87.94 ± 5.49 |
| Model | Training Method | ACC ± Std | AP ± Std | AR ± Std | F1 ± Std |
|---|---|---|---|---|---|
| Proposed (IMFC-Net) | End-to-End | 86.56 ± 2.96 | 85.53 ± 4.09 | 84.07 ± 5.18 | 84.7 ± 3.78 |
| Sequential | 89.09 ± 4.55 | 89.54 ± 3.82 | 86.57 ± 7.63 | 87.94 ± 5.49 |
| Model | ACC ± Std | AP ± Std | AR ± Std | F1 ± Std |
|---|---|---|---|---|
| VGG-16 [19,20,22] | 68.72 ± 7.34 | 66.39 ± 8.94 | 66.51 ± 9.18 | 66.32 ± 8.47 |
| VGG-19 [19,20,22] | 65.82 ± 5.96 | 63.99 ± 6.16 | 62.76 ± 6.61 | 63.29 ± 5.83 |
| DarkNet-53 [32] | 53.13 ± 6.02 | 44.47 ± 4.94 | 40.71 ± 5.08 | 42.4 ± 4.72 |
| NASNet [19,20,33] | 80.48 ± 5.22 | 78.76 ± 6.28 | 76.83 ± 7.73 | 77.68 ± 6.41 |
| ResNet-18 [20,24,35] | 77.38 ± 8 | 77.01 ± 9.42 | 73.6 ± 9.35 | 75.13 ± 8.75 |
| ResNet-50 [19,20,24] | 80.27 ± 6.53 | 79.71 ± 7.62 | 76.61 ± 6.76 | 78.08 ± 6.83 |
| ResNet-101 [24] | 82.18 ± 5.39 | 82.43 ± 7.85 | 77.98 ± 9.29 | 79.92 ± 7.53 |
| DenseNet-201 [19,20,34] | 84.24 ± 2.45 | 84.57 ± 3.66 | 82.1 ± 4.52 | 83.28 ± 3.7 |
| Inception-V3 [23] | 85.77 ± 4.55 | 86.25 ± 5.15 | 81.76 ± 4.76 | 83.85 ± 3.93 |
| MobileNet-V2 [27] | 83.22 ± 3.96 | 81.41 ± 4.38 | 79.81 ± 7.3 | 80.56 ± 5.70 |
| DRE-Net [20] | 85.08 ± 3.12 | 84.75 ± 4.54 | 83.61 ± 4.3 | 84.15 ± 4.09 |
| Proposed (IMFC-Net) | 89.09 ± 4.55 | 89.54 ± 3.82 | 86.57 ± 7.63 | 87.94 ± 5.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sultan, H.; Owais, M.; Choi, J.; Mahmood, T.; Haider, A.; Ullah, N.; Park, K.R. Artificial Intelligence-Based Solution in Personalized Computer-Aided Arthroscopy of Shoulder Prostheses. J. Pers. Med. 2022, 12, 109. https://doi.org/10.3390/jpm12010109
Sultan H, Owais M, Choi J, Mahmood T, Haider A, Ullah N, Park KR. Artificial Intelligence-Based Solution in Personalized Computer-Aided Arthroscopy of Shoulder Prostheses. Journal of Personalized Medicine. 2022; 12(1):109. https://doi.org/10.3390/jpm12010109
Chicago/Turabian StyleSultan, Haseeb, Muhammad Owais, Jiho Choi, Tahir Mahmood, Adnan Haider, Nadeem Ullah, and Kang Ryoung Park. 2022. "Artificial Intelligence-Based Solution in Personalized Computer-Aided Arthroscopy of Shoulder Prostheses" Journal of Personalized Medicine 12, no. 1: 109. https://doi.org/10.3390/jpm12010109
APA StyleSultan, H., Owais, M., Choi, J., Mahmood, T., Haider, A., Ullah, N., & Park, K. R. (2022). Artificial Intelligence-Based Solution in Personalized Computer-Aided Arthroscopy of Shoulder Prostheses. Journal of Personalized Medicine, 12(1), 109. https://doi.org/10.3390/jpm12010109